Abstract
Titin proteins play an essential role in maintaining muscle function and structure. Recent work has implicated the involvement of the novex-3 titin isoform in sarcomere restructuring and disease. Unlike avian and mammalian systems, Xenopus laevis myogenesis is characterized by a wave of primary myogenesis followed by apoptosis of the primary muscles and formation of new muscles by secondary myogenesis. We show here that the Xenopus laevis novex-3 titin isoform (Xtn3) is developmentally expressed throughout the somites, heart, and primary muscles of the developing embryo. Downregulation of Xtn3 expression at tadpole stages appears to coincide with the change in myofiber composition from solely embryonic “fast” fiber types to myofibers containing both “fast” and “slow” fiber types. We demonstrate that Xtn3 is expressed early in the presomitic mesoderm and remains expressed in the somites, ventral myoblasts, and developing jaw muscles through late tailbud stage. Furthermore, we show that Xtn3 is expressed in the cardiac primordia prior to linear heart tube formation and remains expressed in the heart until tadpole stage, at which point it is downregulated in the heart except in discrete patches of cardiac cells. Finally, we demonstrate that Xtn3 transcripts are detectable in adult heart and muscle tissues.
Keywords: Titin, TTN, Novex, Xtn3, Cardiogenesis, Cardiac, Heart, Somite, Muscle, Skeletal muscle, Pharyngeal, Myoblast, Myogenesis, Somitogenesis, Myofibril, Sarcomere, Development, Xenopus
1. Results and discussion
The process of myogenesis in Xenopus laevis appears to be unique compared to mammalian and avian systems in that it is characterized by two completely distinct types of myogenesis (reviewed in Chanoine and Hardy, 2003). Initially, primary myogenesis results in formation of uninucleate myofibers assembled during early neurula embryos that become functional following neurulation (stage 24). These primary muscles are initially composed entirely of one “fast” fiber type, but by late tailbud stage (stage 37), a second “slow” fiber type appears. During metamorphosis the primary myocytes begin a wave of autonomous programmed cell death resulting in a complete replacement of these primary muscles with “adult” multinucleate muscles produced by a process of secondary myogenesis. Furthermore, this process has been shown to involve switching from embryonic to adult muscle protein isoforms by differential mRNA splicing (Hardy et al., 1999; Radice and Malacinski, 1989).
The giant muscle protein titin is the largest known protein in the biological world and the single human titin gene contains 363 exons with a coding potential of 38,138 amino acids. Many splice-variants of titin have been described and full-length isoforms have been shown to span half the sarcomere length from the Z-line to the M-line, serving to link the two regions and stabilize sarcomere length by acting as a molecular spring (reviewed in Granzier et al., 2002). To date, the cloning or expression of Xenopus titin orthologues has not been reported. Recently a novel human titin exon, termed novex-3, was identified within the human titin locus and shown to encode an alternative C-terminal exon resulting in a truncated titin isoform that is too short to reach the A-band (Bang et al., 2001). This isoform was shown to be expressed in human skeletal and cardiac muscle and the protein was shown to bind the signaling molecule, obscurin (Bang et al., 2001). The novex-3 titin/obscurin complex extends in a spring-like manner when sarcomeres are stretched and has been hypothesized to be involved in stretch-initiated sarcomeric restructuring during muscle adaptation and disease (Bang et al., 2001). This observation also lends to the possibility that the novex-3 titin/obscurin contributes to the development and remodeling of embryonic skeletal and cardiac muscles during embryonic myogenesis. Alternatively, this complex may be involved in the process of switching from the embryonic “fast” fiber type to the “slow” fiber type that occurs during late tailbud stage.
