Abstract
Sinocalycanthus chinensis, an endangered species endemic to China, is cultivated as an ornamental landscape tree in China. However, S. chinensis, Chimonanthus species and Calycanthus floridus are difficult to be distinguished in seedling market because of their similar morphological characters. In this study, ISSR (inter-simple sequence repeats) were applied to detect S. chinensis from its closely related species. A unique 748-bp band was found in all accessions of S. chinensis. SCAR (sequence characterized amplified regions) markers were created by cloning and sequencing the specific band, and designing a pair of primers to amplify the band of 748 bp. Diagnostic PCRs were performed using the primer pair with the total DNAs of S. chinensis, Chimonanthus species and C. floridus as templates, with only S. chinensis being able to be amplified. This amplification is not only rapid (results can be obtained in less than 3 h), but is also easy to perform. Hence it is a feasible method for identifying S. chinensis in seedling market.
Keywords: Authentication, Diagnostic PCRs, ISSR, SCAR, Sinocalycanthus chinensis, Specie-specific
INTRODUCTION
Calycanthaceae consists of three genera: Calycanthus, Chimonanthus and Sinocalycanthus. Chimonanthus and Sinocalycanthus are restricted to China, and Calycanthus is distributed in North America (Zhang and Liu, 1998). Sinocalycanthus chinensis Cheng et S.Y. Chang, the only representative in the genus Sinocalycanthus, is a deciduous shrub endemic to China. Both biological and anthropological causes have led to the recent decline of the species, which is now known from only two extant natural populations (Table 1). In China, S. chinensis is classified as second-grade protected wild plants in the Chinese Plant Red Book (Fu, 1992).
Table 1.
Sampling localities and codes of S. chinensis and its 9 close species from Calycanthus and Chimonanthus
| Species | Sources of materials | Sampling code (n) |
| Sinocalycanthus chinensis Cheng et S.Y. Chang | Hangzhou Botanical Garden, Zhejiang | X1 (4) |
| Botanical Garden, Zhejiang University | X2 (2) | |
| Mt. Tianmu, Natural Conservation Area, Zhejiang | X3 (5) | |
| Kunming Institute of Botany, CAS, Yunnan | X4 (1) | |
| ★Lingli, Mt. Tiantai, Zhejiang | X5 (3) | |
| ★Jinguping, Mt. Tiantai, Zhejiang | X6 (3) | |
| ★Mt. Qingliangfeng, Lin’an, Zhejiang | X7 (5) | |
| ★Shunxi, Lin’an, Zhejiang | X8 (4) | |
| Chimonanthus praecox (Linn.) Link | Hangzhou Botanical Garden, Zhejiang | 1 (3) |
| Ch. praecox var. grandiflorus (Lindley) Makino | Hangzhou Botanical Garden, Zhejiang | 2 (2) |
| Ch. praecox var. intermedius Makino | Hangzhou Botanical Garden, Zhejiang | 3 (2) |
| Ch. salicifolius S.Y. Hu | Hangzhou Botanical Garden, Zhejiang | 4 (4) |
| Ch. zhejiangensis M.C. Liu | Hangzhou Botanical Garden, Zhejiang | 5 (3) |
| Ch. campanulatus R.H. Chang et C.S. Ding | Hangzhou Botanical Garden, Zhejiang | 6 (5) |
| Calycanthus floridus Linn. | Raleigh, North Carolina, USA | 7 (1) |
| C. floridus Linn. | Hangzhou Botanical Garden, Zhejiang | 8 (3) |
| C. floridus var. oblongifolius (Nuttall) Boufford et Spongberg | Hangzhou Botanical Garden, Zhejiang | 9 (2) |
Asterisk indicates wild populations of S. chinensis; n: number of specimens tested
Although S. chinensis has very few wild populations, it is now cultivated as an ornamental landscape tree in China due to its high ornamental value with enjoyable white flowers in early summer, with the demand for S. chinensis being increased recently. But for a long period, the identification of this species relies on morphological characteristics. The insufficient ways of visual inspection (Bandana and Mahipal, 2003) tend to cause mistakes, especially when dealing with the vegetative part of similar species or seedlings. This situation exists in S. chinensis seedling market, where some Chimonanthus species were sometimes mistakenly identified as S. chinensis during seedling period, due to morphological similarity. Therefore, it is necessary and urgent to provide an efficient and scientific authenticating method to identify S. chinensis from its related species.
