Abstract
We have isolated the Xenopus orthologue of the T-box gene, Tbx20, and characterized its developmental expression profile. We show that Tbx20 is one of the earliest markers of heart tissue in Xenopus, and is expressed throughout all cardiac tissue during later stages of development. In addition, we also observe expression in the cement gland, the jugular vein, the lung bud, the cloacal aperture, rhombomeres 2, 4, 6 and 8, and in a subset of motor neurons.
Keywords: T-box, Tbx20, Heart development, Xenopus
The T-box family of transcription factors is a large family of proteins required for both early cell fate decisions, such as those necessary for formation of the basic vertebrate body plan, as well as for differentiation and organogenesis. The role of the T-box genes in these processes is emphasized by the observation that T-box genes when mutated give dramatic phenotypes in mouse and zebrafish. Furthermore, T-box genes are implicated in a number of human congenital malformations and are amplified in a subset of cancers (reviewed in Papaioannou 2001; Smith 1999; Wilson and Conlon 2002). The T-box family has recently been shown to comprise approximately 0.1% of genomes as diverse as Caenorhabditis elegans and human and have been identified in a wide variety of chordates from ctenophore to human, while being completely absent in genomes from other phyla (e.g. Arabidopsis thaliana). For many of these genes clear homologues exist, such as Brachyury, which displays a high degree of sequence similarity, expression pattern, and function between a variety of vertebrates including fish, frog, dog and mouse. However, other T-box genes appear to be unique to a particular species. For instance, VegT, a T-box gene thought to be required for endoderm formation in Xenopus, has no apparent homologue or orthologue in mouse or human (reviewed in Wilson and Conlon 2002).
Two sets of clinical studies have provided direct evidence for a role for T-box genes in heart development and differentiation with Tbx1 deleted in patients with the DiGeorge syndrome (Jerome and Papaioannou 2001; Lindsey et al. 2001; Merscher et al. 2001), and Tbx5 often mutated in patients with the congenital heart disease, Holt-Oram Syndrome (Basson et al. 1997; Li et al. 1997). In addition to Tbx1 and Tbx5, recent studies in human, mouse, chick and zebrafish have implicated a third member of the T-box gene family, Tbx12/20, in heart development (Ahn et al. 2000; Carson et al. 2000; Griffin et al. 2000; Iio et al. 2001; Kraus et al. 2001; Meins et al. 2000). To further address the role of Tbx20 in early heart development, we have identified and analyzed the expression of the Tbx20 orthologue in Xenopus laevis.
To isolate X. laevis Tbx20, we designed a set of degenerate primers based on the published Drosophila H15 and C. elegans Tbx12 sequences (Agulnik et al. 1997; Brook and Cohen 1996). These primers were used to isolate a clone from stage 36 X. laevis cDNA. Additional X. laevis sequence was obtained by a second round of amplification using primers derived from mouse, human, and zebrafish Tbx20 sequences (Ahn et al. 2000; Carson et al. 2000; Griffin et al. 2000; Meins et al. 2000). This 928-bp clone was in turn used to screen a cDNA mixed stage (19–26) X. laevis cDNA library (generous gift of Aaron Zorn) by PCR. Partial sequencing and restriction mapping were used to construct a full-length copy of the gene by cloning overlapping fragments into pBluescript II KS (Stratagene). The clone was sequenced from both ends with a minimum of fourfold coverage and shown to contain a 1,741-bp insert with an open reading frame of 441 amino acids (GenBank accession number: AY154394; Fig. 1A). Sequence analysis revealed the clone to have 84% identity with human Tbx20 (Meins et al. 2000), 90% with mouse Tbx12/20 (Carson et al. 2000; Kraus et al. 2001), and 91% with chicken Tbx20 (Fig. 1B; Iio et al. 2001). Based on sequence (Fig. 1), expression analysis (Figs. 2, 3), and current T-box nomenclature, we refer to this gene as the X. laevis orthologue of Tbx20.
Fig. 1.
