Abstract
T-box genes have diverse functions during embryogenesis and are implicated in several human congenital disorders. Here, we report the identification, sequence analysis, and developmental expression patterns of four members of the T-box gene family in the diploid frog Xenopus tropicalis. These four genes—Tbx1, Tbx2, Tbx5, and Tbx20—have been shown to influence cardiac development in a variety of organisms, in addition to their individual roles in regulating other aspects of embryonic development. Our results highlight the high degree of evolutionary conservation between orthologs of these genes in X. tropicalis and other vertebrates, both at the molecular level and in their developmental expression patterns, and also identify novel features of their expression. Thus, X. tropicalis represents a potentially valuable vertebrate model in which to further investigate the functions of these genes through genetic approaches.
Keywords: atrium, branchial, cardiac, cement, DiGeorge, ear, eye, frontonasal, heart, Holt-Oram, hypaxial, pharyngeal, placode, proctodeum, profundal, pronephric, Silurana, T-box, Tbx1, Tbx2, Tbx5, Tbx20, T-domain, trigeminal, tropicalis, ventricle, Xenopus
INTRODUCTION
DNA binding transcription factors encoded by several members of the T-box gene family have been shown to have both cell-autonomous and non–cell-autonomous roles in controlling the development of the heart during embryogenesis (Plageman and Yutzey, 2005; Stennard and Harvey, 2005). These roles appear to be conserved during evolution, and in some cases, their importance is highlighted by the association between mutations in these factors and the incidence of human congenital heart defects (Packham and Brook, 2003; Ryan and Chin, 2003; Mandel et al., 2005). In addition, the same genes have been shown to be required for the proper development of other tissues and organs, such as the eye (Tbx5; Koshiba-Takeuchi et al., 2000) and ear (Tbx1; Piotrowski et al., 2003; Vitelli et al., 2003; Liao et al., 2004; Raft et al., 2004; Moraes et al., 2005), while other T-box genes have key roles in regulating early embryonic patterning (Showell et al., 2004).
Xenopus is a valuable model organism in which to investigate the molecular and genetic regulation of organogenesis in general and heart development in particular, and reverse genetic approaches recently have been developed to isolate mutant alleles in specific genes of interest in the diploid frog Xenopus (Silurana) tropicalis. In comparison with the zebrafish (Danio rerio), Xenopus cardiac morphology is more similar to that of humans, including septation of the atrium into left and right chambers (Hu et al., 2000; Mohun et al., 2000). Also, the accessibility of the embryo throughout development and the high fecundity of the frog are significant advantages over the mouse, both in embryological analysis and in genetic screening.
The genes analyzed here—Tbx1, Tbx2, Tbx5, and Tbx20—are all known to play important roles in regulating normal cardiac development. TBX1 lies within a critical region of human chromosome 22 (22q11.2) that is deleted in patients with DiGeorge syndrome, and loss of Tbx1 function in the mouse mimics the severe morphological defects of the outflow tract of the heart that are seen in DiGeorge patients (Jerome and Pappaioannou, 2001; Lindsay et al., 2001). Similarly, mutations in the human TBX5 gene are associated with Holt–Oram syndrome, affecting atrial and ventricular septation, the cardiac conduction system and the development of the upper limbs (Basson et al., 1997; Li et al., 1997). Tbx5 has been shown to act in concert with Tbx20 at the molecular level to control cardiac morphogenesis (Brown et al., 2005). Conversely, Tbx5 and Tbx2 appear to function within distinct domains of the developing heart, contributing to the patterning of the early heart tube and its subsequent morphological regionalization. Several recent studies have also demonstrated a requirement for Tbx20 function for proper regulation of Tbx2 expression within the developing heart (Cai et al., 2005; Singh et al., 2005; Stennard et al., 2005). As a preliminary step in investigating the molecular basis of their developmental roles through genetic analysis in the emerging model organism Xenopus tropicalis, we have identified cDNA clones containing full-length coding sequences corresponding to these four T-box genes, determined the structure of their genomic loci in silico, and characterized their spatial expression patterns over a wide range of stages during embryogenesis. Our results demonstrate the high degree of sequence conservation of T-box gene orthologs in X. tropicalis and highlight both conserved and previously undescribed aspects of their embryonic expression.
RESULTS AND DISCUSSION
Sequence Analysis of Xenopus tropicalis T-Box Gene Orthologs
cDNA clones corresponding to Tbx1, Tbx2, and Tbx20 were identified by BLAST searches within a database of Xenopus tropicalis expressed sequence tags and a clone containing the translation initiation codon was obtained and sequenced for each gene. A cDNA encoding the X. tropicalis ortholog of Tbx5 was cloned by reverse transcriptase-polymerase chain reaction (RT-PCR) and sequenced. These cDNA sequences were used to search the X. tropicalis draft genome sequence (DoE Joint Genome Institute) for genomic scaffolds containing the corresponding loci. The cDNA sequences were then mapped onto the genomic locus sequences, and the exon/intron boundaries were identified based on consensus sequences for eukaryotic splice donor and acceptor sites (Mount, 1982).
All four Xenopus tropicalis cDNA clones exhibit a very high degree of sequence identity when compared with their Xenopus laevis orthologs, particularly within their coding regions. The 1,389-nucleotide (nt) open reading frame within the 3,065-bp Tbx1 cDNA is 94% identical to that of Xenopus laevis Tbx1 (GenBank accession no. AF526274; 89% identity in untranslated regions). The degrees of identity and similarity between the conceptually translated Xenopus tropicalis Tbx1 coding sequence and several vertebrate orthologs are shown in Table 1. The results of our analysis of the genomic Tbx1 locus are shown in Figure 1a.
