Abstract
Cytoplasmic aminoacyl-tRNA synthetases of higher eukaryotes acquired extra peptides in the course of their evolution. It has been thought that these appendices are related to the occurrence of the multiprotein complex consisting of at least eight different tRNA synthetase polypeptides. This complex is believed to be a signature feature of metazoans. In this study, we used multiple sequence alignments to infer the locations of the peptide appendices from human cytoplasmic tRNA synthetases found in the multisynthetase complex. The selected peptide appendices ranged from 22 aa of aspartyl-tRNA synthetase to 267 aa of methionyl-tRNA synthetase. We then made genetic constructions to investigate interactions between all 64 combinations of these peptides that were individually fused to nonsynthetase test proteins. The analyses identified 11 (10 heterologous and 1 homologous) interactions. The six peptide-dependent interactions paralleled what had been detected by crosslinking methods applied to the isolated multisynthetase complex. Thus, small peptide appendices seem to link together different synthetases into a complex. In addition, five interacting pairs that had not been detected previously were suggested from the observed peptide-dependent complexes.
Keywords: peptide appendices, yeast two-hybrid assay, interaction map
Proteins are molecular fossils that may help to unravel the history and mechanism of evolution. Aminoacyl-tRNA synthetases (ARSs) are one of the ancient proteins evolved to decode genetic information to amino acids. Although all of the tRNA synthetases catalyze the same chemical reactions, these enzymes have accumulated a wide range of sequence and structural diversity throughout evolution (1). In particular, the tRNA synthetases of higher eukaryotes have acquired a few features that are not present in those of other organisms. Most intriguing is the presence of noncatalytic peptide appendices, the roles of which are yet to be elucidated. It has been thought that these peptides are involved in protein–protein interactions among the tRNA synthetases that form the multiprotein complex. This complex is another characteristic of the cytoplasmic tRNA synthetases of higher eukaryotes (2–4) and contains at least eight different tRNA synthetase polypeptides [Glu-Pro-tRNA synthetase (EPRS), IRS, LRS, MRS, QRS, RRS, KRS, and DRS; refs. 5–8], as well as three nonsynthetase proteins of 43 kDa, 38 kDa, and 18 kDa (5–11).
Although these complexes have been known for more than two decades, the structural organization and interactions between the components have not been well understood. Structural analysis with electron microscopy has shown that the complex forms an elongated U shape (8). The complex structure has been studied extensively by stepwise dissociation of the components by using nonionic detergent (12, 13), changing salt concentration (14), and chemical crosslinking (15, 16). The multi-ARS complex has been proposed to consist of three subdomains. The base subdomain consists of EPRS, IRS, and LRS, which are the enzymes of higher molecular weights. There are two arm subdomains on the top of the base subdomain. Subdomain I contains a dimer of DRS and monomers of MRS and QRS; subdomain II is made of dimers of KRS and RRS (16).
Most of the structural analyses on the multi-ARS complex have relied on biochemical approaches, as listed above. Because these methods address only the physical relationships of the protein components, other approaches are necessary to determine the molecular mechanisms and the peptide regions involved in the assembly or maintenance of the complex. We thought that a genetic approach would be suitable to investigate these questions. A yeast two-hybrid system has proven useful to analyze protein–protein interactions (17). This method was employed previously to analyze the interactions of the repeated motifs of EPRS with the C-terminal repeated motifs of IRS (18), as well as with the N-terminal extension of RRS (19). The interaction between the repeats of EPRS and IRS was confirmed further by biochemical and biophysical methods (19). Interactions of p38 with the other ARS components recently have been reported by using two-hybrid analysis (11).
The role of the peptide appendices in the formation of the multi-ARS complex has been studied in a few cases such as the N-terminal sequences of DRS (20, 21), RRS (22, 23), and KRS (6). However, systematic studies on the structural features and molecular interactions of these peptides in eukaryotic ARSs are needed to understand their functional significance. In the present study, we focused on the peptide appendices deduced from the complex-forming ARSs. We investigated whether these regions actually are involved in protein–protein interactions among the complex-forming ARSs and, if so, how they are connected to each other. Finally, the interactions between the unique peptides of complex-forming tRNA synthetases determined by the genetic method in this work were compared with those previously obtained by biochemical methods.
MATERIALS AND METHODS
Determination of Unique Peptide Appendices.
Sequences of the complex-forming human tRNA synthetases were aligned with those of the corresponding enzymes from various organisms by using a macaw program (24) to determine the unique peptide appendices present in each of the complex-forming human tRNA synthetases. Secondary structures of the determined peptide appendices were predicted by using the gor, dpm, gibrat, and homologue methods installed in the antheprot program (25) and the paircoil programs (26). The previous reports on the peptide appendices also have been consulted to determine the peptide appendices, especially, in the cases of DRS (20, 21), RRS (22, 23), QRS (27), IRS (28), and EPRS (29). These peptide regions were predicted to be predominantly α-helices and were used for the interaction mapping.
