Abstract
High resolution atomic force microscopy is a powerful tool to characterize nanoscale morphological features of protein amyloid fibrils. Comparison of fibril morphological properties between studies has been hampered by differences in analysis procedures and measurement error determination used by various authors. We describe a fibril morphology analysis method that allows for quantitative comparison of features of amyloid fibrils of any amyloidogenic protein measured by atomic force microscopy. We have used tapping mode atomic force microscopy in liquid to measure the morphology of fibrillar aggregates of human wild-type α-synuclein and the disease-related mutants A30P, E46K, and A53T. Analysis of the images shows that fibrillar aggregates formed by E46K α-synuclein have a smaller diameter (9.0 ± 0.8 nm) and periodicity (mode at 55 nm) than fibrils of wild-type α-synuclein (height 10.0 ± 1.1 nm; periodicity has a mode at 65 nm). Fibrils of A30P have smaller diameter still (8.1 ± 1.2 nm) and show a variety of periodicities. This quantitative analysis procedure enables comparison of the results with existing models for assembly of amyloid fibrils.
The misfolding of proteins is at the heart of many human diseases (see, for example, (1–4)). In Parkinson's disease, the misfolding and subsequent aggregation of the natively unfolded protein α-synuclein is an essential factor (5). In familial forms of Parkinson's disease, three point mutations in the α-synuclein gene have been identified that result in single amino acid substitutions in the α-synuclein protein: A30P, E46K, and A53T (6–8). Differences in the aggregation kinetics and/or aggregate morphology among the mutant proteins may yield new insights into the general process of protein misfolding and aggregation.
The nanoscale morphology of amyloid fibrils can be visualized with atomic force microscopy (AFM) or electron microscopy (see, for example, (9,10)). The aggregation of α-synuclein into mature fibrils has been described by models based on AFM images (11) and is supposed to proceed through various stages, each with characteristic morphological features such as typical fibril heights and periodicities.
Although there are many studies on the morphology of fibrillar aggregates of various amyloidogenic proteins, there has been little uniformity in the analysis and description of fibril characteristics (11). In this Letter, we describe a quantitative analysis procedure that is applicable to atomic force micrographs of amyloid fibrils originating from any amyloidogenic protein. The procedure (Fig. 1) takes into account the limitations of the physical process of imaging a biological sample with atomic force microscopy, such as tip-sample convolution.
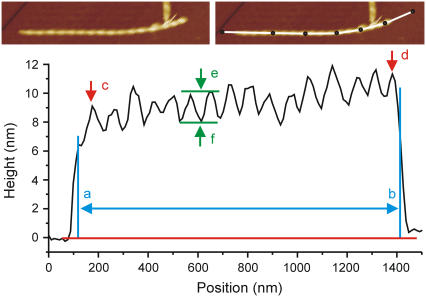
FIGURE 1.
Amyloid fibril characterization procedure applied to an E46K α-synuclein fibril. The top panel shows an AFM topography image of an α-synuclein fibril (left panel) with a manually drawn (ImageJ (12)) segmented line profile (right panel) on the curved fibril. The bottom panel shows the corresponding height along the profile. The length ab is considered the fibril length (a and b are placed just inside the ramp-up and ramp-down, which are ignored because of tip-sample convolution). The reported fibril height is the average height between a and b. The periodicity of the fibril is calculated as the distance cd divided by, in this case, 15 periods. The “typical” peak and trough heights are indicated by e and f. Finally, the height difference ef is the measured modulation depth (see Fig. 4 for comments on the interpretation). There is a certain subjectivity in the choice of which peaks to consider “typical”, due to the limited spatial resolution of the image (in this case, the lateral resolution is 11 nm/pixel) and due to possible peak height variations in the fibril itself.