TABLE 1.
Degrees of Identity and Similarity Between Xenopus tropicalis T-domain Proteins and Several Vertebrate Orthologs
| Xenopus laevis | Danio rerio | Gallus gallus | Mus musculus | Homo sapiens | |
|---|---|---|---|---|---|
| Tbx1 | 97% (98%) | 80% (87%) | N/A | 71% (79%) | 69% (77%) |
| Tbx2 | 96% (97%) | 77% (84%) | N/A | 70% (77%) | 70% (77%) |
| Tbx5 | 95% (96%) | 64% (73%) | 81% (87%) | 78% (85%) | 78% (85%) |
| Tbx20 | 97% (98%) | 85% (92%) | 90% (95%) | 90% (95%) | N/A |
Fig. 1.
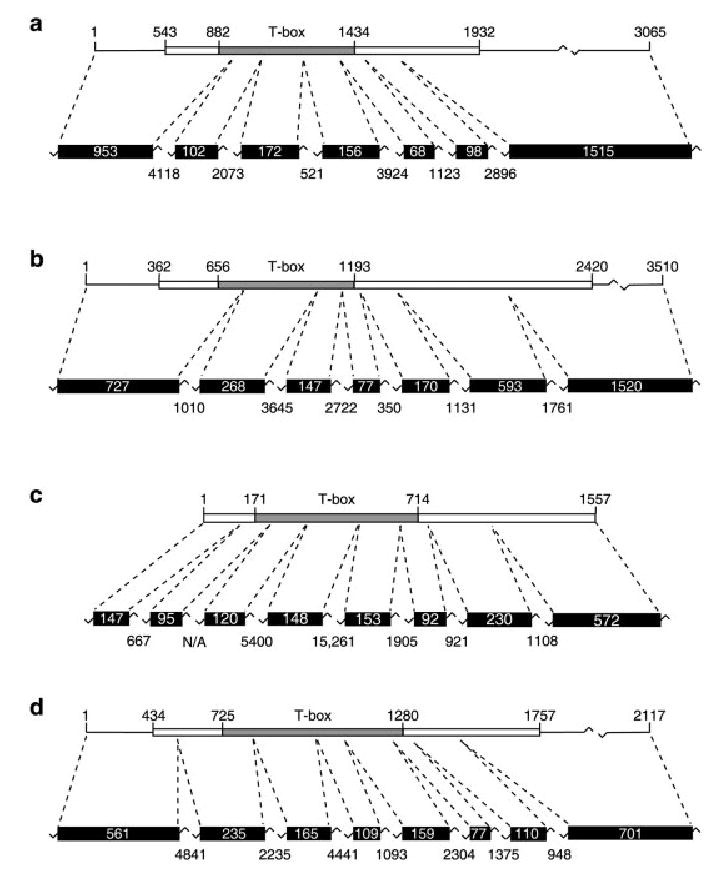
Genomic locus structure of Tbx1, Tbx2, Tbx5, and Tbx20 in Xenopus tropicalis. Tbx1 (a), Tbx2 (b), Tbx5 (c), and Tbx20 (d) cDNAs and their corresponding genomic loci are shown in diagrammatic form (not to scale). Coding regions of each cDNA are shown (boxes) together with their nucleotide positions and the position of the T-box (defined by alignment of the encoded proteins with the T-domain of Xbra) is also indicated. The exons corresponding to the cDNA sequences are shown together with their sizes (in base pairs) plus those of the intervening introns. Note that, as the size of the first exon of each gene is predicted based on the available cDNA sequence, the sizes of these exons may be underestimated here.
The 3,510-bp Tbx2 cDNA identified here contains a 2,055-nt open reading frame with 94% identity to Xenopus laevis Tbx2 (GenBank accession no. AB032941; 86% identity in untranslated regions). Table 1 shows the degrees of identity and similarity between conceptually translated Xenopus tropicalis Tbx2 and vertebrate orthologs. Mapping of the Tbx2 cDNA sequence to the available genome sequence identified a 14,129-bp region containing the complete cDNA sequence divided among seven exons (Fig. 1b).
Xenopus tropicalis Tbx5 is encoded by a 1,557-nt open reading frame. Alignment of this sequence with the Xenopus laevis Tbx5 cDNA (GenBank accession no. AF133036) identified 93% nucleotide sequence identity between the coding regions of the two orthologs. The Xenopus tropicalis Tbx5 cDNA encodes a product exhibiting a high degree of evolutionary conservation among vertebrate species (Table 1). Results obtained from in silico analysis of the Tbx5 genomic locus are shown in Figure 1c.
The Tbx20 cDNA clone obtained consists of 2,117 bp, containing a 1,320-nt open reading frame with 93% sequence identity to that of Xenopus laevis Tbx20 (GenBank accession no. AY154394; 75% identity in untranslated regions). Table 1 shows the degree of sequence identity and similarity between conceptually translated Xenopus tropicalis Tbx20 and its orthologs in other vertebrates. The Tbx20 sequence was found to be divided among eight exons within a 19,354-bp region of a single genomic scaffold (Fig. 1d).