Subcloning of Human ARSs for Two-Hybrid Assay.
The gene coding for the N-terminal 22 aa (T5–E26) of human DRS was synthesized chemically. The other cDNAs encoding the unique ARS peptides (Fig. 1) were isolated by PCR by using the specific primers and template cDNAs. The templates used were pM116 for KRS (30), pM182 for RRS (this study), pM191 for QRS (this study), pM184 for MRS (H.M. and K.S., unpublished work), and pM208 for LRS (H.M. and K.S., unpublished work). The pM182 that contains the N-terminal part of RRS was constructed by cloning of the 5′ rapid amplification of cDNA ends/PCR product with human fetal brain (Clontech). The pM191 carrying the full-length cDNA of human QRS was constructed by cloning the PCR product of the published sequence (27). The forward and backward primers contained EcoRI and SalI sites, respectively, so that the PCR products could be cloned into pLexA and pJG4–5 vectors (19, 31). For the deletion mapping of the interaction motifs in RRS and DRS, the corresponding primers were synthesized and used to isolate cDNA fragments. These fragments were subcloned into LexA or B42 fusion vectors as described above. The B42 plasmid containing the full DRS was cleaved with EcoRI and NcoI to delete the N-terminal part of the DRS protein. The staggered ends of the digested plasmid were filled with Klenow fragment and self-ligated. The resulting plasmid thus expressed the C-terminal 192 aa from M309 to P500 of DRS and was used for the interaction mapping. The deletion constructs of IRS and EPRS have been described (18). The sequences of the constructs were confirmed, and expression of the LexA and B42 hybrid proteins was tested by immunoblotting with monoclonal antibodies specific to LexA and hemagglutinin (which was tagged between B42 and the fused polypeptides), respectively (data not shown). The alkaline phosphatase-conjugated secondary antibody was used to detect the hybrid proteins by using disodium 3–{4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo-[3.3.1.1.3,7]decan]-4-yl}phenylphosphate according to the manufacturer’s instructions (Boehringer Mannheim).
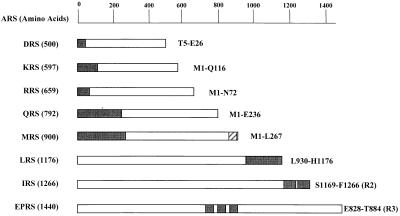
Figure 1.
Peptide appendices in human complex-forming ARSs. A group of eight complex-forming human ARS polypeptides are listed according to their amino acid lengths. The unique peptide appendices of ARSs are indicated by black boxes, and their locations used for the interaction studies are described. The C-terminal extension of IRS contains two repeats of about 90 aa, and the internal linker region of EPRS consists of three repeats of 57 aa. IRS-R2 and EPRS-R3 indicate the second and third units of their repeats, respectively. Although a single copy of the motif homologous to the repeats of EPRS is also present in Q841–K897 of MRS (indicated as a hatched box), it was not used for the experiment, because the same result as the motif of EPRS was expected (18).
Determination of Interaction.
Interactions between the unique peptide regions of the complex-forming ARSs were determined by the peptide-dependent induction of the reporter genes, LEU2 and lacZ. The LexA- and B42-hybrid proteins were coexpressed in yeast strain EGY48 (31), and positive interactions between the two-hybrid proteins were detected by cell growth on the leucine-depleted synthetic yeast medium containing 2% (wt/vol) galactose and by the formation of blue colony on the same synthetic medium containing 5 mM X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside), 2% (wt/vol) galactose, and 2% (wt/vol) raffinose (18, 31).
RESULTS
Determination of Peptides Appendices in Complex-Forming ARSs.
Sequences of eight human ARS polypeptides found in the multi-ARS complex were compared with those of other species (lower eukaryotes, eubacteria, and archaebacteria) to identify the peptide appendices that are unique to higher eukaryotes (data not shown). In addition to these sequence comparisons, we also took account of the continuity of the predicted secondary structures and inferred the higher eukaryote-specific domains of these enzymes whose locations and lengths are summarized in Fig. 1.
ARSs of smaller molecular weights, including DRS (500 aa; ref. 20), KRS (597 aa; ref. 30), RRS (659 aa; ref. 22, 32), QRS (792 aa; ref. 27), and MRS (900 aa; ref. 33), commonly contain the unique N-terminal peptides, although their sequences and lengths are different from each other. In contrast, ARSs of larger molecular weights, including LRS (1,176 aa; H.M. and K.S., unpublished work) and IRS (1,266 aa; ref. 28), contain the unique peptides in their C-terminal extensions. In particular, the C-terminal extension of IRS contains two tandem repeats of about 90 aa. The bifunctional tRNA synthetase (EPRS; 1,440 aa) is the largest polypeptide containing three unique repeats of 57 aa in the noncatalytic linker region between the two catalytic domains (29).