We applied the quantitative image analysis procedure to AFM images (made in tapping mode and in liquid) of fibrils of wild-type and mutant α-synuclein formed after 72 h of aggregation (13). The analysis reveals grossly similar morphology but varying heights and periodicities among the disease-related mutants (Table 1). Fibrils from A30P and E46K α-synuclein appear smaller in height and correspondingly lower in peak height, trough height, and modulation depth than wild-type fibrils. Height measurements of wild-type α-synuclein fibrils in this study are consistent with those measured by others. We measure 10.0 ± 1.1 nm average height for the fibrils formed by wild-type α-synuclein, as compared to 11 ± 2 nm (14) and 9.8 ± 1.2 nm (11).
TABLE 1.
Quantitative comparison of fibril morphology of disease-related α-synuclein mutants
| Protein variant | N fibrils | Fibril height (nm) | Typical trough height (nm) | Typical peak height (nm) | Modulation depth (nm) | Periodicity (nm) |
|---|---|---|---|---|---|---|
| α-Synuclein wt | 35 (22) | 10.0 ± 1.1 | 9.3 ± 1.3 | 11.4 ± 1.2 | 2.1 ± 0.6 | 81 ± 24 (mode 65 nm)† |
| α-Synuclein A30P | 26 (21) | 8.1 ± 1.2 | 7.4 ± 1.4 | 8.9 ± 1.4 | 1.7 ± 0.6 | 103 ± 20 |
| α-Synuclein E46K | 26 (17) | 9.0 ± 0.8 | 7.7 ± 1.0 | 9.8 ± 1.1 | 2.0 ± 0.5 | 76 ± 34 (mode 55 nm) |
| α-Synuclein A53T | 0* |
N fibrils is the number of fibrils analyzed for a given mutant, with the number of fibrils classified as periodic in brackets. All fragments were profiled and characterized individually as described in Fig. 1. Those fibril fragments not classified as periodic were irregular in height. Height values are averaged over all N fibrils; the other parameters are averaged over the periodic fibrils. The standard deviation is used as the measurement error.
In our experiment, α-synuclein A53T did not aggregate into fibrils but into large amorphous aggregates (>1 μm diameter) that could not be characterized.
For a more accurate representation of the range of periodicities, see the histogram in Fig. 3.
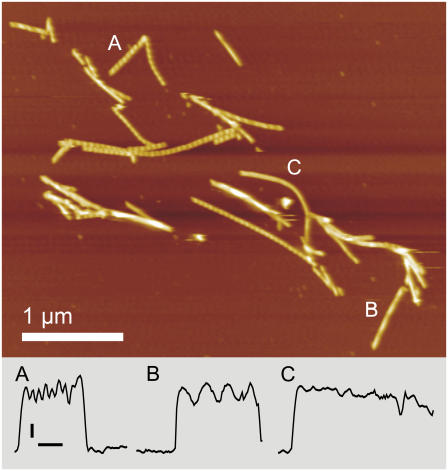
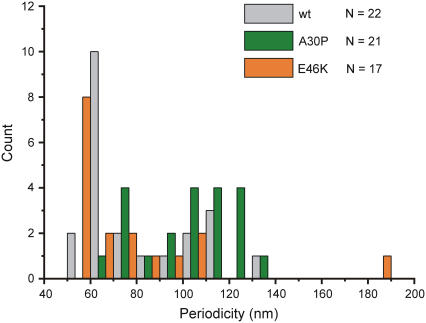
Most E46K fibrils display a periodicity of around 55 nm (as in Fig. 2 A), but much larger periodicities (Fig. 2 B) or unperiodic fibrils (Fig. 2 C) are also observed. The periodicities of E46K and wild-type α-synuclein fibrils have modes at 55 and 65 nm, respectively, and fibrils formed by A30P α-synuclein display a wider variety of periodicities (Fig. 3).
FIGURE 2.
Tapping mode atomic force micrograph of E46K α-synuclein in liquid after 72 h of aggregation. The color scale represents 17.6 nm in height. The fibril fragments marked A, B, and C are profiled along their length in the bottom panel indicating the range of morphologies observed in E46K fibrils. Vertical scale bar 2 nm; horizontal scale bar, 200 nm.
FIGURE 3.