Analysis of Tbx1 Expression During Embryogenesis
Tbx1 function is required for normal heart development in vertebrates. It is thought to act indirectly, influencing the differentiation of migrating cardiac neural crest cells by regulating the expression of one or more intercellular signals emanating from Tbx1-expressing cells in the pharyngeal endoderm and the mesenchymal core of the pharyngeal arches (Kochilas et al., 2005). The cardiac neural crest cells contribute to the formation of the outflow tract of the heart and the development of this region is severely affected in DiGeorge patients and in mouse models of the syndrome. Initial analysis of the phenotype of a hypomorphic Tbx1neo allele in the mouse suggests that the observed alignment and septation defects of the outflow tract are independent, thus underscoring the value of analyzing more subtle alleles in addition to single gene knockouts and larger deletions in vertebrate models (Xu et al., 2005). To determine the spatial patterns of Tbx1 mRNA expression during the course of X. tropicalis embryogenesis, whole-mount in situ hybridization was performed. At the earliest stage analyzed, stage 10.5 (early gastrula), no expression of Tbx1 was detected. In early neurulae (stage 13), regionally restricted expression was clearly detected in a broad anterior domain surrounding the anterior end of the mediodorsal groove of the neural plate (Fig. 2a). Within this broad ectodermal domain, two bilateral patches of strong Tbx1 expression were detected flanking the mediodorsal groove (Fig. 2a,c). These patches marked the posterior boundary of the Tbx1 expression domain. In late neurulae (stage 19), strong expression was detected in the anterior ectoderm (Fig. 2d–f). This expression domain appeared to largely exclude the central nervous system, commonly defined by the expression of pan-neural markers such as the neural cell adhesion molecule (N-CAM; Eagleson et al., 1995). Expression was not detected in the developing eye anlagen and cement gland and was only weakly detected in the region of the neural tube posterior to the eye anlagen. Instead, expression of Tbx1 was found to immediately abut these regions of the ectoderm. As at stage 13, two distinct bilateral regions of strong staining were observed within the Tbx1 expression domain at stage 19, extending as approximate dorsoventral stripes in the ectoderm on either side of the anterior central nervous system. It is unclear whether this Tbx1 expression domain corresponds to the location of the proposed primordium of the ectodermal (neurogenic) placodes (Schlosser and Northcutt, 2000; Schlosser and Ahrens, 2004). At early tail bud stage (stage 25), Tbx1 was found to be expressed in three distinct areas within the pharyngeal region and in the ventral region of each otic vesicle (Fig. 2g,h). At stage 33, expression within the otic vesicles extended further laterally (Fig. 2n). However, in subsequent stages (stages 40, 47) expression remained restricted to the ventral and lateral regions of the vesicles. This finding differs from the pattern reported for X. laevis, in which Tbx1 appeared to be expressed throughout the vesicles (Ataliotis et al., 2005).
Fig. 2.
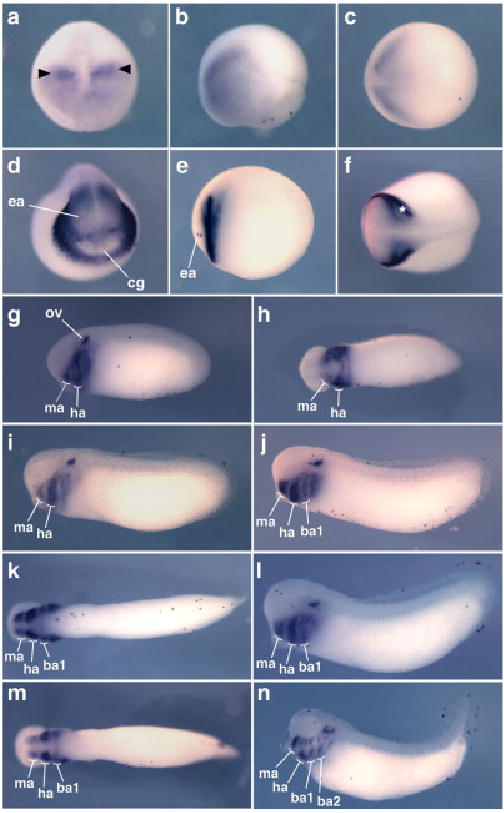
Expression pattern of Tbx1 in Xenopus tropicalis. The results of in situ hybridizations for Tbx1 expression from early neurula to late tail bud stages are shown (embryos uncleared). Except for the anterior views shown in a and d, all embryos are oriented with anterior to the left. a– c: Stage 13 is shown in anterior (a), lateral (b), and dorsal (c) views. Bilateral patches of stronger expression are indicated in a) by arrowheads. d–f: Stage 19 is shown in anterior (d), lateral (e), and dorsal (f) views. Bilateral stripes of stronger expression are indicated in f) by an asterisk. g–n: Tbx1 expression through tail bud stages is shown as follows: stage 25 lateral (g) and ventral (h), stage 26 lateral (i), stage 27 lateral (j) and ventral (k), stage 28 lateral (l) and ventral (m), stage 33 lateral (n). ba1, first branchial arch; ba2, second branchial arch; cg, cement gland; ea, eye anlagen; ha, hyoid arch; ma, mandibular arch; ov, otic vesicle.
