Abstract
Many proteins expressed in Escherichia coli cells form inclusion bodies that are neither refoldable nor soluble in buffers. Very surprisingly, we recently discovered that all 11 buffer-insoluble protein fragments/domains we have, with a great diversity of cellular function, location, and molecular size, could be easily solubilized in salt-free water. The circular dichroism (CD) and NMR characterization led to classification of these proteins into three groups: group 1, with no secondary structure by CD and with narrowly-dispersed but sharp 1H-15N heteronuclear single quantum correlation (HSQC) peaks; group 2, with secondary structure by CD but with HSQC peaks broadened and, consequently, only a small set of peaks detectable; and group 3, with secondary structure by CD and also well-separated HSQC peaks. Intriguingly, we failed to find any protein with a tight tertiary packing. Therefore, we propose that buffer-insoluble proteins may lack intrinsic ability to reach or/and to maintain a well-packed conformation, and thus are trapped in partially-folded states with many hydrophobic side chains exposed to the bulk solvent. As such, a very low ionic strength is sufficient to screen out intrinsic repulsive interactions and, consequently, allow the hydrophobic clustering/aggregation to occur. Marvelously enough, it appears that in pure water, proteins have the potential to manifest their full spectrum of structural states by utilizing intrinsic repulsive interactions to suppress the attractive hydrophobic clustering. Our discovery not only gives a novel insight into the properties of insoluble proteins, but also sheds the first light that we know of on previously unknown regimes associated with proteins.
INTRODUCTION
Proteins are important functional players that implement the most difficult but essential tasks in living cells (1). However, one commonly-encountered problem in protein research and application is their insolubility (2). For example, in most structure genomics initiatives it has been estimated that ∼35–50% of the proteins expressed in Escherichia coli cells were in inclusion bodies, most of which were not refoldable in buffer systems (3,4) with currently-available methods. In fact, it is extensively found that many proteins misfold and subsequently aggregate when overproduced in microorganisms (4), although it remains unknown whether these proteins will misfold in vivo. Despite extensive attempts to reduce formation of inclusion bodies in E. coli cells by fusing with a highly-soluble tag, coexpressing folding catalysts and chaperones, reducing culture temperature, and modifying culture media, these approaches do not always work (5). This observation implies that the intrinsic property of a protein may be responsible for its insolubility. Therefore, structural characterization of insoluble proteins will provide valuable insights and consequently offer rationales for enhancing protein solubility. Unfortunately, to date no general method is available to solubilize these proteins without addition of detergents or/and denaturants such as sodium dodecyl sulfate, urea, and guanidine hydrochloride at high concentrations.
Very unexpectedly, we recently discovered that several buffer-insoluble Nogo-66 fragments could be easily solubilized in water at high concentrations. In particular, this discovery allowed us to determine the NMR structure and dynamics of Nogo-60 and, subsequently, to obtain a critical rationale to further design the structured and buffer-soluble Nogo-54 (6). This result inspired me to explore the general question of whether other buffer-insoluble proteins also could be solubilized in water. To address this, we have initiated a systematic study on all 11 buffer-insoluble protein fragments/domains we currently have, which are very diverse in cellular function, location, and molecular size. To our great surprise, again they could be dissolved in salt-free water at high concentrations. This discovery therefore offers us an unprecedented possibility to characterize them by circular dichroism (CD) and NMR 1H-15N heteronuclear single quantum correlation (HSQC) spectroscopy.
MATERIALS AND METHODS
Selection of protein fragments/domains
All protein fragments/domains studied here were fused with a His-tag for affinity purification. The His-tag is composed of 16 residues with a sequence of MHHHHHHSSG LVPRGS and a molecular mass of 1.8 kD. The molecular mass values stated below already included the contribution of the His-tag.
Fragments of the Nogo-66 extracellular domain: Nogo proteins are myelin-associated transmembrane proteins that play a critical role in inhibiting central nervous system (CNS) axonal regeneration. Nogo is composed of three splicing variants, namely the 1192-residue Nogo-A, 373-residue Nogo-B, and 199-residue Nogo-C. Despite their significant size differences, all three Nogo variants contain a conserved extracellular domain with ∼66 residues that is capable of inhibiting neurite growth and inducing growth cone collapse. In this study, four differentially-truncated fragments of the Nogo-66 domain were included, namely, Nogo-66, with a MM of 9.3 kD spanning residues 1055–1120 of Nogo-A (AAM64248); Nogo-60, with a MM of 8.7 kD over residues 1055–1114; Nogo-(21-60), with a MM of 6.3 kD over residues 1075–1114; and Nogo-54, with a MM of 8.0 kD over residues 1055–1108. Only one cysteine is presented in these fragments, and therefore no disulfide bridge is expected to form.
Fragments of the attachment glycoprotein (G) of Nipah virus: the G-protein of Nipah virus is a 602-residue viral surface protein that was shown to mediate viral entry by interacting with ephrinB ectodomains. In this study, two dissected fragments of the G-protein were included, namely Niv-44, with a MM of 6.7 kD, spanning residues 430–473; Niv-86, with a MM of 11.6 kD, spanning residues 402–487 of the Nipah G-protein (AAF73957). There is no cysteine residue in the two fragments.