Distribution of periodicities in fibrils of disease-related α-synuclein mutants. Wild-type and E46K α-synuclein fibrils have modes at 65 and 55 nm, respectively. A30P shows a wider variety of periodicities. Histogram bin size is 10 nm.
Quantitative discussion of morphological features of objects studied by AFM requires consideration of the influence of scanning system properties on the resulting image. One of the main limiting factors in deriving fibril properties from AFM images is tip-sample convolution, as evidenced by the large difference between the apparent width and the height of fibrils that are expected to be approximately round in cross section.
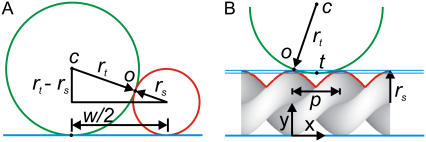
To assess the reliability of the measured parameters, we use a simple one-dimensional model for tip-sample convolution (Fig. 4). From Fig. 4 A, it follows that the tip radius is rt = w2/16rs, and the lowest measurable trough height can be calculated based on Fig. 4 B (see Supplementary Material). In the case of our α-synuclein fibrils, the trough height is overestimated in the images since the tip radius is large relative to the periodicity. This complicates attributing the observed periodicity to a supposed helical structure of the fibrils. We consider the measurements of the periodicity to be reliable since they were found to be equivalent for fibrils in any orientation with respect to the scan direction, and because the tip radius does not affect the periodicity observed but only the modulation depth, as shown in Fig. 4 B.
FIGURE 4.
Simple one-dimensional models for tip-sample convolution for helical fibrils. Fig. 4 A allows calculation of the tip radius assuming the tip (green) is spherical and the sample (red) has a spherical cross section (see Supplementary Material). Fig. 4 B models the apparent trough height for a tip with given radius scanning along the length of a double helical fibril. rt, tip radius; rs, sample radius; w, apparent width; o, contact point; c, tip center; t, apparent trough position; and p, periodicity.
In the hierarchical assembly model of α-synuclein fibril formation, as proposed by Khurana et al. (11), the trough height would be yt = (3/2)d, where d is the protofibril diameter (equal to the radius of the mature fibril rs). We can now compare the trough height values as modeled with the tip-sample convolution model, as modeled with the hierarchical assembly model, and as actually measured, for two fibrils with a different periodicity (Table 2). Based on this comparison, we conclude that the morphological features we observe are those one would expect of fibrils formed by twisted protofibrils, but we note that the lateral resolution of our images is not high enough to prove unambiguously that this model is indeed correct.
TABLE 2.
Comparison of measured versus modeled trough heights
| Fibril | rs (nm) | w (nm) | p (nm) | rt* (nm) | yt* (nm) | yt† (nm) | yt‡ (nm) |
|---|---|---|---|---|---|---|---|
| A | 4.5 | 90.7 | 56.2 | 114.3 | 7.5 | 6.8 | 7.0 |
| B | 4.75 | 91.2 | 186.4 | 109.4 | 7.2 | 7.1 | 7.0 |
Fibrils A and B refer to the fibrils highlighted in Fig. 2. Sample radius rs is half the peak height; w was measured from the image as the average width of 17 line profiles perpendicular to the fibril; p, periodicity; rt, tip radius; yt, trough height *from the convolution model; †from the hierarchical assembly model; ‡as measured.
This study introduces a quantitative morphological analysis procedure of fibrillar aggregates of amyloidogenic proteins. We show that high-resolution atomic force microscopy under sample-favorable imaging conditions (imaging under buffer, low tapping amplitude (∼4 nm), and low interaction force), combined with detailed image analysis, allows nanoscale comparison of fibrils formed by human wild-type α-synuclein and disease-related α-synuclein mutants. The fibrils formed by the disease-related mutants have a similar overall morphology, but a smaller diameter and a different periodicity than fibrils formed by wild-type α-synuclein. The observed fibril morphology is compatible with the hierarchical assembly model of α-synuclein fibrillization described in Khurana et al. (11). We emphasize that great care should be taken in deriving structural models from AFM data. We believe that the careful quantitative approach to determining fibril morphological characteristics reported here should be used more widely, and will be of increasing importance considering the growing number of studies that use advanced scanning probe microscopies for nanometer scale characterization of fibrils.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
The authors thank Kirsten van Leijenhorst for protein expression and purification, and Kees van der Werf for expert advice on atomic force microscopy.