Expression of Tbx1 orthologs in the pharyngeal region is broadly conserved among vertebrate species. Between stages 25 and 33, the elaboration of the expression pattern of Tbx1 in this region of the X. tropicalis embryo reflects the morphogenesis of the pharyngeal arches. In this region, the cells expressing Tbx1 lay beneath the overlying epidermis. At stages 25 and 26, expression was detected in the mandibular and hyoid arches (Fig. 2g–i) and in a third domain corresponding to the future branchial arches, posterior to the hyoid arch. At stage 27, at which the first branchial arch becomes fully formed, Tbx1 expression was detected in four distinct pharyngeal domains—the mandibular, hyoid, and first branchial arches and a more posterior branchial region (Fig. 2j,k). By stage 33, expression was also detected in the second branchial arch (Fig. 2n). At this stage, Tbx1 appears to mark distinct dorsal and ventral regions within the hyoid, first branchial, second branchial, and forming third branchial arches.
Analysis of Tbx2 Expression During Embryogenesis
In situ hybridization showed that, as in X. laevis (Hayata et al., 1999), Tbx2 is expressed ventrally in X. tropicalis early gastrulae (stage 10.5; Fig. 3a,b). However, in contrast to the reported expression in X. laevis, Tbx2 is expressed most strongly in the outer layer of ectodermal cells in X. tropicalis (Fig. 3c). In dissected whole-mount embryos and in cryosectioned embryos (Fig. 3c), very faint staining was observed in the underlying ventral mesoderm. At the late gastrula stage (stage 12), expression appeared to be up-regulated consistently in a small group of cells clustered around the ventral edge of the closing blastopore (Fig. 3d). At the early neurula stage (stage 13), four regions of ectodermal expression were clearly detected. Strong staining was observed in the developing cement gland (Fig. 3e) and in a U-shaped domain around the proctodeum at the posterior of the embryo (Fig. 3g). Two bilateral patches of expression were seen in the head, at the edge of the neural plate, in the region of the future neurogenic placodes caudal to the eye anlagen (Fig. 3f,g). It is unclear whether this domain includes both the profundal–trigeminal placodal area and the dorsolateral placodes. In Xenopus, the dorsolateral placodes give rise to the lateral line placodes and the otic placodes at later stages (Schlosser and Northcutt, 2000). Finally, a diffuse pattern of Tbx2-positive cells was seen in the ventral epidermis (Fig. 3h,j–l). At stage 19 (late neurula), expression persists in the cement gland, the ventral epidermis, the proctodeal region, the lens placodes, and in a broad placodal area caudal to the eye anlagen (Fig. 3h–l). In addition, expression was detected in the dorsal root ganglia of the future spinal cord (Fig. 3i). At stage 21/22, Tbx2 expression was seen in a wishbone-shaped group of cells situated dorsal and caudal to each developing optic vesicle (Fig. 3m), corresponding to the cranial (profundal and trigeminal) ganglia. Expression was found to persist in these cells through tail bud and into early tadpole stages (Fig. 3n–r). From stage 21/22 onward, the bilateral expression of Tbx2 in the ectodermal placodes became restricted primarily to the otic placode and the developing otic vesicles. Unlike Tbx1, Tbx2 was found to be expressed throughout the otic vesicles, and this expression was detected at all subsequent stages analyzed (stages 24 to 40). At stage 24, additional staining was observed in the precursors of the hypaxial muscles and the pronephric duct in the trunk, in the developing branchial arches, and in the primordium of the heart (Fig. 3o,p). A small group of cells within the telencephalon is also stained at this stage (Fig. 3o–r). In stage 29 embryos, expression was clearly detected in the frontonasal process (Fig. 3q). Tbx2 continues to be expressed in the same regions of the embryo at stage 33, although its expression becomes clearly regionalized in the looping heart. A higher level of expression was clearly detected in the ventricle compared with the atrium, as reported in other organisms.
Fig. 3.
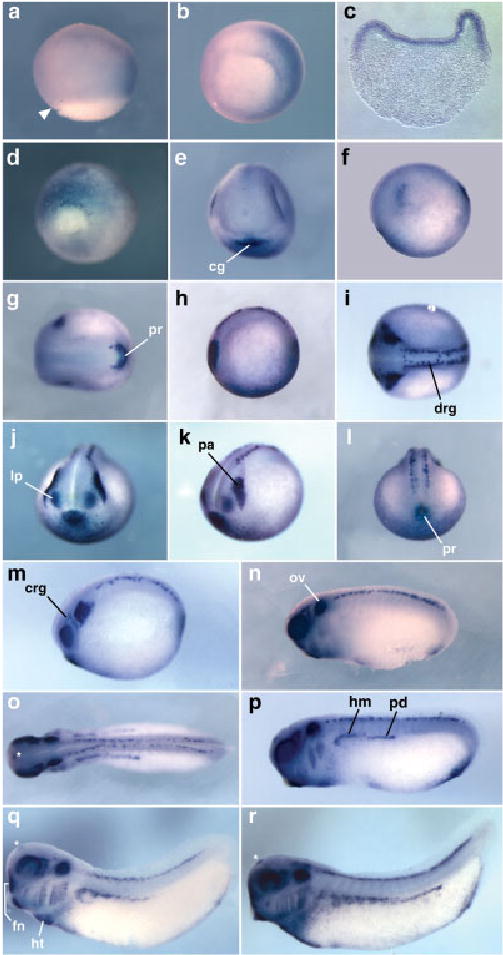
Expression pattern of Tbx2 in X. tropicalis. In situ hybridization results are shown for Tbx2 (embryos uncleared). a– c: Expression at early gastrula (stage 10.5) is shown in lateral (a) and vegetal (b) views of whole-mount embryos and in transverse section (c; ventral to the right). In both a and b, the embryo is oriented with dorsal to the left and the dorsal blastopore lip is indicated by an arrowhead in a. d: A vegetal view of a late gastrula (st12) is shown, ventral side uppermost. e–r: Expression at early neurula (stage 13; e– g) late neurula (stage 19; h,l), and tail bud stages 21/22 (m), 25 (n), 26 (o,p), 29 (q), and 33 (r) are also shown. Expression in the forebrain (telencephalon) at tail bud stages is indicated by an asterisk (o–r). Except for anterior (j) and posterior (d,l) views, all embryos are oriented with anterior to the left. cg, cement gland; crg, cranial ganglia; drg, dorsal root ganglia; fn, frontonasal process; hm, hypaxial muscle; ht, heart tube; lp, lens placode; ov, otic vesicle; pa, placodal area; pd, pronephric duct; pr, proctodeum.