Hemagglutinin receptor binding domain (RBD): the 152-residue RBD was dissected from the 561-residue hemagglutinin of the Avian Influenza A virus, with a MM of 19.1 kD, spanning residues 126–277 (AAT73276.1). There is only one cysteine residue and therefore no disulfide is expected to form.
SH3 domains of the human Nck2 (hNck2) protein: based on our previous study, the first SH3 domain of the 381-residue human Nck2 protein is totally insoluble (7). Therefore, in this study, we included the first SH3 domain, with a MM of 8.8 kD, spanning residues 5–62 of the human Nck2 (AAC04831).
Wrch1 Cdc42-like domain: Wrch1 protein is a 270-residue new member of the Cdc42 superfamily, unique in having a Cdc42-like domain but no GTPase activity. In our previous study (7), we investigated the binding interaction between Nck2 SH3 domains and the Wrch1 N-terminus (highly soluble). However, we found that the Wrch1 Cdc42-like domain was totally buffer-insoluble. As such, we include the 173-residue Cdc42-like domain here with a MM of 20.7 kD over residues 52–225 of the human Wrch1 (BAD92748).
The Claudin 1 extracellular domain: the entire first ectodomain of the 211-residue human Claudin 1 was selected, namely Claudin-51, with a MM of 7.4 kD spanning residues 31–81 of the human Claudin 1 (AAK20945). There are two cysteines in the sequence, and thus they are expected to form one intramolecular disulfide bridge.
Dissected domains of Nogo receptor (NgR): NgR is a 473-residue myelin protein GPI-anchored onto the cell membrane, which transmits signals to inhibit CNS axonal regeneration by interacting Nogo-66 or other binding-partner proteins. Here, we cloned two NgR fragments, one composed only of the 319-residue Nogo-66 binding domain, with a MM of 36.8 kD, spanning residues 26–344 (AAG53612). Another covered the entire 421-residue ectodomain of the NgR receptor, with only signal peptide and GPI-anchor sequences removed, having a MM of 47 kD, spanning residues 26–446. It is expected that the N-terminal NgR will have five disulfide bridges formed, whereas the entire NgR ectodomain will have six disulfide bridges.
Cloning and expression
Except for DNA fragments encoding the hNck SH3 domains, which were obtained by polymerase chain reaction-based de novo gene synthesis with E. coli preferred codons (7), all other fragments were polymerase chain-reacted out from cDNA templates by designed primers. The obtained DNA segments were subsequently cloned into the His-tagged expression vector pET32a (Novagen, Madison, WI), as previously described.
Recombinant proteins were overexpressed in the E. coli bacterial strain BL21 cells. Briefly, the cells were cultured at 37°C to reach an OD600 of 0.6, and then isopropyl-ß-D-thiogalactopyranoside was added to a final concentration of 1 mM to induce the recombinant protein expression for 4 h at 37°C. Except for Nogo-54, the rest of the proteins were found to be totally in inclusion bodies and, as such, were first purified by Ni2+-affinity chromatography under denaturing conditions in the presence of 8 M urea. Subsequently, 100 mM dithiothreitol was added to affinity-purified proteins in the 8 M urea for half an hour to reduce the possibility of forming disulfide bridges if those proteins contained cysteine residues. Depending on their molecular size, these proteins were finally purified by the reverse-phase high-performance liquid chromatography (HPLC) on a semipreparative C18, C8, or C4 column (Vydac, Hesperia, CA) and then lyophilized. For NMR isotope labeling, recombinant proteins were prepared by growing the cells in the M9 medium with the addition of (15NH4)2SO4 for 15N labeling (7).
Sample preparation and CD, NMR experiments
Except for Nogo-54, which was soluble in both water and buffer, all other protein fragments/domains were insoluble in buffer. Therefore, they were dissolved in deionized water (Milli-Q, Millipore, Billerica, MA) with addition of an aliquot of 5 mM NaOH to adjust pH and of 10% D2O for spin-lock.
CD experiments were performed on a J-810 spectropolarimeter (Jasco, Tokyo, Japan) equipped with a thermal controller, as described previously (7). The far-UV CD spectra were collected in a wide range of peptide concentrations at 20°C, using a 1-mm path length cuvette with a 0.1-nm spectral resolution. The near-UV CD spectra were collected at a protein concentration of ∼200 μM in the absence and in the presence of 8 M urea. Data from five independent scans were added and averaged.
HSQC NMR experiments were acquired on an 800-MHz Bruker Avance spectrometer (Bruker Daltonics, Billerica, MA) equipped with pulse-field gradient units at 293 K, as described previously (8). NMR data were processed with NMRPipe (9) and analyzed with NMRView (10). The three-dimensional structures were displayed and drawn using MolMol software (11).
RESULTS
In this study, we have included all 11 buffer-insoluble protein fragments/domains currently in our laboratory. They are composed of extracellular protein domains, including those dissected from Nogo-66, Nipah virus G-protein, Claudin, avian flu hemaglutinin, and Nogo receptor (NgR), as well as cytoplasmic protein domains including the first Nck2 SH3 and Wrch1 Cdc42-like domains. DNA fragments encoding the above 11 proteins plus Nogo-54 were successfully cloned into expression vector pET32a (Novagen) as His-tagged forms and subsequently expressed in E. coli BL21 cells. Except for Nogo-54, the 11 proteins were all found in inclusion bodies and could not be refolded by either fast dilution or dialysis. Nevertheless, we purified them first by affinity and then by reverse-phase HPLC chromatography for further CD and NMR assessment in salt-free water.