SCAR (sequence characterized amplified regions) markers are developed with a pair of longer primers (usually the extended sequence of a RAPD (random amplified polymorphic DNA) primer) that has a specific sequence of approximately 20 bases. Compared with universal primers, unique primers for special regions prevent site-competition among primers, and make the results less sensitive to reaction conditions and more reproducible by increasing the specificity (Hernández et al., 1999). Reliable SCAR markers have already been successfully derived from RAPD fragments in Lettuca, Triticum, and Agrostis (Paran and Michelmore, 1993; Hernández et al., 1999; Elizabeth et al., 2003). ISSR (inter-simple sequence repeats) are presented to detect differences between SSRs (simple sequence repeats) (Zietkiewicz et al., 1994). No sequence information is required prior to analysis, and only a minute amount of DNA is needed (Welsh and McClelland, 1990; Williams et al., 1990). Compared with the low fidelity of RAPD in some circumstances (Micheli et al., 1994), the longer primers and higher annealing temperature of ISSR can provide more reproducibility and stability (Esselman et al., 1999; Camacho and Liston, 2001; Zou et al., 2001).
Here we use ISSR (inter-simple sequence repeats) method to detect specific fragments among sample species, and converse the ISSR fragments into SCAR markers. Stable specific SCAR markers can help us to distinguish S. chinensis individuals from its closely related species.
MATERIALS AND METHODS
Plant materials and DNA extraction
Fresh leaves of S. chinensis and its closely related species were sampled from natural populations and introduced populations (Table 1). Species of Parakmeria lotungensis (Chun et Txoong) La, Illicium henryi Diels and Chloranthus henryi Hemsley were also selected as contrast species. Two to five individuals of each species were tested, except C. floridus Linn. with only one specimen. Genomic DNA was extracted using the modified CTAB (hexadecyltrinethyl-ammoniu bromide) method (Doyle, 1991). DNA concentration for all samples was estimated using a DNA ladder of known concentration and a spectrophotometer at 260 and 280 nm. Working stocks of DNA were then prepared based on both estimates. Total DNA was solvated in 0.1×TE for further use.
ISSR-PCR amplification
Fifty-two primers (UBC primer set No. 9, Biotechnology Laboratory, University of British Columbia, Vancouver, Canada, http://www.ubc.ca/) were screened initially to identify well amplified (polymerase provided by Shanghai Sangon Biotechnology Co. Ltd., Shanghai, China), polymorphic bands against all plants used in this study. Out of 52 primers tested, eleven primers, which gave the strongest, clearest and most reproducible bands, were then chosen for further study (Table 2). PCR reaction mix (25 μl) contained 60 ng template DNA, 1.2 U of Taq DNA polymerase, 1.5 mmol/L MgCl2, 0.25 mmol/L each of dATP, dTTP, dCTP, and dGTP, 0.4 μmol/L primer. Amplification was performed in a thermal cycler (GeneAmp® PCR System 9700, Applied Biosystems, Foster City, USA) with profile: 94 °C, 5 min; 45 cycles of 94 °C 1 min, 52 °C 45 s and 72 °C 2 min; 72 °C, 5 min. A negative contrast with no DNA was included in each PCR run. The amplification was repeated at least twice. PCR products were separated according to size on 1.5% agarose gels in a 0.5×TBE buffer, stained with ethidium bromide (0.5 μg/ml), visualized with ultraviolet light and photographed. DNA Marker DL2000 (TaKaRa Biotechnology Co. Ltd., Dalian, China) was used.
Table 2.
Eleven ISSR primers amplifying clear and reproducible banding pattern in the study
| Primer code | Sequence (5′~3′) |
| UBC 807 | (AG)8T |
| UBC 811 | (GA)8C |
| UBC 817 | (CA)8A |
| UBC 818 | (CA)8G |
| UBC 825 | (AC)8T |
| UBC 842 | (GA)8CG |
| UBC 852 | (TC)8GA |
| UBC 854 | (TC)8AG |
| UBC 857 | (AC)8GG |
| UBC 879 | CTT CAC TTC ACT TCA |
| UBC 880 | GGA GAG GAG AGG AGA |
Cloning and sequencing of specific ISSR fragments
The desired band only amplified in S. chinensis was excised from agarose gel, purified by GENECLEAN II Kit (BIO 101, Inc. Carlsbad, USA) according to the manufacturer’s protocols, and cloned into PUCm-T vector (Sangon, Shanghai, China). Sequencing of the cloned PCR product was carried out with BigDye Terminator-Sequencing kit (Perkin-Elmer, Inc. Norfolk, Connecticut, USA). Resulting sequence was edited by using SEQUENCHER software (v. 4.0.5 Gene Codes Corporation, Ann Arbor, MI, USA).