Analysis of Tbx5 Expression During Embryogenesis
The expression pattern of Tbx5 was analyzed at developmental stages from mid-gastrula (stage 11) to early tadpole (stage 40). No expression was detected at stage 11. In late neurulae (stage 19), a gradient of Tbx5 expression was present within the eye anlagen, with higher levels dorsally (Fig. 4a,b). At this stage, two small patches of cells on either side of the embryo were also stained, corresponding to regions within the migrating bilateral heart primordia. At early tail bud stage (stage 25), this pattern of expression was maintained in a dorsal region of each developing eye (Fig. 4c) and in the heart primordia, located ventrally (Fig. 4c,d). In stage 26 embryos, the Tbx5-expressing cells of the heart primordia were seen to converge at the ventral midline (Fig. 4f), while expression was also detected in two bilateral groups of cells continuous with and extending dorsally from the heart primordia. These cells likely correspond to the progenitors of the right and left branches of the sinus venosus and common cardinal veins (Nieuwkoop and Faber, 1967; Horb and Thomsen, 1999; Brown et al, 2005). In other organisms, Tbx5 has been shown to play an important role in eye development, particularly in guiding the projection of neurons between the retina and tectum (Koshiba-Takeuchi et al., 2000). In X. tropicalis, expression in the dorsal region of the eye was found to be maintained until early tadpole stages (stage 40), although its expression becomes greatly restricted between stages 33 and 40 (Fig. 4h,i). At stage 31/32, strong expression was detected in the posterior region of the heart tube in cleared embryos (Fig. 4g). After looping of the heart, a higher level of expression was detected in the ventricle (situated ventrally and offset to the left side of the embryo) than in the atrium (Fig. 4j). The regional differences in the expression of Tbx5 within the hearts of X. tropicalis tadpoles were seen consistently in both whole-mount and sectioned embryos. Transverse sections through the heart at stages after heart looping showed expression of Tbx5 in the ventricular myocardium, while staining was not detected in the atrial region of the heart (Fig. 6b).
Fig. 4.
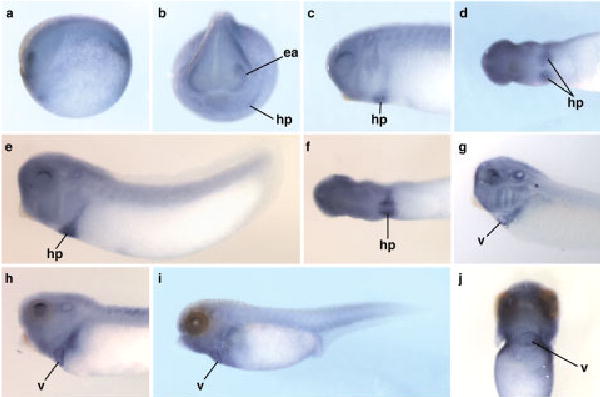
Expression pattern of Tbx5 in Xenopus tropicalis. The expression pattern of Tbx5 detected by in situ hybridization between late neurula and early tadpole stages is shown (uncleared except for g). a–j; Stages are as follows: stage 19 (a,b), stage 25 (c,d), stage 26 (e,f), stage 31/32 (g; cleared), stage 33 (h), and stage 40 (i,j). Except for the anterior view in b and the ventral view in j, embryos are oriented with anterior to the left. Anterior is to the top in j. ea, eye anlagen, hp, heart primordium, v, ventricle.
Fig. 6.
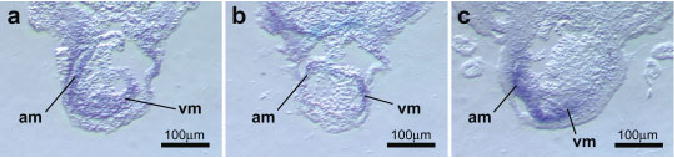
Cardiac expression of Tbx2, Tbx5, and Tbx20. The in situ hybridization patterns of Tbx2, Tbx5, and Tbx20 in the forming cardiac chambers were examined in transverse sections through the tadpole heart after looping. a: Tbx2 expression was seen in the myocardium of both the atrial (am) and ventricular (vm) regions of the looped heart at stage 36. b: Expression of Tbx5 was restricted primarily to the developing ventricular myocardium at stage 38. c: In contrast to Tbx5, high levels of Tbx20 expression were seen in the atrial region but not in the ventricle (stage 40). Original magnification, ×100.