Nogo-66 fragments
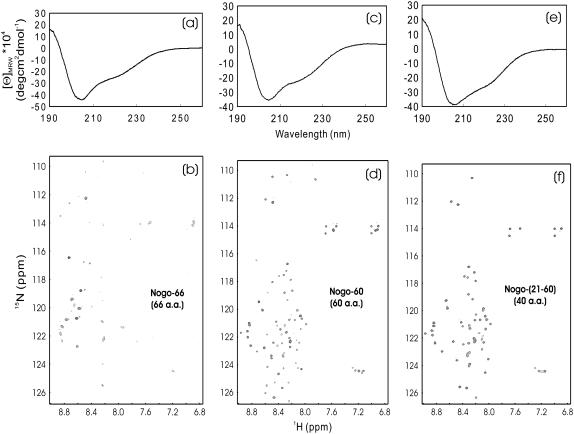
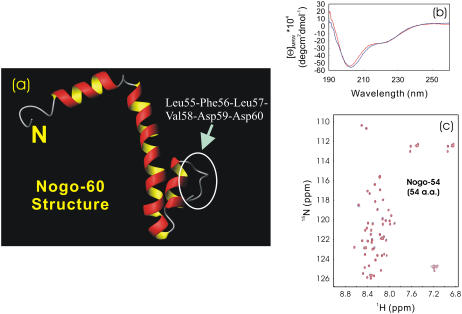
The entire Nogo extracellular domain consists of 66 residues, usually called Nogo-66. Previously, Nogo-66 was found to be totally insoluble in buffer and, as such, only Nogo-40, with C-terminal 26 residues removed, was studied with the presence of 50% trifloroethanol TFE (12). However, we now found that Nogo-66, along with its truncated forms, all could be solubilized in salt-free water. As seen in Fig. 1, whereas Nogo-66 had a far-UV CD spectrum typical of helical structure (Fig. 1 a), only five HSQC peaks resulting from non-His-tag residues were detectable (Fig. 1 b). This observation was attributed to conformational exchanges on the microsecond to millisecond timescale (6). On the other hand, although two truncated forms, namely Nogo-60 and Nogo-(21-60) were again only soluble in salt-free water, they both have helical CD spectra (Fig. 1, c and e) and well-separated HSQC spectra, with almost all nonproline residues detectable (Fig. 1, d and f). This discovery thus led to our previous determination of the solution structure and dynamics of Nogo-60 (Fig. 2 a), as well as further design of Nogo-54, which was structured and soluble in both salt-free water and buffer (6). A detailed comparison of the CαH conformational shifts indicated that Nogo-54 had a structure almost identical to the corresponding region of Nogo-60 (M. Li and J. Song, unpublished). Also, it is worthwhile to point out that the structures of Nogo-54 in salt-free water and buffer were essentially the same, as is clearly evident from its superimposable CD (Fig. 2 b) and HSQC (Fig. 2 c) spectra.
FIGURE 1.
CD and HSQC characterization of Nogo-66 fragments. (a and b) Far-UV CD and NMR HSQC spectra of Nogo-66; (c and d) Nogo-60; and (e and f) Nogo-(21-60). All three peptides were only soluble in salt-free water (pH 4.0), with protein concentrations of ∼100 μM for Nogo-66, and ∼200 μM for Nogo-60 and Nogo-(21-60). NMR spectra were acquired on a Bruker Avance 800 MHz NMR spectrometer at 20°C.
FIGURE 2.
Conformational properties of Nogo-54. (a) NMR structure of Nogo-60 previously determined (PDB ID: 2G31) in salt-free water. Based on this structure, the buffer-soluble Nogo-54 was thus designed by deleting the C-terminal six unstructured residues (circles), which constitute an exposed hydrophobic patch. (b and c) Far-UV CD and NMR HSQC spectra of Nogo-54 in salt-free water (blue) and in 10 mM phosphate buffer (red) at pH 4.2 and 20°C. The protein concentrations of the NMR samples were ∼120 μM in both water and buffer.
Viral surface protein fragments
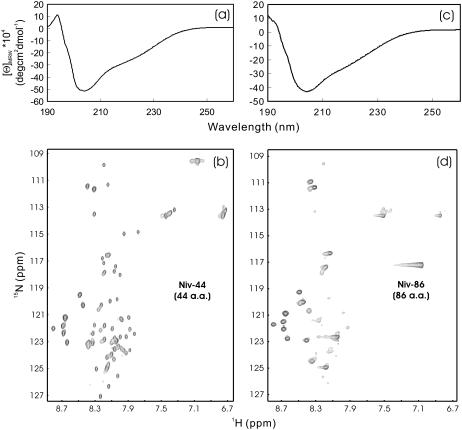
The G-protein of Nipah virus is a 602-residue viral surface protein which was recently demonstrated to mediate viral entry by interacting with ephrinB ectodomains. Previously we have cloned two G-protein fragments but found them to be totally insoluble in buffer and thus not open to study. However, both fragments Niv-44 and Niv-88 could be easily solubilized in salt-free water. As seen in Fig. 3 a, the far-UV CD spectrum indicated that the 44-residue Niv-44 definitely was not unstructured but instead it might have a helical and/or loop conformation. On the other hand, HSQC peaks for almost all residues were detectable (Fig. 3 b), thus recommending its suitability for further high-resolution NMR study. On the other hand, although the 86-residue Niv-86 with sequence extensions at both termini of Niv-44 still owned a similar CD spectrum (Fig. 3 a), only ∼24 non-His-tag HSQC peaks were detectable (Fig. 3 b), indicating that the introduction of additional residues resulted in an increase of μs-ms conformational exchange or/and dynamic aggregation.