The motifs homologous to the repeats of EPRS have been found as a single copy in the C-terminal region of MRS (Fig. 1, hatched box), as well as in the N-terminal extensions of GRS, HRS, and WRS that have not been found in the multi-ARS complex. We previously found that the motif in the N-terminal extension of GRS also interacted with the C-terminal repeats of IRS as the repeated motif in the EPRS (18), although its interaction strength was weaker than that of the three repeats of EPRS. Thus, we did not include the C-terminal motif of MRS in the interaction test, because it would give the same result as the single repeated unit of EPRS. The cDNA fragments encoding the unique peptide appendices were isolated by PCR and used for the yeast two-hybrid analyses. The ARS peptides were expressed as fusion proteins with LexA and B42 to test their interactions in a reciprocal manner (31). Sequences and expression of the hybrid proteins were confirmed by DNA sequencing and immunoblotting, respectively (data not shown).
Interactions of Peptide Appndices.
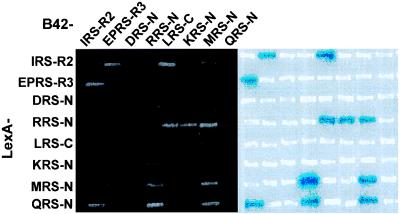
Interactions between pairs of the human ARS peptide appendices were investigated for 64 combinations in the two-hybrid assay by monitoring the simultaneous induction of lacZ and LEU2 reporter genes. Expression of the LEU2 and lacZ genes was detected by cell growth on the leucine-depleted medium and the formation of blue colonies on the X-Gal containing medium, respectively. The results determined by the two reporter systems were consistent with each other as shown in Fig. 2.
Figure 2.
Interactions between peptide appendices of human ARSs determined by the yeast two-hybrid system. The peptide appendices of human ARSs were expressed as LexA and B42 fusion proteins in yeast. The positive interactions were determined by cell growth on leucine-depleted medium (Left), as well as by the formation of blue colonies on medium containing X-Gal (Right). Cells were incubated for 3 days at 30°C. In the cases of EPRS and IRS, only single copies (EPRS-R3 and IRS-R2) of their repeated sequences were used for the experiment (see Fig. 1). N and C indicate the N- and C-terminal extensions of the complex-forming ARSs, respectively.
Of 64 combinations (28 heterologous pairs in two directions and 8 homologous pairs), 8 heterologous (I–EP, I–L, I–Q, R–L, R–K, R–Q, R–M, and Q–M) and 1 homologous (M–M) pairs had positive interactions. Most of the peptides (except for DRS-N and KRS-N) showed the ability to interact with more than a single peptide. For instance, the N-terminal 267-aa extension of human MRS interacted not only with two heterologous peptides, RRS-N and QRS-N, but also with its homologous partner (Fig. 2). Similarly, RRS-N interacted with LRS-C, KRS-N, QRS-N, and MRS-N. The C-terminal appendix of IRS interacted with EPRS-R, LRS-C, and QRS-N. LRS-C interacted with IRS-C and RRS-N. The interaction between IRS and EPRS, but not the interaction of IRS-C with LRS-C and QRS-N, was reported previously (18).
Multiple Interactions of N-terminal Extension in RRS.
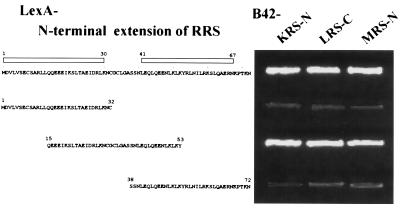
The 72-aa N-terminal extension of RRS fused to LexA showed interactions with three different peptides of LRS, KRS, and MRS fused to B42 (Fig. 2). RRS-N was predicted to form helices from M1 to K30 and from L41 to N67 (data not shown). To determine the peptide region of RRS involved in the interactions with the heterologous peptides, this extension was divided further into three fragments, M1 to C32, Q15 to Y53, and S38 to N72. All three peptide fragments were stably expressed in yeast as fusion proteins with LexA (data not shown). The peptide spanning from Q15 to Y53 interacted with all of the three tested peptides (Fig. 3), suggesting that the interactions would be localized in a small peptide motif. We currently do not know whether these three peptides interact with RRS-N simultaneously or competitively.
Figure 3.
Determination of interacting motif in RRS. The peptide fragments of the N-terminal extension in RRS were tested for interactions with KRS, LRS, and MRS. The helical regions in the N-terminal extension predicted by the antheprot program are marked by open boxes. The N-terminal extensions of KRS and MRS and the C-terminal extension of LRS were used for the experiments. Cells were grown on leucine-depleted yeast synthetic medium for 3 days at 30°C.