Analysis of Tbx20 Expression During Embryogenesis
The expression pattern of X. tropicalis Tbx20 was analyzed in embryos between stages 13 (neural plate stage) and 40 (early tadpole). Although expressed weakly in the developing cement gland as early as stage 13 in X. laevis (Brown et al., 2003), we did not detect expression at this stage in X. tropicalis. At late neurula stage (stage 20), Tbx20 was strongly expressed in the developing cement gland and in the bilateral heart primordia (Fig. 5a–c). Expression in the cement gland was found to decrease from stage 25 onward, while expression continued to be strongly detected in the developing heart. In the heart-forming region at stage 25, a single domain of expression was detected, corresponding to the heart field formed by fusion of the bilateral heart primordia (Fig. 5e). This fusion of the Tbx20-expressing domains appears to occur earlier in X. tropicalis than in X. laevis. Notably, this pattern of expression differs considerably from that of Tbx5, in which fusion of the bilateral precardiac expression domains begins at around stage 26 (see above). In addition to this cardiac expression, two small domains of expression were observed in the hindbrain (rhombencephalon) at this stage, corresponding to the second and fourth rhombomeres. At stage 29/30, expression persisted in these regions and also was weakly detected in a more posterior region of the hind-brain (Fig. 5f). The hindbrain expression of Tbx20 was found to be up-regulated in embryos at subsequent stages and, as in more anterior regions, was detected in distinct paired subdomains (Fig. 5g,h,j,k,m,n). At stage 33, when heart looping is initiated, Tbx20 was found to be broadly expressed in the heart tube, with strong staining detected in the ventricle, atrium, and both branches of the sinus venosus (inflow tract) (Fig. 5g,i). Thus, the expression domain of Tbx20 in the developing chambers of the heart tube only partially overlaps that of Tbx5. This finding is consistent with the patterns of Tbx20 expression reported in other vertebrates (Ahn et al., 2000; Kraus et al., 2001; Brown et al., 2003; Plageman and Yutzey, 2004; Yamagishi et al., 2004). During heart looping (stage 36), Tbx20 was expressed at a higher level in the atrium than in the ventricle (Fig. 5l). This regional difference in the expression level of Tbx20 was maintained in early tadpole stage embryos (stage 40) and was clearly seen both in whole embryos and in transverse sections through the heart (Figs. 5o, 6c).
Fig. 5.
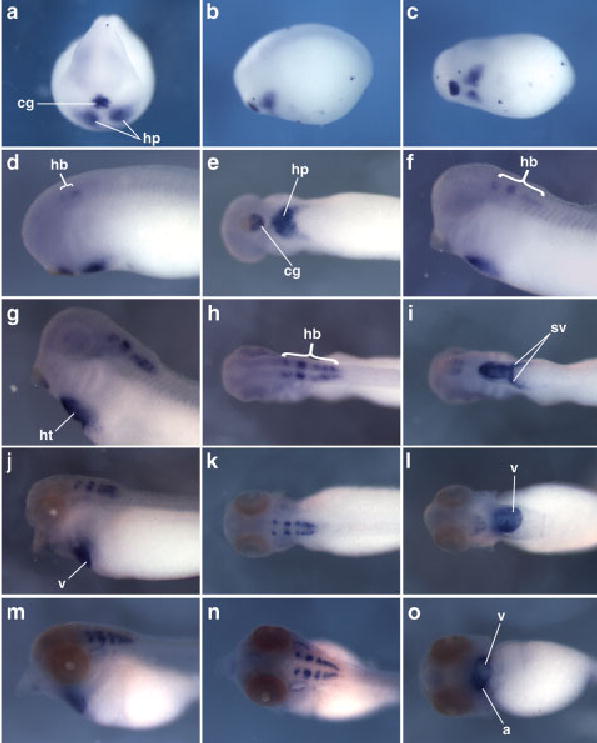
Expression pattern of Tbx20 in Xenopus tropicalis.The expression pattern of Tbx20 detected by in situ hybridization between late neurula and early tadpole stages is shown (embryos uncleared). a– o: Stages are as follows: stage 20 (a– c), stage 25 (d,e), stage 29/30 (f), stage 33 (g–i), stage 36 (j–l), and stage 40 (m– o). Ventral views are shown in i, l, and o. Except for the anterior view in a, embryos are oriented with anterior to the left. a, atrium; cg, cement gland; hb, hindbrain; hp, heart primordium; ht, heart tube; sv, sinus venosus; v, ventricle.
EXPERIMENTAL PROCEDURES
Identification and Isolation of cDNA Clones
cDNA clones TNeu106g11, TGas050k23, and TTpA031n09, encoding X. tropicalis orthologs of Tbx1, Tbx2, and Tbx20, respectively, were identified by searching a database of X. tropicalis expressed sequence-tagged clones derived from oligo-dT primed cDNA libraries specific to several developmental stages (www.sanger.ac.uk; Gilchrist et al., 2004). Specifically, nucleotide sequences from the 5′ ends of the coding regions of the corresponding X. laevis orthologs (Tbx1 GenBank accession no. AF526274; Tbx2 Gen-Bank accession no. AB023815; Tbx20 GenBank accession no. AY154394) were used to BLAST search (Altschul et al., 1990) for X. tropicalis clones containing the predicted translation start codon and, therefore, were likely to contain full-length cDNAs. The clones were obtained (MRC Geneservice), and the cDNA inserts were sequenced to 4× coverage. A cDNA encoding Tbx5 was cloned by low-stringency RT-PCR, using total RNA template from stage 13–20 X. tropicalis embryos. Primers were designed based on sequences flanking the X. laevis Tbx5 coding sequence (forward, 5′-GAAGATCTATGGCGGACACAGAGGAGGCT-3′; reverse, 5′-GAGAGATCTACGCTGTTTTCATTCCAGTCTGG-3′). The resulting product was cloned into pcDNA3.1 (Invitrogen Corp.). All cDNA sequences were deposited in GenBank (Tbx1 accession no. DQ124205; Tbx2 accession no. DQ124206; Tbx5 accession no. DQ124207; Tbx20 accession no. DQ124208).