FIGURE 3.
CD and HSQC characterization of Nipah G-protein fragments. (a and d) Far-UV CD and HSQC NMR spectra of the Nipah G fragment Niv-44 (44 residues) and Niv-86 (86 residues) at pH 3.8 and 20°C. The protein concentrations of the NMR samples were ∼460 μM for Niv-44 and ∼340 μM for Niv-86.
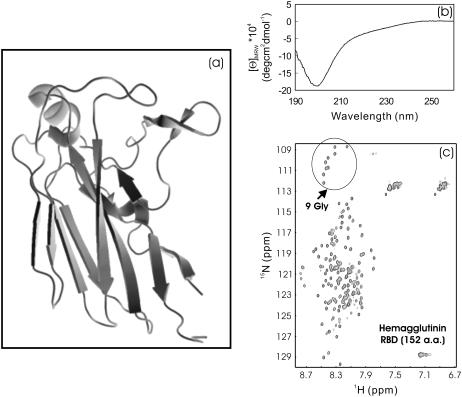
We previously isolated the RBD from the 561-residue avian influenza hemagglutinin in an attempt to solve its NMR structure and to screen binding ligands. Unfortunately, although RBD has a well-defined β-structure (Fig. 4 a) in the context of the entire hemagglutinin, the isolated RBD was found to be totally insoluble. Amusingly, this 152-residue domain was again soluble in salt-free water and had a CD spectrum characteristic of an unstructured protein (Fig. 4 b). In agreement with this, the RBD had a narrowly-dispersed HSQC spectrum with almost all nonproline residues detectable. In particular, all nine Gly residues (including one from His-tag) were clearly-resolved (Fig. 4 c), thus indicating that the protein is highly unfolded in salt-free water. This result also revealed that the RBD is not an autonomous folding unit, needing other parts of hemagglutinin to maintain its three-dimensional structure as determined by x-ray crystallography (13).
FIGURE 4.
CD and HSQC characterization of viral hemagglutinin RBD. (a) Three-dimensional structure of the RBD in the context of the entire hemagglutinin molecule (PDB code 1JSN). (b and c) Far-UV CD and NMR HSQC spectra of the receptor binding domain (152 residues) dissected from the 561-residue hemagglutinin of the Avian Influenza A virus at pH 6.2 and 20°C. The protein concentration of the NMR sample was ∼400 μM. The HSQC peaks for nine Gly residues (including one from His-tag) are indicated by the arrow.
Intracelluar protein domains
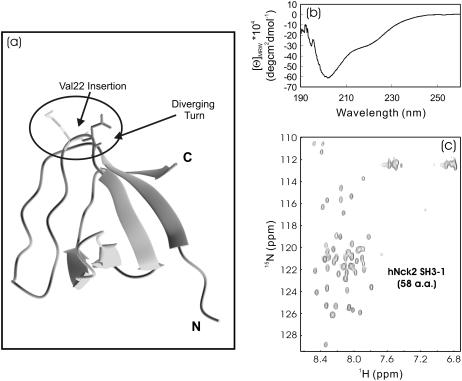
Recently, we determined the NMR structures of the second and third hNck2 SH3 domains but found that the first SH3 domain (SH3-1) with the sequence derived from AAC04831 was totally insoluble (12). On the other hand, the hNck2 SH3-1 derived from another hNck2 protein sequence was soluble and its NMR structure was reported (Fig. 5 a) (14). Comparison of the two SH3-1 sequences revealed that in our SH3-1 protein, an additional Val residue was presented at the diverging turn linking the RT-loop to the second β-strand, and consequently, this insertion resulted in the insolubility of our 58-residue hNck2 SH3-1. However, this SH3-1 domain was again soluble in salt-free water but appeared to be highly denatured, as indicated by its CD (Fig. 5 b) and HSQC (Fig. 5 c) spectra. In particular, its HSQC spectrum had very narrow dispersions (∼0.7 ppm over proton and ∼19 ppm over 15N dimensions), with peaks detectable for almost all nonproline residues. Very unexpectedly, with our extensive heteronuclear NMR characterization, this denatured SH3 domain was just mapped out to have a native-like topology with significantly limited backbone motions but meanwhile, its secondary structure shifts were demonstrated to be severely abolished (J. Liu and J. Song, unpublished).
FIGURE 5.
CD and HSQC characterization of the first Nck2 SH3 domain. (a) The NMR structure of the first human Nck2 SH3 domain (PDB code 2B86). (b and c) Far-UV CD and NMR HSQC spectra of the first SH3 domain (58 residues) of the human Nck2 protein (AAC04831) at pH 6.2 and 20°C. The protein concentration of the NMR sample was ∼200 μM.