1.1. Sequence comparisons
Xtn3, the partial Xenopus novex-3 titin isoform (GenBank Accession No.: DQ241814) was initially identified in a screen for differentially expressed cardiac genes as an expressed-sequence tag (EST; GenBank Accession No.: CB564652) expressed in the developing heart. This EST was subsequently sequenced and a BLAST search revealed that the EST is highly similar to the human truncated titin isoform, novex-3 titin. The partial Xenopus novex-3 titin EST consists of 2771 nucleotides, encodes for 544 putative amino acids, and appears to lie completely within the putative novex-3 exon as identified by comparisons with human novex-3 titin (Fig. 1A). Furthermore, this EST appears to align with a single exon within the titin gene in the Xenopus tropicalis genome (JGI, ver. 4.1, scaffold 163). Protein alignments performed against a predicted chick titin (GenBank Accession No.: XM_421979), predicted mouse novex-3 titin (GenBank Accession No.: XM_619766), predicted cow novex-3 titin (GenBank Accession No.: XM_617216), and human novex-3 titin (GenBank Accession No.: NM_133379) reveal identities in the range of 39–48% (Figs. 1A and B). Furthermore, all three immunoglobulin cell adhesion molecule (IGcam) domains are conserved between human and frog, as identified by the NCBI conserved domain search (cd#: cd00931.2; Fig. 1A). Thus, the partial Xenopus titin clone is likely the Xenopus orthologue of human novex-3 titin and we refer to this transcript as Xtn3 for Xenopus titin novex 3.
Fig. 1.
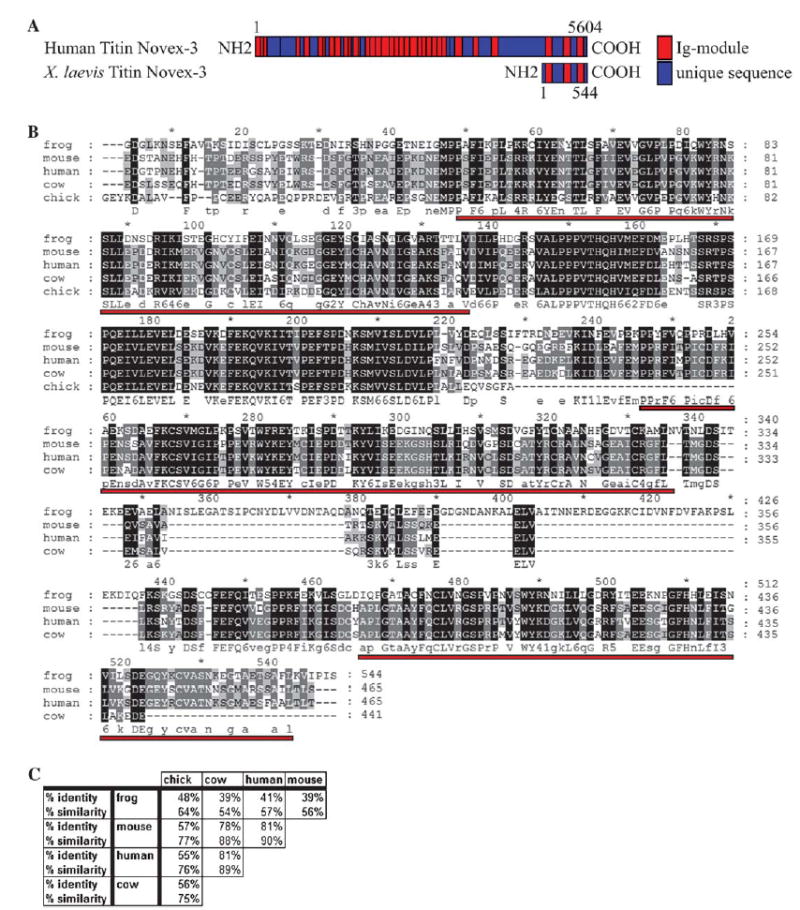
XTN3 is a conserved titin isoform. (A) Schematic depicting human novex-3 titin and the partial XTN3 proteins, including positions of conserved Ig domains and amino acid lengths. (B) Protein alignment of Xenopus laevis XTN3, a predicted chick titin, a predicted mouse novex-3 titin, a predicted cow titin, and human novex-3 titin. Alignment was performed using the GeneDoc program. Red underlines indicate conserved Ig domains as identified by the NCBI conserved domain search. (C) Table showing degree of protein identity and similarity between the putative novex-3 titin orthologues.