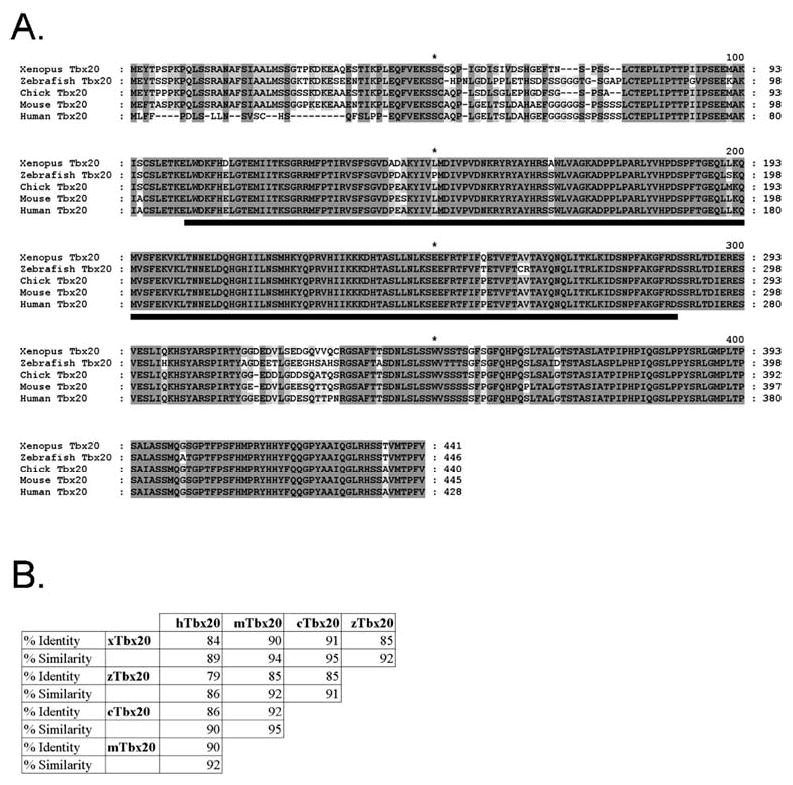
A, B Comparative sequence analysis of TBX20. Analysis was performed with the GeneDoc program. A Alignment of vertebrate TBX20 proteins. Fully conserved amino acids including those containing conservative substitutions are shaded in dark gray. Lighter shading represents lower conservation. The conserved T-box domain is underlined in black. B Vertebrate TBX20 amino acid conservation. The percentage of identical amino acid residues (identity) and the percentage of conservative substitutions and identical residues (similarity) are given for comparison between TBX20 proteins. TBX20 prefixes: x, Xenopus; z, zebrafish; c, chicken; m, mouse; h, human
Fig. 2.
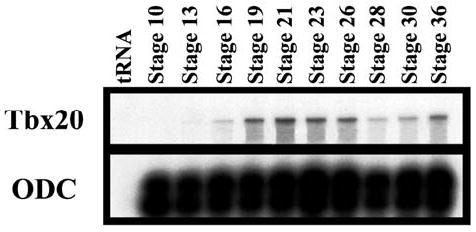
Temporal expression of Xenopus laevis Tbx20 as detected by RNase protection assay. Ornithine decarboxylase (ODC) is included in the lower panel as an internal loading control
Fig. 3.