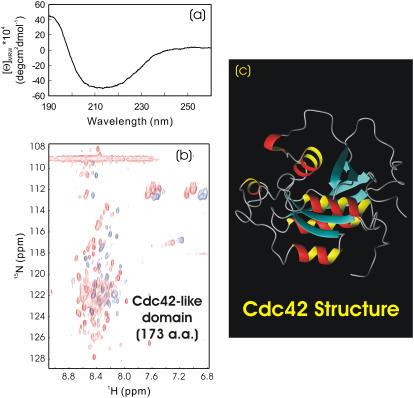
On the other hand, the 173-residue Wrch1 Cdc42-like domain solubilized in salt-free water showed a CD spectrum for a mixture of helix, β-sheet, and random coil (Fig. 6 a). CD spectral decomposition (www.embl-heidelberg.de/∼andrade/k2d.html) further revealed that the Wrch1 Cdc42 domain contained ∼15% α-helix; ∼33% β-sheet, and 52% random coil, very similar to the secondary structure patterns observed for its homolog, the human Cdc42 protein (15) (Fig. 6 c). This strongly implied that the Wrch1 Cdc42-like domain already assumed a native-like secondary structure in salt-free water. On the other hand, it had an HSQC spectrum with a narrow dispersion (∼0.6 ppm over proton and ∼22 ppm over 15N dimensions) and with only ∼30 broad NMR peaks detectable (Fig. 6 b). This phenomenon has been extensively observed for the molten globule states due to the loose side-chain packing (16–19). To assess the degree of the side-chain packing, we collected its near-UV and HSQC spectra in the absence and presence of 8 M urea. As expected, there was no significant difference between the near-UV spectra in salt-free water and 8 M urea (spectra not shown), indicating that the tight side-chain packing was largely disrupted even without 8 M urea. Nevertheless, addition of 8 M urea did result in a dramatic increase of detectable HSQC peaks (Fig. 6 b), indicating that besides the native-like secondary structure, Cdc42-like domain also owned a native-like topology, as previously demonstrated on α-lactalbumin and CHABII molten globules (16–19). These results together strongly suggested that although salt-free water offered the media for the Wrch1 Cdc42-like domain to be soluble and manifest its intrinsic structural propensity, it was somehow trapped in the molten-globule state with a native-like secondary structure and tertiary topology, but without a tight side-chain packing. To further transform it into a protein with a buffer solubility and tight side-chain packing, the introduction of a binding partner or sequence mutations was highly demanded (4).
FIGURE 6.
CD and HSQC characterization of Wrch1 Cdc42-like domains. (a and b) Far-UV CD and HSQC NMR spectra of the Cdc42-like domain (174 residues) of the Wrch1 protein in the absence (blue) and presence (red) of 8 M urea at pH 6.0 and 20°C. The protein concentration of the NMR sample was ∼120 μM. (c) NMR structure of the human Cdc42 (PDB code 1AJE), having ∼60% sequence identity with the Wrch1 Cdc42-like domain studied here.
Extracellular receptor domains without disulfide formation
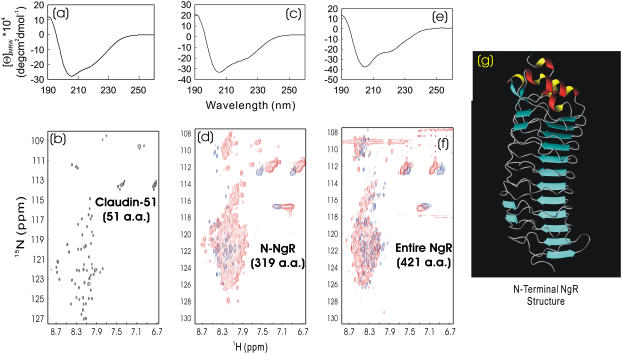
The most unusual group in our current study is the disulfide-free extracellular receptor domains from human Claudin 1 and Nogo-receptors (NgR). In the native proteins, disulfide bridges were needed to maintain their structural integrity: one disulfide was expected for Claudin-51, five for N-terminal NgR, and six for the entire NgR. In our previous investigation, the three proteins lacking disulfide bridge(s) were totally insoluble. Nevertheless, to our great surprise, in this study, they were all soluble in salt-free water. The 51-residue Claudin-51 had a CD spectrum of a helical conformation (Fig. 7 a) and an HSQC spectrum with a narrow dispersion but with almost all nonproline residues detectable (Fig. 7 b). Currently we are focusing on determining its NMR structure in salt-free water.
FIGURE 7.
CD and HSQC characterization of extracellular domains of transmembrane receptors. (a and b) Far-UV CD and NMR HSQC spectra of the first extracellular domain (51 residues) of the human Claudin 1 at pH 3.8 and ∼200 μM. (c and d) Far-UV CD and HSQC NMR spectra of the N-terminal domain (318 residues) of the human Nogo-66 receptor (NgR) in the absence (blue) and presence (red) of 8 M urea at pH 4.2 and 20°C. (e and f) Far-UV CD and HSQC NMR spectra of the entire extracelluar domain (420 residues) of the same NgR in, respectively, the absence (blue) and presence (red) of 8 M urea at pH 6.2 and 20°C. The protein concentrations of the NMR samples were ∼120 μM for both N-NgR and NgR. (g) Crystallographic structure of the NgR N-terminal domain (1OZN), which adopts a typical leucine-rich repeat fold. The protein samples used here were all disulfide-free.