1.2. Expression pattern
Human novex-3 titin consists of 5604 amino acids and 16,812 nucleotides. Due to the large size of the titin gene and the many titin isoforms, it was not feasible to clone the full-length version of Xtn3, thus the partial Xtn3 EST was used for whole-mount in situ hybridization analyses. The novex-3 exon is contained only in the novex-3 titin isoform mRNA, thus the partial Xtn3 probe is specific for this titin isoform. Expression analyses of Xtn3 reveal that this transcript is initially detected during the onset of neurulation (stage 14/15) throughout the developing presomitic mesoderm. At this stage, Xtn3 is expressed in a narrow anterior domain which flares laterally toward the posterior region of the presomitic mesoderm (Fig. 2A). This pattern mirrors that of the presomitic mesoderm as it constricts in a wave from anterior to posterior during somite formation. By mid-neurula stage (stage 19) Xtn3 expression becomes more restricted as the forming somites develop into vertical stripes (Figs. 2B and C). In higher magnifications of tailbud stage embryos (stage 35) it is apparent that the somitic expression lies within a thin stripe down the center of the somite as well as within a lower intensity stripe along the anterior and posterior borders of individual somites (inset, Fig. 2H). The somitic expression continues through development until tadpole stage at which point Xtn3 expression is almost completely undetectable (Fig. 2N). By stage 37, Xtn3 also becomes expressed in the migrating ventral myoblasts, which will eventually become the ventral abdominal muscles (Figs. 2J and L). Also at this stage Xtn3 expression becomes apparent in the developing facial and jaw muscles (Figs. 2J and L). Expression in the migrating heart primordia was first detected prior to fusion of the primordia at the midline (stage 26; Figs. 2D and E). Cardiac expression persists throughout all stages of heart development until tadpole stages, at which point Xtn3 expression is almost completely lost (Figs. 2N–P). By tadpole stage (stage 46), only weak expression is detected in the jaw muscles. Staining of the adult