In Silico Analysis
To identify genomic sequence scaffolds corresponding to Tbx1, Tbx2, Tbx5, and Tbx20, the corresponding cDNA sequences were used to search the X. tropicalis draft genome sequence (versions 2.0 and 3.0) using the BLAST algorithm (Altschul et al., 1990; DoE Joint Genome Institute). Pairwise sequence alignments and analyses of sequence conservation of conceptually translated proteins were performed using GeneDoc (www.psc.edu/biomed/genedoc).
Embryo Collection and In Situ Hybridization
X. tropicalis embryos were collected after natural single-pair mating between animals from a partially inbred (F6) line (NASCO). Males and females were preprimed with 10 U of human chorionic gonadotropin (hCG; SIGMA) 20 hr before being primed with an additional 200 U. One hour after priming, males and females were paired and allowed to mate for approximately 5 hr in shallow water at 25°C. Embryos and unfertilized eggs from successful matings were collected, treated with 2% cysteine hydrochloride to remove their jelly coat, and sorted. Embryos were cultured at 25°C in sterilized water from our aquatic system and staged according to criteria set out in the Normal Table of Xenopus laevis (Nieuwkoop and Faber, 1967).
A 908-bp KpnI-XhoI fragment of the Tbx1 EST clone was subcloned into pBluescript-KS, and this construct was linearized with Acc65I to generate a template for in situ hybridization probe synthesis. Template for Tbx2 probe synthesis was produced by linearizing the full-length cDNA clone described above using HindIII. The Tbx5 cDNA was cut from pcDNA3.1-Tbx5 by NotI-SpeI digest and sub-cloned into pBluescript-KS to generate a probe template construct that was subsequently linearized with NotI. To generate a template for Tbx20 probe synthesis, a 565-bp SalI-NotI fragment from the Tbx20 EST clone was subcloned into pBluescript-KS, and the construct was linearized using SalI. In situ hybridizations were performed according to a standard protocol (Sive, 2000) with the following exceptions: Fixed embryos were devitellinized by enzymatic treatment with collagenase A (Roche Applied Science), proteinase K, and hyaluronidase (SIGMA) (Islam, 1996). No further proteinase K treatment was performed. Embryos were prehybridized overnight (approximately 15 hr), and the RNase treatment step before antibody incubation was omitted (Khokha et al., 2002). After staining with BM Purple alkaline phosphatase substrate (Roche Diagnostics), embryos were re-fixed in 1× MEM salts containing 10% formamide and then dehydrated in methanol.
Where necessary, embryos were cleared in 2:1 benzyl benzoate:benzyl alcohol (SIGMA). Embryos were photographed on a Leica M-series stereomicroscope (Leica Microsystems Ltd.) using the Spot Advanced image capture system (Diagnostic Instruments, Inc.) and edited using Photoshop 7.0 (Adobe Systems, Inc.).
Cryosectioning
For cryosectioning, embryos were embedded in gelatin using a method modified from Stern and Holland (Stern and Holland, 1993). After in situ hybridization, embryos were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) and incubated overnight at 4°C in 30% sucrose/PBS (w/v). The embryos were then pre-warmed to 38°C before being transferred to 15% sucrose/PBS containing 7.5% gelatin (~300 Bloom; SIGMA) at 38°C. Embryos were incubated in gelatin for a minimum of 30 min before being transferred to specimen molds (Tissue-Tek; Sakura Finetek USA, Inc.). Embedded embryos were stored at 4°C before cryosectioning. Sections were taken at a thickness of 20 μm. Gelatin was rinsed from the sections using PBS at 38°C before mounting in aqueous mounting medium (Faramount; DakoCytomation).
Acknowledgments
Expressed sequence tag (EST) clones were produced by Cambridge University, the Wellcome Trust Sanger Institute, and Wellcome Trust/Cancer Research UK Institute Xenopus tropicalis EST Project. The authors thank Dr. Victoria K. Graham (Duke University) and Sarah Goetz (UNC–Chapel Hill) for advice on cryosectioning; Dr. Eva Anton (UNC–Chapel Hill) for the use of equipment; and Misty Hurt (University of Virginia), Shruti Nagaraj and Shauna Vasilatos (UNC–Chapel Hill) for technical assistance. F.L.C. was supported by NIH/NHLBI grants.
Footnotes
This article was accepted for inclusion in Developmental Dynamics 235#1–Cardiovascular Special Issue.
Grant sponsor: NIH/NHLBI; Grant number: HL075256-01; Grant number: HL083965-01.