The 319-residue N-terminal NgR contained a leucine-rich repeat fold plus two C-terminal helical regions. As shown in Fig. 7 c, the disulfide-free N-NgR had a CD spectrum for a helical protein. As seen in the N-NgR structure (20) (Fig. 7 g), although the leucine-rich repeat fold contained many short β-strands in addition to helical segments, they usually gave rise to helix-like CD spectra probably due to the unique solenoid-like fold they adopt (21,22). Therefore, if judged from its CD spectrum (Fig. 7 c), it might be concluded that the native-like secondary structure might already form in the disulfide-free N-NgR. On the other hand, similar to that observed for the Wrch1 Cdc-42 domain (Fig. 6 b), N-NgR had an HSQC spectrum with a narrow dispersion (∼0.7 ppm over proton and ∼17 ppm over 15N dimensions) and very broad NMR peaks (Fig. 7 b). Only ∼45 HSQC peaks were detectable, clearly indicating that the disulfide-free N-NgR underwent intermediate conformational exchanges or/and dynamic aggregation, largely owing to its dynamic side-chain packing (16–19). Addition of 8 M urea resulted in no significant change in the near-UV spectra (spectra not shown), again indicating that the side-chain packing was largely disrupted. However, introduction of 8 M urea did lead to the appearance of many new HSQC peaks, similar to those observed on the Wrch1 Cdc42-like domain above (Fig. 6 b). Moreover, the entire 421-residue NgR with a 103-residue C-terminal extension also behaved in a manner similar to N-NgR (Fig. 7, e and f), although the structure of the extended 103 residues remained completely unknown. These results together suggested that although the disulfide-free N- and entire NgR proteins could be dissolved in salt-free water, they were still trapped in the molten globule states with both a native-like secondary structure and tertiary topology, but without a tight side-chain packing, largely due to the complete absence of essential disulfide bridges. In this regard, it appeared that although the disulfide formation is not essential for folding, it is vital for maintaining protein stability and tight side-chain packing (23–25).
DISCUSSIONS
Water bears a variety of unique properties, many of which still remain poorly-understood. It is universally accepted that life on Earth would not be possible without water, and consequently, the existence of water on other planets in the universe is a sign that extraterrestrial life might originate and evolve there (26). On Earth, almost every activity in living cells occurs in water-based media, but mostly with the presence of salt ions, so it is really a great surprise to find that all 11 buffer-insoluble protein fragments/domains could be solubilized in salt-free water. Usually, to enhance protein solubility, the common method is to add more agents such as detergents, denaturants, or arginine (27) into the solution. Strikingly, this study shows that all 11 buffer-insoluble proteins could be solubilized in aqueous solution by removing salt ions in water. However, based on our experience, it seems that “double purity” is needed to get buffer-insoluble proteins dissolved in salt-free water. In other words, not only does the purity of water matter, but the purity of the protein is also essential. It appears that proteins have to be highly purified from other molecules such as lipid by affinity chromatography followed by reverse-phase HPLC purification. Probably, the existence of these molecules even in a tiny amount may block proteins from optimistically interacting with water by sticking to the exposed hydrophobic patches of proteins. For example, it seemed impossible to directly dissolve buffer-insoluble proteins out of lyophilized inclusion bodies with salt-free water.
This finding offered us the possibility to gain the first systematic insight into the structural properties of previously-deserted proteins. Based on CD and NMR HSQC assessment, the 11 protein fragments/domains can be categorized into three groups. The first group has no secondary structure, as judged by CD, and has narrowly-dispersed HSQC spectra with sharp peaks. This group includes 152-residue viral hemagglutinin RBD and 58-residue hNck2 first SH3 domain. The second group possesses secondary structure based on CD assessment, but broadened HSQC peaks. Consequently, only a small set of HSQC peaks was detectable. This group is composed of 66-residue Nogo-66; 86-residue Niv-86; 174-residue Wrch1 Cdc42-like domain; 319-residue N-NgR; and 421-residue entire NgR. The third group has secondary structure based on CD assessment, but, again, narrowly-dispersed HSQC spectra. However, these have well-separated HSQC peaks and thus were suitable for further high-resolution NMR study. This group consists of Nogo-60 (60 residues); Nogo-(21-60) (40-residues); Niv-44 (44 residues) and Claudin-51 (51 residues). Very strikingly, we failed to find any protein fragment/domain with a tight tertiary packing, as indicated by the possession of the near-UV signal or/and well-dispersed HSQC spectrum. This observation strongly implies that proteins are insoluble in buffers, probably because they lack an intrinsic propensity to reach or/and maintain the well-packed native state and consequently were trapped in the molten-globule or highly-disordered states, which have a tremendous tendency to aggregation in the presence of salt ions. However, such buffer-insoluble proteins may just account for a portion of denatured and partially-folded proteins, because it is well known that many denatured, partially-folded, as well as “intrinsically-unfolded”, proteins are soluble in buffer systems. In fact, the solubility of many intrinsically-unfolded proteins was found to be high because they usually have relatively low hydrophobicity.