heart shows very little Xtn3 expression globally. However, discrete groups of Xtn3-expressing cells can be detected near the surface of the myocardium (Figs. 2O and P). Thus, it appears that a global downregulation of Xtn3 corresponds with fully differentiated muscle and completion of heart development, although some expression remains in adult tissues.
Fig. 2.
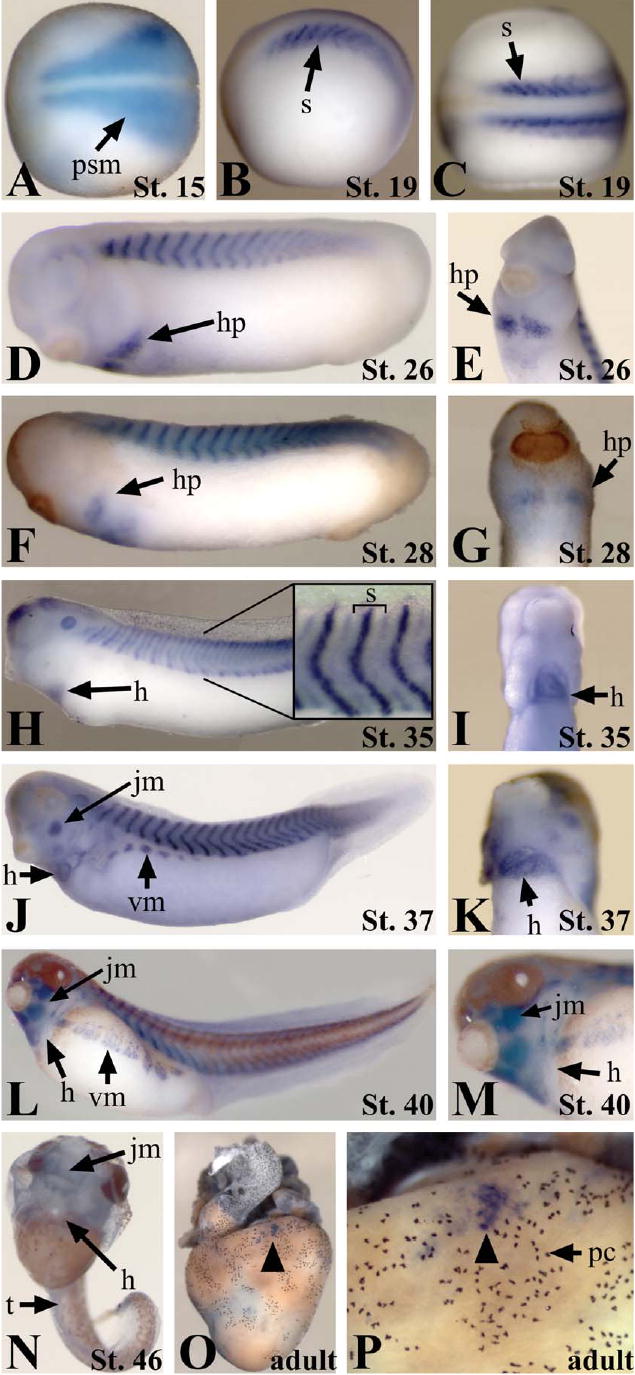
Xtn3 is developmentally expressed throughout the somites, heart, and jaw myoblasts from early neurula to late tailbud stages. (A–P) In situ hybridization showing expression of the Xtn3 at the indicated stages. (A–C) Neurula stage embryos. Anterior is to the left. (A and C) Dorsal views. (B) Lateral view. (D–M) Tailbud stage embryos. (E, G, I, K, and M) Ventral anterior views. Anterior is up. (H) Inset shows higher magnification of somite region. Bracket indicates one individual somite. (N) Tadpole stage embryo. Note the lack of staining in trunk and heart. Some light staining remains in facial muscles. (O and P) Adult heart lacking global Xtn3 expression. (P) A magnification of (O). Black arrowhead indicates isolated group of Xtn3-expressing cells. h, heart; hp, heart primordia; jm, jaw myoblasts; psm, presomitic mesoderm; pc, pigment cells; vm, ventral myoblasts; s, somites; t, trunk.
Histological sectioning of embryos stained by whole-mount in situ hybridization revealed that Xtn3 is initially expressed more highly in the dorsal medial lip and medial regions of the developing myotome during neurula stage (stage 19; Fig. 3A). Xtn3 expression then becomes expanded throughout the entire dorsal/ventral axis of the somitic domain during later stages of myotome development (Figs. 3A, C, E, and G). Cardiac expression is detected exclusively in the myocardium, consistent with the defined role of titin as a sarcomeric structural protein (Figs. 3D–H). Xtn3 appears to be expressed in a gradient during looping stages of cardiogenesis, with higher levels in the ventral cardiac regions (Figs. 3F and H). Sectioning further demonstrates Xtn3 expression throughout the developing jaw muscles in the head (Fig. 3I).
Fig. 3.