References
- Ahn DG, Ruvinsky I, Oates AC, Silver LM, Ho RK. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95:253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ataliotis P, Ivins S, Mohun TJ, Scambler PJ. XTbx1 is a transcriptional activator involved in head and pharyngeal arch development in Xenopus laevis. Dev Dyn. 2005;232:979 –991. doi: 10.1002/dvdy.20276. [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30 –35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Brown DD, Binder O, Pagratis M, Parr BA, Conlon FL. Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev Genes Evol. 2003;212:604 –607. doi: 10.1007/s00427-002-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Martz SN, Binder O, Goetz SC, Price BMJ, Smith JC, Conlon FL. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagleson G, Ferreiro B, Harris WA. Fate of the anterior neural ridge and the morphogenesis of the Xenopus forebrain. J Neurobiol. 1995;28:146 –158. doi: 10.1002/neu.480280203. [DOI] [PubMed] [Google Scholar]
- Gilchrist MJ, Zorn AM, Voigt J, Smith JC, Papalopulu N, Amaya E. Defining a large set of full-length clones from a Xenopus tropicalis EST project. Dev Biol. 2004;271:498 –516. doi: 10.1016/j.ydbio.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Hayata T, Kuroda H, Eisaki A, Asashima M. Expression of Xenopus T-box transcription factor, Tbx2 in Xenopus embryo. Dev Genes Evol. 1999;209:625–628. doi: 10.1007/s004270050297. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. Tbx5 is essential for heart development. Development. 1999;126:1739 –1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000;260:148 –157. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Islam N, Moss T. Enzymatic removal of vitelline membrane and other protocol modifications for whole mount in situ hybridization of Xenopus embryos. Trends Genet. 1996;12:459. doi: 10.1016/0168-9525(96)99990-4. [DOI] [PubMed] [Google Scholar]
- Jerome LA, Pappaioannou VE. Di-George syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286 –291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn AM, Harland RM, Grammer TC. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499 –510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Kochilas L, Liao J, Merscher-Gomez S, Kucherlapati R, Morrow B, Epstein JA. 2005. New insights into the role of Tbx1 in the DiGeorge mouse model. In: Artman M, Benson DW, Srivastava D, Nakazawa M, editors. Cardiovascular development and congenital malformations: molecular and genetic mechanisms. Malden, MA: Blackwell Publishing.
- Koshiba-Takeuchi K, Takeuchi JK, Matsumoto K, Momose T, Uno K, Hoepker V, Ogura K, Takahashi N, Nakamura H, Yasuda K, Ogura T. Tbx5 and the retinotectum projection. Science. 2000;287:134 –137. doi: 10.1126/science.287.5450.134. [DOI] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene tbx20. Mech Dev. 2001;100:87–91. doi: 10.1016/s0925-4773(00)00499-8. [DOI] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, Epstein JA, Brown MC, Adams J, Morrow BE. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13:1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haplo-insufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Mandel EM, Callis TE, Wang D-Z, Conlon FL. Transcriptional mechanisms of congenital heart disease. Drug Discovery Today: Disease Mechanisms. 2005;2:33–38. [Google Scholar]
- Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. The morphology of heart development in Xenopus laevis. Dev Biol. 2000;218:74 –88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- Moraes F, Novoa A, Jerome-Majewska LA, Papaioannou VE, Mallo M. Tbx1 is required for proper neural crest migration and to stabilize spatial patterns during middle and inner ear development. Mech Dev. 2005;122:199 –212. doi: 10.1016/j.mod.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Mount S. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J, editors. 1967. Normal table of Xenopus laevis (Daudin) Amsterdam: North-Holland Publishing Co.
- Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet. 2003;12(Spec1):R37–R44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Ahn DG, Schilling TF, Nair S, Ruvinsky I, Geisler R, Rauch GJ, Haffter P, Zon LI, Zhou Y, Foott H, Dawid IB, Ho RK. The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development. 2003;130:5043–5052. doi: 10.1242/dev.00704. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem. 2004;279:19026 –19034. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. T-box genes and heart development: putting the “T” in heart. Dev Dyn. 2005;232:11–20. doi: 10.1002/dvdy.20201. [DOI] [PubMed] [Google Scholar]
- Raft S, Nowotschin S, Liao J, Morrow BE. Suppression of neural fate and control of inner ear morphogenesis by Tbx1. Development. 2004;131:1801–1812. doi: 10.1242/dev.01067. [DOI] [PubMed] [Google Scholar]
- Ryan K, Chin AJ. T-box genes and cardiac development. Birth Defects Res C Embryo Today. 2003;69:25–37. doi: 10.1002/bdrc.10001. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271:439 –466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Northcutt RG. Development of neurogenic placodes in Xenopus laevis. J Comp Neurol. 2000;418:121–146. [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. Dev Dyn. 2004;229:201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- Sive H, Grainger RM, Harland RM. 2000. Early development of Xenopus laevis - a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Stern CD, Holland PWH, editors. 1993. Essential developmental biology: a practical approach. Oxford: IRL Press at Oxford University Press.
- Vitelli F, Viola A, Morishima M, Pramparo T, Baldini A, Lindsay E. TBX1 is required for inner ear morphogenesis. Hum Mol Genet. 2003;12:2041–2048. doi: 10.1093/hmg/ddg216. [DOI] [PubMed] [Google Scholar]
- Xu H, Morishima M, Baldini A. 2005. Tbx1 and DiGeorge syndrome: a genetic link between cardiovascular and pharyngeal development. In: Cardiovascular development and congenital malformations: molecular and genetic mechanisms. Malden, MA: Blackwell Publishing.
- Yamagishi T, Nakajima Y, Nishimatsu S, Nohno T, Ando K, Nakamura H. Expression of tbx20 RNA during chick heart development. Dev Dyn. 2004;230:576 –580. doi: 10.1002/dvdy.20076. [DOI] [PubMed] [Google Scholar]