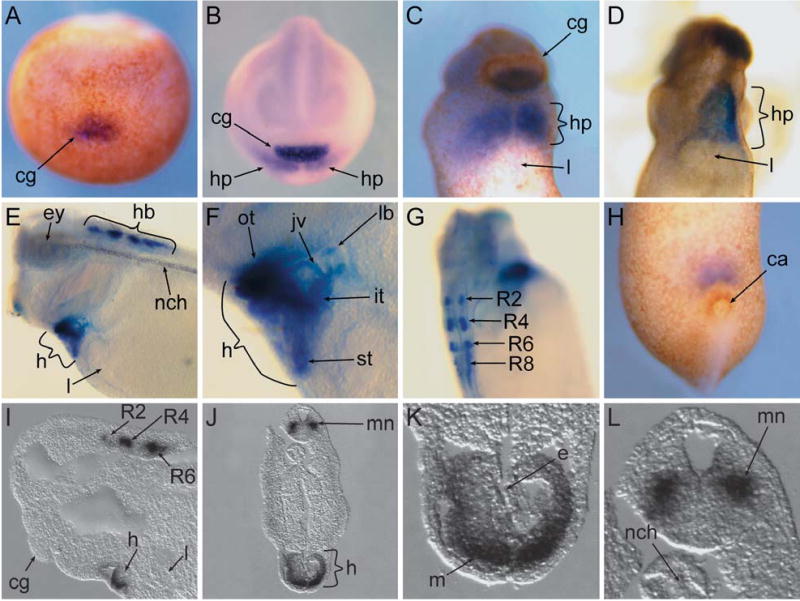
A–L Expression of Tbx20 during Xenopus laevis development as detected by whole-mount in situ hybridization. A Anterior view of a stage-13 embryo. Dorsal side is facing up. B Anterior view of a stage-16 embryo. Dorsal side is facing up. C Ventral view of the anterior portion of a stage-27 embryo. D Ventral view of the anterior portion of a stage-32 embryo. E Lateral view of the anterior portion of a stage-35 embryo. F Higher magnification of E. G Dorsolateral view of a stage-35 embryo. H Ventral view of the posterior portion of a stage-27 embryo. I–L Sections of stage-35 embryos. I Anterior view of a parasagittal section. J Transverse section through the anterior region. K, L Higher magnifications of J. ca Cloacal aperture, cg cement gland, e endocardium, ey eye, h heart, hb hindbrain, hp heart primordium, it inflow tract, jv jugular vein, l liver, lb lung bud, m myocardium, mn motor neuron, nch notochord, ot outflow tract, st septum transversum, R2–R8 rhombomeres 2, 4, 6, and 8
We determined the onset and relative levels of Tbx20 during X. laevis development by RNase protection analysis using a probe derived from the 3′ Tbx20 on staged embryos from early gastrula (stage 10) to early tadpole (stage 36; Fig. 2). This probe does not contain sequences within the putative T-box domain and therefore is assumed to be Tbx20-specific. Ornithine decarboxylase (ODC) was used as an internal loading control and tRNA was used as a negative control. Tbx20 transcripts are first detected at low but consistent levels by early neural stage (stage 16) with expression then increasing by stage 19 and remaining relatively constant until a sharp drop off at later neurula stages (stages 23–28). However, there is a sharp increase in expression between early (stage 30) and mid-tadpole stages (stage 36; Fig. 2). RT-PCR analysis shows no maternal expression as judged in unfertilized eggs and early gastrula embryos (stage 10; data not shown).
To determine the spatial pattern of expression we performed whole-mount in situ hybridizations on staged X. laevis from early gastrula (stage 10) to mid-tadpole (stage 40). Consistent with RNase protection and RT-PCR analysis, we first detect Tbx20 expression by in situ hybridization at late gastrula stages in the region of the most anterior developing cement gland (stage 13; Fig. 3A) and, at slightly later stages, in the heart field (stage 16; Fig. 3B). Tbx20 is expressed in the heart field before fusion of the primordium along the ventral midline. Thus, together with Tbx5 (Horb and Thomsen 1999) and the Nkx paralogues (Newman and Krieg 1998), Tbx20 is one of the earliest markers of X. laevis cardiac tissue. Expression of Tbx20 in the heart gradually increases during development (compare Fig. 3B with D) and by mid-tadpole stage (stage 35), expression is found throughout the cardiac region (Fig. 3E, F, I, K) including the atrial and ventricular tissue, the inflow and outflow tract, and the septum transversum (Fig. 3F), while being completely absent from more posterior tissues such as the liver (Fig. 3C–E, I). In addition, Tbx20 is expressed in both tissue layers of the heart, with relatively high levels in the myocardial layer and lower levels in the endocardial layer (Fig. 3K). Therefore, Tbx20 is expressed at the same time and in many regions of the heart that also express the heart markers Tbx5 and Nkx2.5 (Horb and Thomsen 1999; Tonissen et al. 1994).