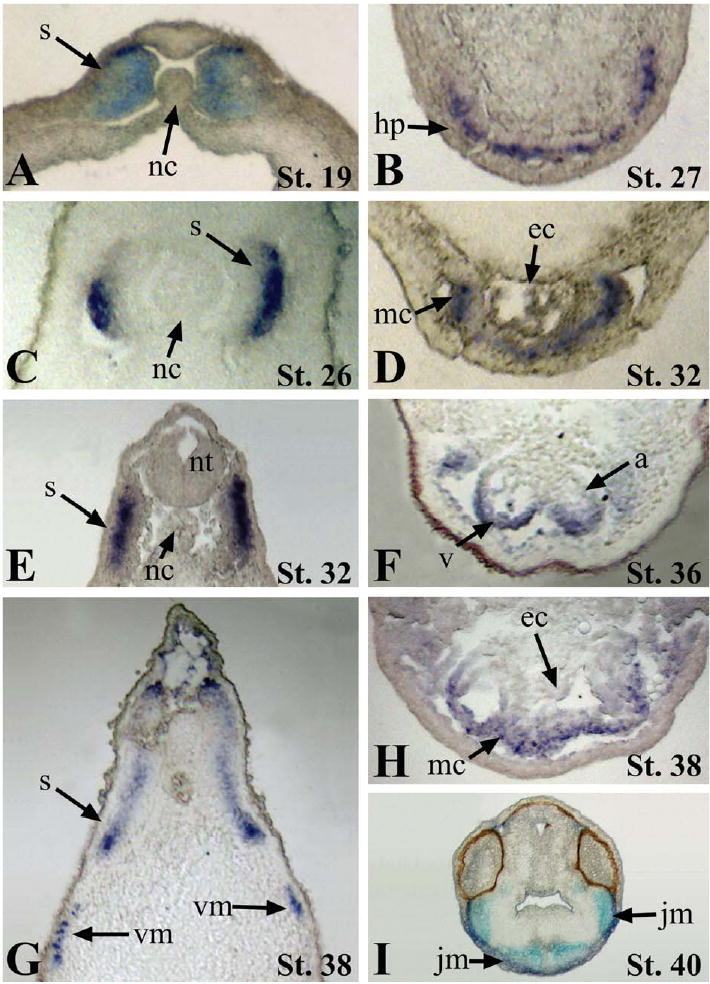
Histology of Xtn3 expression. Paraffin embedded embryos were sectioned at a depth of 16 μm after in situ hybridization. (A, C, E, and G) Transverse sections through developing somites. (B, D, F, and H) Transverse sections through developing cardiac region. (I) Transverse section showing jaw myoblasts in the head. a, atrium; ec, endocardium; hp, heart primordia; jm, jaw myoblasts; mc, myocardium; nc, notochord; nt, neural tube; s, somites; v, ventricle; vm, ventral myoblasts.
To test whether Xtn3 transcripts are detectable in adult heart and muscle tissues, RT-PCR was performed on RNA isolated from St.13, St.38, adult heart and adult muscle tissues. Xtn3 transcripts were present in St.38, adult heart and adult muscle tissues, as were transcripts for myosin heavy chain α (MHCα) and muscle actin (Fig. 4). In contrast, the heart marker Tbx20 was only detectable in St.38 embryos and in adult heart tissue (Fig. 4). Thus, it appears that while Xtn3 may be down-regulated at tadpole stages as assayed by whole-mount in situ hybridization, Xtn3 transcripts can be detected by sensitive methods in adult muscle tissues.
Fig. 4.
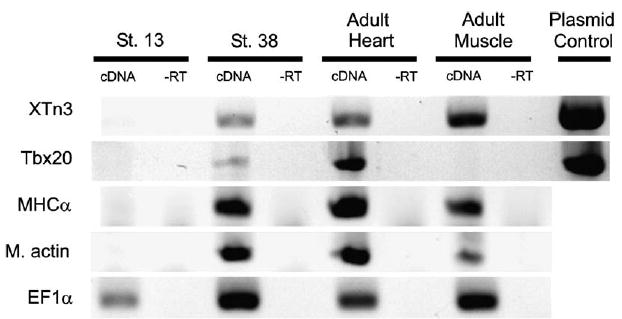
Xtn3 is expressed in adult heart and skeletal muscle. RNA from St.13 embryos, St.38 embryos, adult heart, and adult muscle tissue was isolated and 100 ng of RNA subjected to RT-PCR. Xtn3 RT-PCR shows expression in St.38, adult heart and adult muscle tissues. “−RT” controls lack reverse transcriptase to show lack of genomic contamination. Tbx20 primers serve as controls for heart tissue, MHCα and muscle actin serve as controls for muscle tissue, and EF1α serves as loading control.