Since the proteins we investigated here hold a significant diversity of cellular function, location, molecular sizes, and other properties, it would be reasonable to contemplate that, if not all, at least a substantial portion of previously abandoned proteins can be solubilized in salt-free water for further study, as exemplified on Nogo-60 (6). On the other hand, it appears extremely challenging to fully understand the molecular mechanism underlying the solubilization of buffer-insoluble proteins by salt-free water. Currently, in most cases, protein aggregation is considered to be dominated by intermolecular hydrophobic clustering and salt has significant effects on this process (2,28–31). The salt ion is thought to have both “salting-in” and “salting-out” effects on protein solubility. In other words, salts at a low concentration enhance protein solubility, whereas salts at a high concentration reduce solubility. Interestingly, it was recently reported that under some conditions, in particular when the protein molecules bear a significant number of net charges, only “salting-out” effects could be observed and, consequently, proteins should have the highest solubility in aqueous solution with the near-zero ionic strength (32,33). Theoretically, it was proposed that the net interaction between two charged particles such as protein molecules is the balance between the repulsive electrostatic contribution and the attractive van der Waals contribution. Therefore, the repulsive electrostatic interactions between two charged protein molecules will be reduced by increasing ionic strength (32). To explain our results, we propose here that buffer-insoluble proteins represent a portion of denatured and partially-folded proteins that lack intrinsic ability to reach or/and maintain a well-packed native conformation, and consequently, their hydrophobic side chains are highly exposed to the bulk solvent. As such, a very low ionic strength is sufficient to screen out intrinsic repulsive interactions such as electrostatic interaction, and consequently, the hydrophobic clustering/aggregation will rapidly occur. Most interestingly, our current results seem to reveal the marvelous fact that when proteins were originally selected to be functional players for life on Earth, they might have been offered the potential to manifest their full spectrum of structural states, ranging from the denatured to native states, by utilizing intrinsic repulsive interactions to suppress attractive hydrophobic clustering in pure water.
In summary, our study seems to suggest that if not all, at least a substantial fraction of buffer-insoluble proteins can be dissolved in salt-free water to manifest their intrinsic conformational states without going to significant aggregation. This finding may bear significant implications not only in practical applications, but also in our understanding of fundamental regimes of proteins.
Acknowledgments
We thank Professor Walter Englander at the University of Pennsylvania for his kind and critical comments on salt effects on protein stability and solubility.
This study is supported by Biomedical Research Council of Singapore (BMRC) grant R-183-000-097-305 and a BMRC Young Investigator Award, R-154-000-217-305, to J. Song).
References
- 1.Pace, C. N., S. Trevino, E. Prabhakaran, and J. M. Scholtz. 2004. Protein structure, stability and solubility in water and other solvents. Philos. Trans. R. Soc. Lond. B Biol. Sci. 59:1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi, E. Y., S. Krishnan, T. W. Randolph, and J. F. Carpenter. 2003. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm. Res. 20:1325–1336. [DOI] [PubMed] [Google Scholar]
- 3.Christendat, D., A. Yee, A. Dharamsi, Y. Kluger, M. Gerstein, C. H. Arrowsmith, and A. M. Edwards. 2000. Structural proteomics: prospects for high throughput sample preparation. Prog. Biophys. Mol. Biol. 73:339–345. [DOI] [PubMed] [Google Scholar]
- 4.Pedelacq, J. D., E. Piltch, E. C. Liong, J. Berendzen, C. Y. Kim, B. S. Rho, M. S. Park, T. C. Terwilliger, and G. S. Waldo. 2002. Engineering soluble proteins for structural genomics. Nat. Biotechnol. 20:927–932. [DOI] [PubMed] [Google Scholar]
- 5.Tsumoto, K., M. Umetsu, I. Kumagai, D. Ejima, J. S. Philo, and T. Arakawa. 2004. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 20:1301–1308. [DOI] [PubMed] [Google Scholar]
- 6.Li, M., J. Liu, and J. Song. 2006. Nogo goes in the pure water: solution structure of Nogo-60 and design of the structured and buffer-soluble Nogo-54 for enhancing CNS regeneration. Protein Sci. 15:1835–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu, J., M. Li, X. Ran, J. Fan, and J. Song. 2006. Structural insight into the binding diversity between the human Nck2 SH3 domains and proline-rich proteins. Biochemistry. 45:7171–7184. [DOI] [PubMed] [Google Scholar]
- 8.Ran, X., and J. Song. 2005. Structural insight into the binding diversity between the Tyr phosphorylated human ephrinBs and Nck2 SH2 domain. J. Biol. Chem. 280:19205–19212. [DOI] [PubMed] [Google Scholar]