In the cement gland, the most anterior neural ectodermal tissue, Tbx20 is gradually restricted to the ventral half of the gland by stage 27 (Fig. 3C), and expression decreases during neurula and early tadpole stages (Fig. 3E, I) such that, by stage 40, Tbx20 can no longer be detected in the tissue (data not shown). In addition to the cement gland and heart, high levels of Tbx20 expression are found in the external jugular vein, the lung bud (Fig. 3F), and the cloacal aperture (Fig. 3H), very low levels in the retina and transient low levels in the notochord (Fig. 3E). Similar to reports in mouse, chick, and zebrafish (Ahn et al. 2000; Carson et al. 2000; Iio et al. 2001; Kraus et al. 2001), we also observe expression in rhombomeres 2, 4, 6, and 8 (Fig. 3E–G, I), and as shown by transverse and parasagittal sections through stage-35 embryos, in a subset of motor neurons emerging from these rhombomeres (Fig. 3I, J, L). However, in contrast to the mouse, we never detect Tbx20 expression in the liver (Fig. 3C–E, I; Kraus et al. 2001). Therefore, although Tbx20 displays a very high degree of sequence conservation across species, only a subset of tissues, such as the rhombomeres, show a conservation of expression, while other sites of expression appear to be unique to X. laevis, such as the lung bud and jugular vein.
Acknowledgments
We would like to thank Amanda Marshburn for technical assistance and Aaron Zorn for the Xenopus cDNA library. B.A.P. is supported in part by research grant no. 1-FY02-26 from the March of Dimes Birth Defects Foundation, D.D.B. by a NSF Graduate Research Fellowship, and F.L.C. by the American Heart Association.
Footnotes
Edited by C. Desplan
References
- Agulnik SI, Ruvinsky I, Silver LM. Three novel T-box genes in Caenorhabditis elegans. Genome. 1997;40:458–464. doi: 10.1139/g97-061. [DOI] [PubMed] [Google Scholar]
- Ahn DG, Ruvinsky I, Oates AC, Silver LM, Ho RK. Tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95:253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc Straceski J, et al. Mutations in human Tbx5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Brook WJ, Cohen SM. Antagonistic interactions between wingless and decapentaplegic responsible for dorsal-ventral pattern in the Drosophila leg. Science. 1996;273:1373–1377. doi: 10.1126/science.273.5280.1373. [DOI] [PubMed] [Google Scholar]
- Carson CT, Kinzler ER, Parr BA. Tbx12, a novel T-box gene, is expressed during early stages of heart and retinal development. Mech Dev. 2000;96:137–140. doi: 10.1016/s0925-4773(00)00376-2. [DOI] [PubMed] [Google Scholar]
- Griffin KJ, Stoller J, Gibson M, Chen S, Yelon D, Stainier DY, Kimelman D. A conserved role for H15-related T-box transcription factors in zebrafish and Drosophila heart formation. Dev Biol. 2000;218:235–247. doi: 10.1006/dbio.1999.9571. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. Tbx5 is essential for heart development. Development. 1999;126:1739–1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- Iio A, Koide M, Hidaka K, Morisaki T. Expression pattern of novel chick T-box gene, Tbx20. Dev Genes Evol. 2001;211:559–562. doi: 10.1007/s00427-001-0187-y. [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx20. Mech Dev. 2001;100:87–91. doi: 10.1016/s0925-4773(00)00499-8. [DOI] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Lindsey EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scrambler PJ, et al. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Meins M, Henderson DJ, Bhattacharya SS, Sowden JC. Characterization of the human TBX20 gene, a new member of the T-Box gene family closely related to the Drosophila H15 gene. Genomics. 2000;67:317–332. doi: 10.1006/geno.2000.6249. [DOI] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, et al. TBX1 is responsible for cardiovascular defects in velo-cardiofacial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Newman CS, Krieg PA. tinman-related genes expressed during heart development in Xenopus. Genetics. 1998;22:230–238. doi: 10.1002/(SICI)1520-6408(1998)22:3<230::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE. T-box genes in development: from hydra to humans. Int Rev Cytol. 2001;207:1–70. doi: 10.1016/s0074-7696(01)07002-4. [DOI] [PubMed] [Google Scholar]
- Smith J. T-box genes: what they do and how they do it. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- Tonissen KF, Drysdale TA, Lints TJ, Harvey RP, Krieg PA. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- Wilson V, Conlon FL. The T-box genes. Genome Biol. 2002;3(6):reviews 3008. doi: 10.1186/gb-2002-3-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]