This work is part of the research programme of the Stichting voor Fundamenteel Onderzoek der Materie, which is financially supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek.
References
- 1.Uversky, V., and A. Fink. 2004. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 1698:131–153. [DOI] [PubMed] [Google Scholar]
- 2.Dobson, C. M. 2003. Protein folding and misfolding. Nature. 426:884–890. [DOI] [PubMed] [Google Scholar]
- 3.Soto, C. 2003. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4:49–60. [DOI] [PubMed] [Google Scholar]
- 4.Forman, M. S., J. Q. Trojanowski, and V. M. Y. Lee. 2004. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 10:1055–1063. [DOI] [PubMed] [Google Scholar]
- 5.Lundvig, D., E. Lindersson, and P. H. Jensen. 2005. Pathogenic effects of α-synuclein aggregation. Brain Res. Mol. Brain Res. 134:3–17. [DOI] [PubMed] [Google Scholar]
- 6.Polymeropoulos, M. H., C. Lavedan, E. Leroy, S. E. Ide, A. Dehejia, A. Dutra, B. Pike, H. Root, J. Rubenstein, R. Boyer, E. S. Stenroos, S. Chandrasekharappa, A. Athanassiadou, T. Papapetropoulos, W. G. Johnson, A. M. Lazzarini, R. C. Duvoisin, G. Di Iorio, L. I. Golbe, and R. L. Nussbaum. 1997. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 276:2045–2047. [DOI] [PubMed] [Google Scholar]
- 7.Zarranz, J. J., J. Alegre, J. C. Gomez-Esteban, E. Lezcano, R. Ros, I. Ampuero, L. Vidal, J. Hoenicka, O. Rodriguez, B. Atares, V. Llorens, E. Gomez Tortosa, T. del Ser, D. G. Munoz, and J. G. de Yebenes. 2004. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55:164–173. [DOI] [PubMed] [Google Scholar]
- 8.Kruger, R., W. Kuhn, T. Muller, D. Woitalla, M. Graeber, S. Kosel, H. Przuntek, J. T. Epplen, L. Schols, and O. Riess. 1998. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat. Genet. 18:106–108. [DOI] [PubMed] [Google Scholar]
- 9.Hoyer, W., D. Cherny, V. Subramaniam, and T. Jovin. 2004. Rapid self-assembly of α-synuclein observed by in situ atomic force microscopy. J. Mol. Biol. 340:127–139. [DOI] [PubMed] [Google Scholar]
- 10.Choi, W., S. Zibaee, R. Jakes, L. C. Serpell, B. Davletov, R. Anthony Crowther, and M. Goedert. 2004. Mutation E46K increases phospholipid binding and assembly into filaments of human α-synuclein. FEBS Lett. 576:363–368. [DOI] [PubMed] [Google Scholar]
- 11.Khurana, R., C. Ionescu-Zanetti, M. Pope, J. Li, L. Nielson, M. Ramirez-Alvarado, L. Regan, A. L. Fink, and S. A. Carter. 2003. A general model for amyloid fibril assembly based on morphological studies using atomic force microscopy. Biophys. J. 85:1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasband, W. S. 1997. –2006. ImageJ. http://rsb.info.nih.gov/ij/.
- 13.See Supplementary Material for protein expression, purification and aggregation, and AFM imaging conditions.
- 14.Hoyer, W., T. Antony, D. Cherny, G. Heim, T. M. Jovin, and V. Subramaniam. 2002. Dependence of α-synuclein aggregate morphology on solution conditions. J. Mol. Biol. 322:383–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.