- 9.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, B. A., and R. A. Blevins. 1994. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 4:603–614. [DOI] [PubMed] [Google Scholar]
- 11.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55. [DOI] [PubMed] [Google Scholar]
- 12.Li, M., J. Shi, Z. Wei, F. Y. Teng, B. L. Tang, and J. Song. 2004. Structural characterization of the human Nogo-A functional domains. Solution structure of Nogo-40, a Nogo-66 receptor antagonist enhancing injured spinal cord regeneration. Eur. J. Biochem. 271:3512–3522. [DOI] [PubMed] [Google Scholar]
- 13.Stevens, J., O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and L. A. Wilson. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 312:404–410. [DOI] [PubMed] [Google Scholar]
- 14.Park, S., K. Takeuchi, and G. Wagner. 2006. Solution structure of the first SRC homology 3 domain of human nck2. J. Biomol. NMR. 34:203–208. [DOI] [PubMed] [Google Scholar]
- 15.Feltham, J. L., V. Dotsch, S. Raza, D. Manor, R. A. Cerione, M. J. Sutcliffe, G. Wagner, and R. E. Oswald. 1997. Definition of the switch surface in the solution structure of Cdc42Hs. Biochemistry. 36:8755–8766. [DOI] [PubMed] [Google Scholar]
- 16.Schulman, B. A., P. S. Kim, C. M. Dobson, and C. Redfield. 1997. A residue-specific NMR view of the non-cooperative unfolding of a molten globule. Nat. Struct. Biol. 4:630–634. [DOI] [PubMed] [Google Scholar]
- 17.Song, J., P. Bai, L. Luo, and Z. Y. Peng. 1998. Contribution of individual residues to formation of the native-like tertiary topology in the alpha-lactalbumin molten globule. J. Mol. Biol. 280:167–174. [DOI] [PubMed] [Google Scholar]
- 18.Song, J., N. Jamin, B. Gilquin, C. Vita, and A. Menez. 1999. A gradual disruption of tight side-chain packing: 2D 1H-NMR characterization of acid-induced unfolding of CHABII. Nat. Struct. Biol. 6:129–134. [DOI] [PubMed] [Google Scholar]
- 19.Wei, Z., and J. Song. 2005. Molecular mechanism underlying the thermal stability and pH induced unfolding of CHABII. J. Mol. Biol. 348:205–218. [DOI] [PubMed] [Google Scholar]
- 20.He, X. L., J. F. Bazan, G. McDermott, J. B. Park, K. Wang, M. Tessier-Lavigne, Z. He, and K. C. Garcia. 2003. Structure of the Nogo receptor ectodomain: a recognition module implicated in myelin inhibition. Neuron. 38:177–185. [DOI] [PubMed] [Google Scholar]
- 21.Fralish, G. B., B. Dattilo, and D. Puett. 2003. Structural analysis of yoked chorionic gonadotropin-luteinizing hormone receptor ectodomain complexes by circular dichroic spectroscopy. Mol. Endocrinol. 17:1192–1202. [DOI] [PubMed] [Google Scholar]
- 22.Stumpp, M. T., P. Forrer, H. K. Binz, and A. Pluckthun. 2003. Designing repeat proteins: modular leucine-rich repeat protein libraries based on the mammalian ribonuclease inhibitor family. J. Mol. Biol. 332:471–487. [DOI] [PubMed] [Google Scholar]
- 23.Song, J., B. Gilquin, N. Jamin, E. Drakopoulou, M. Guenneugues, M. Dauplais, C. Vita, and A. Menez. 1997. NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin α/β motif and its hydrophobic side chain packing. Biochemistry. 36:3760–3766. [DOI] [PubMed] [Google Scholar]
- 24.Luo, Y., and R. L. Baldwin. 1999. The 28–111 disulfide bond constrains the α-lactalbumin molten globule and weakens its cooperativity of folding. Proc. Natl. Acad. Sci. USA. 96:11283–11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redfield, C., B. A. Schulman, M. A. Milhollen, P. S. Kim, and C. M. Dobson. 1999. Alpha-lactalbumin forms a compact molten globule in the absence of disulfide bonds. Nat. Struct. Biol. 6:948–952. [DOI] [PubMed] [Google Scholar]
- 26.Transcript of final discussion session for Meeting Issue. 2004. ‘The molecular basis of life: is life possible without water?’. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:1323–1328. [Google Scholar]
- 27.Ishibashi, M., K. Tsumoto, M. Tokunaga, D. Ejima, Y. Kita, and T. Arakawa. 2005. Is arginine a protein-denaturant? Protein Expr. Purif. 42:1–6. [DOI] [PubMed] [Google Scholar]
- 28.Baldwin, R. L. 1996. How Hofmeister ion interactions affect protein stability. Biophys. J. 71:2056–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neagu, A., M. Neagu, and A. Der. 2001. Fluctuations and the Hofmeister effect. Biophys. J. 81:1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos, C. H., and R. L. Baldwin. 2002. Sulfate anion stabilization of native ribonuclease A both by anion binding and by the Hofmeister effect. Protein Sci. 11:1771–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, H. X. 2005. Interactions of macromolecules with salt ions: an electrostatic theory for the Hofmeister effect. Proteins. 61:69–78. [DOI] [PubMed] [Google Scholar]
- 32.Retailleau, P., M. Ries-Kautt, and A. Ducruix. 1997. No salting-in of lysozyme chloride observed at low ionic strength over a large range of pH. Biophys. J. 73:2156–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruckenstein, E., and I. Shulgin. 2006. Effect of salts and organic additives on the solubility of proteins in aqueous solutions. Adv. Colloid Interface Sci. In press. [DOI] [PubMed]