Primer design and SCAR-PCR
Specific primers were designed from the sequence of the specific DNA fragments using Oligo 6.0 (Molecular Biology Insights, Inc. Cascade, CO, USA), and synthesized by Sangon of Shanghai. SCAR-PCR amplification was performed with the same reaction mixture (containing 0.2 μmol/L of the upper and lower primer), in the same thermal cycler as described above, using a cycle of 5 min at 94 °C; 45 cycles of 1 min at 94 °C, 45 s at 65 °C, 30 s at 72 °C; and 1 cycle of 5 min at 72 °C. Amplification products were resolved electrophoretically by 1.5% TBE agarose gels and proved to be 748 bp.
RESULTS
Screening the specific ISSR marker
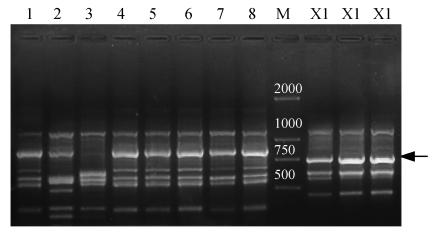
Seventeen accessions consisting of 6 species and 4 varieties (Table 1) were chosen to develop species-specific SCAR markers of S. chinensis. After ISSR-PCRs for screening against the DNAs of the 17 accessions, we found that the bright, consistent and specific band existed only in S. chinensis accessions. As seen in Fig.1, ISSR primer UBC 811 provided an approximately 750-bp band unique to the species. The species-specific band could be obtained in both wild populations and introduced populations of the species, and was selected for conversion into species-specific SCAR markers.
Fig. 1.
PCR profiles of UBC 811. Arrow indicates the specific band
Lanes: M: Weight marker (bp); 1: Ch. praecox; 2: Ch. praecox var. grandiflorus; 3: Ch. praecox var. intermedius; 4: Ch. salicifolius; 5: Ch. zhejiangensis; 6: Ch. campanulatus; 7: C. floridus; 8: C. floridus var. floridus; X1: S. chinensis
Conversion of ISSR marker into SCARs
The approximately 750-bp band amplified from 8 accessions of S. chinensis was cloned and sequenced to obtain a 748-bp sequence (GenBank accession number: bankit755109 DQ321501). It was noted that the first and the last seventeen bases of the 748-bp sequence corresponded to the initial sequence of UBC 811 primer. A pair of 25-bp SCAR primers was designed from the sequence (Table 3), with the best places of the upper primer being 28 bp from 5′ of the sequence, and the lower primer being 25 bp from 3′ of the sequence. Due to consideration to reduce the secondary structure between primers, the SCAR primer pair contained no bases of the UBC 811 primer. The reaction system and PCR conditions of diagnostic PCR are described in materials and methods. The annealing temperature was tested from 60 °C to 65 °C, with the best annealing temperature of these species-specific primers determined to be 65 °C. The SCAR primer pair was named ZJUFY01/ZJUFY02 (Table 3).
Table 3.
Species-specific SCAR primer pair derived from cloned ISSR band of S. chinensis. Optimal annealing temperature for SCAR primer is noted
| ISSR primer | SCAR primer | 5′~3′ sequence | Annealing temperature (°C) |
| UBC 811 | ZJUFY01 | CATCATATACATTGTCAGGTGCTAC | 65 |
| ZJUFY02 | GTCCCATATTCAACTGTGTTAAAGT |
Testing designed SCAR primers
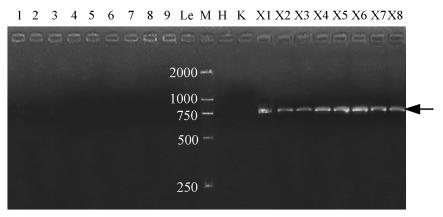
The SCAR primer pair was then used to amplify sample DNAs listed in Table 1. The 748-bp fragment was only amplified in all accessions of S. chinensis (Fig.2). Besides, ZJUFY01 and ZJUFY02 were also tested with three species from close families of S. chinensis (namely P. lotungensis, I. henryi, and C. henryi), but produced nothing (Fig.2). A bright, unique and easily identifiable band could be observed in the banding patterns of all S. chinensis individuals collected from different localities, whereas no band was found in any other related species, genera and families (Fig.2). So that the SCAR primer pair (ZJUFY01/ZJUFY02) designed in this study proved to be a species-specific marker of S. chinensis.
Fig. 2.