2. Experimental procedures
The Xtn3 EST clone was obtained from Open Biosystems (IMAGE ID: 5537366). The Xtn3-pCMV-SPORT6 plasmid was linearized using SalI and digoxigenin-conjugated in situ hybridization probes were synthesized using T7 polymerase. Embryos were collected and fixed in MEMFA for 1.5 h at room temperature. Whole-mount in situ hybridization was performed as previously described (Harland, 1991). Embryos were paraffin-embedded using a Tissue-Tek II Tissue Embedding Center and sectioned at a depth of 16 μm on a Microm HM 340 E microtome. Protein alignments were performed using the GeneDoc program.
RNA for RT-PCR was extracted from 30 St.13 embryos, 30 St.38 embryos, 500 mg adult heart tissue, and 500 mg adult thigh muscle tissue using the RNeasy Kit (QIAgen). First-strand cDNA synthesis was performed using Superscript II Kit (Invitrogen) and random hexamers. PCRs with Taq polymerase (Promega) were performed according to established protocol using 2 mu;l of the resulting cDNA. MHCα, m. actin, and EF1αprimers sequences were obtained from the website of Eddie DeRobertis. Primers: Xtn3: forward – AGT TGA TAA TAC CTT CAA TGC CAC, reverse – AGA TTA TTG CAT GAA ACC CTG C; Tbx20: forward – GAG AAT GTT TCC TAC AAT CCG, reverse – TTC CCT CTC AAT ATC AGT GAG; m. actin: forward – GCT GAC AGA ATG CAG AAG, reverse – TTG CTT GGA GGA GTG TGT; MHCα: forward – GCC AAC GCG AAC CTC TCC AAG TTC CG, reverse – GGT CAC ATT TTA TTT CAT GCT GGT TAA CAG G; EF1α: forward – CAG ATT GGT GCT GGA TAT GC, reverse – ACT GCC TTG ATG ACT CCT AG.
Acknowledgments
This work is supported by a grant from the NIH/NHLBI HL075256-01. D.D.B. is funded by a NSF graduate research fellowship. We wish to thank Kathleen Christine, Jeanine LaRocque, Erika Paden, and Dr. Chris Showell for critical reading of the manuscript.
References
- Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Chanoine C, Hardy S. Xenopus muscle development: from primary to secondary myogenesis. Dev Dyn. 2003;226:12–23. doi: 10.1002/dvdy.10206. [DOI] [PubMed] [Google Scholar]
- Granzier H, Labeit D, Wu Y, Labeit S. Titin as a modular spring: emerging mechanisms for elasticity control by titin in cardiac physiology and pathophysiology. J Muscle Res Cell Motil. 2002;23:457–471. doi: 10.1023/a:1023458406346. [DOI] [PubMed] [Google Scholar]
- Hardy S, Hamon S, Cooper B, Mohun T, Thiebaud P. Two skeletal alpha-tropomyosin transcripts with distinct 3′ UTR have different temporal and spatial patterns of expression in the striated muscle lineages of Xenopus laevis. Mech Dev. 1999;87:199–202. doi: 10.1016/s0925-4773(99)00148-3. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole mount method for Xenopus embryos. Methods Cell Biol. 1991;36:675–685. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Radice GP, Malacinski GM. Expression of myosin heavy chain transcripts during Xenopus laevis development. Dev Biol. 1989;133:562–568. doi: 10.1016/0012-1606(89)90058-4. [DOI] [PubMed] [Google Scholar]