Banding pattern of S. chinensis (showing a distint and reproducible band) and its closely related species and families (showing no positive marker) with the designed SCAR primers (ZJUFY01/ZJUFY02)
Lanes: M: Weight marker (bp); 1: Ch. praecox; 2: Ch. praecox var. grandiflorus; 3: Ch. praecox var. intermedius; 4: Ch. salicifolius; 5: Ch. zhejiangensis; 6: Ch. campanulatus; 7: C. floridus; 8: C. floridus var. floridus; 9: C. floridus var. oblongifolius; Le: P. lotungensis; H: I. henryi; K: C. henryi; X1-X8: S. chinensis: X1: Hangzhou Botanical Garden; X2: Botanical Garden, Zhejiang Univ.; X3: Mt. Tianmu, Natural Conservation Area, Zhejiang; X4: Kunming Institute of Botany, CAS, Yunnan; X5: Lingli, Mt. Tiantai, Zhejiang; X6: Jinguping, Mt. Tiantai, Zhejiang; X7: Mt. Qingliangfeng, Lin’an, Zhejiang; X8: Shunxi, Lin’an, Zhejiang
DISCUSSION AND CONCLUSION
S. chinensis has been studied extensively in China because it is an endangered species of high ornamental value, and is also a traditional Chinese herbal medicine. The molecular method we reported here is confirmed to be a credible and convenient solution for identifying S. chinensis, other than morphological method, especially during seedling period. A minute amount of genomic DNA is needed. DNA quality is not as important as in RAPD fingerprinting analysis. The longer length (25 bp) and higher temperature (65 °C) of SCAR primers make the reaction more specific, reproducible and stable. Therefore, the work we did here proved SCAR marker to be an efficient and available way for S. chinensis species identification. It also throws light on the authentication of Chinese herbal medicine. Besides, we hope the information here would contribute to the conservation of the endangered species.
Acknowledgments
The authors are grateful to Mr. Hu Shaoqing (Hangzhou Botanical Garden of Zhejiang, China) for assistance in collection of specimens.
Footnotes
Project (No. G2000046806) supported by the National Basic Research Program (973) of China
References
- 1.Bandana D, Mahipal S. Molecular detection of cashew husk (Anacardium occidentale) adulteration in market samples of dry tea (Camellia sinensis) Planta Med. 2003;69(9):882–884. doi: 10.1055/s-2003-43211. [DOI] [PubMed] [Google Scholar]
- 2.Camacho FJ, Liston A. Population structure and genetic diversity of Botrychium pumicola (Ophioglossaceae) based on ISSR. Am J Bot. 2001;88(6):1065–1070. [PubMed] [Google Scholar]
- 3.Doyle JJ. DNA Protocols for Plants—CTAB Total DNA Isolation. In: Hewitt GM, Johnston A, editors. Molecular Techniques in Taxonomy. Berlin, Germany: Springer-Verlag; 1991. pp. 283–293. [Google Scholar]
- 4.Elizabeth AS, Michael DC, Geunhwa J. Development of species-specific SCAR markers in Bentgrass. Crop Sci. 2003;43:345–349. [Google Scholar]
- 5.Esselman EJ, Li JQ, Crawford DJ, Windus JL, Wolfe AD. Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Placeae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Mol Ecol. 1999;8(3):443–443. doi: 10.1046/j.1365-294X.1999.00585.x. [DOI] [Google Scholar]
- 6.Fu LG. Chinese Plant Red Book. Beijing: Science Press; 1992. (in Chinese) [Google Scholar]
- 7.Hernández P, Martin A, Dorado G. Development of SCARs by direct sequencing of RAPD products: a practical tool for the introgression and marker-assisted selection of wheat. Mol Breed. 1999;5(3):245–253. doi: 10.1023/A:1009637928471. [DOI] [Google Scholar]
- 8.Micheli MR, Bova R, Pascale E, D′Ambrosio E. Reproducible DNA fingerprint with the random amplified polymorphic DNA (RAPD) method. Nucl Acids Res. 1994;22:1921–1922. doi: 10.1093/nar/22.10.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paran I, Michelmore RW. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet. 1993;85(8):985–993. doi: 10.1007/BF00215038. [DOI] [PubMed] [Google Scholar]
- 10.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucl Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey S. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang RH, Liu HE. Wax Shrubs in World (Calycanthaceae) Beijing, China: China Science and Technology Press; 1998. (in Chinese) [Google Scholar]
- 13.Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20(2):176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]
- 14.Zou YP, Ge S, Wang XD. Molecular Markers in Systemic and Evolutionary Botany. Beijing, China: Science Press; 2001. (in Chinese) [Google Scholar]