Interaction of the N-Terminal Extension of DRS with Repeated Sequences of IRS.
It was surprising that human DRS-N did not interact with any of the tested peptides in the two-hybrid assay (Fig. 2). The homologous N-terminal extension of rat DRS was shown to be necessary to associate the enzyme to the multi-ARS complex by comparing the behaviors of the full-size and N-terminal truncated DRS (34, 35). We thus expected that the human DRS-N would play a similar role by interacting with other peptide appendices.
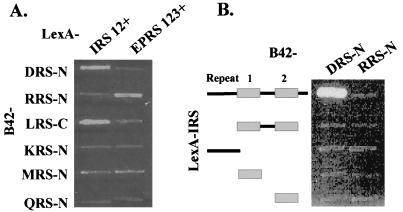
It reminded us that RRS-N interacted with the three repeats of EPRS but not with its single unit (19). Among the eight polypeptides of tRNA synthetases found in the multi-ARS complex, two tRNA synthetases, EPRS and IRS, contain repeated sequences (Fig. 1). If DRS-N interacts with one of these repeats, this interaction would be missed, because only single repeated units of the two enzymes were used to obtain the data shown in Fig. 2. We thus tested whether the repeated sequences of EPRS and IRS create a different pattern of interaction with other ARS peptides, including DRS-N. Although the EPRS repeats interacted with RRS as shown previously (19), the IRS repeats interacted with DRS-N (Fig. 4A). The interaction of DRS-N with IRS was refined further by using the C-terminal peptides of IRS containing different numbers of repeats (Fig. 4B). Interaction was shown only with the C-terminal peptide of IRS containing the two repeats and flanking sequence, further supporting the idea that the sequence repetition and flanking sequences are important to determine the interaction.
Figure 4.
Interactions of ARSs with the repeated sequences in IRS and EPRS. (A) Interactions of the peptide appendices in the complex-forming human ARSs were tested with the C-terminal region of IRS containing two repeats and flanking sequences (IRS–R12+ from E966 to F1,266) and the internal linker region of EPRS containing three repeats and flanking sequences (EPRS–R123+ from V573 to K889; ref. 15). (B) The N-terminal motifs of DRS and RRS were tested for interaction with the C-terminal region of IRS containing different repeats and flanking sequences (from top to bottom: E966–F1,266; S1,078–F1,266; E966–S1,077; S1,078–G1,161; and S1,169–F1,266). Incubation conditions were the same as described in Fig. 3.
Heterologous Interactions of the N- and C-Terminal Regions of DRS.
Although our results showed that DRS-N interacts with IRS (Fig. 4), recent two-hybrid analysis using the full-length DRS suggested its interaction with EPRS (11). This result suggested that the N-terminal extension and core domain of DRS may have a different interaction partner. We tested whether the N- and C-terminal regions of DRS would interact respectively with IRS and EPRS. As expected, DRS-N interacted with IRS-C, whereas the C-terminal 192-aa peptide of DRS interacted only with the EPRS peptide containing the repeats and flanking sequence (Fig. 5). No interaction of DRS with other ARSs was observed (11). These results suggest that the interactions of the core bodies of ARSs may not necessarily be the same as those of the peptide appendices.
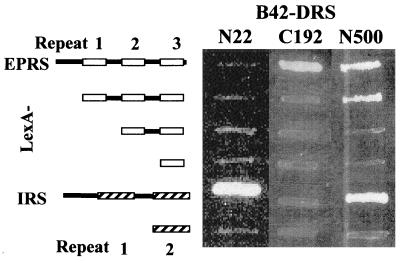
Figure 5.
Heterologous interactions of the N- and C-terminal regions of DRS. The full-length DRS (N500) and its N-terminal (N22; T5–E26) and C-terminal (C192; M309–P500) fragments were tested for the interaction with various fragments of EPRS (from top to bottom: V573–K889; D677–T884; E750–T884; and E828–T884) and IRS (from top to bottom: E966–F1,266 and S1,169–F1,266).
Interaction Map.
The neighboring pairs of the complex-forming ARSs were analyzed extensively by chemical crosslinking methods by using cis-dichloroammineplatinum (II), ethyleneglycobis(succinimydylsuccinate), and 4-succinimydyl-oxycarbonyl-α-(2-pyridyldithio)toluene. Because the chemical reactivity and structures of these crosslinkers were different, they identified different pairs of proteins. In the present work, the interactions between the complex-forming ARSs were investigated by using a genetic approach. Genetic analyses are useful to determine a molecular interaction, whereas biochemical methods address the physical proximity of the molecules. In addition, we focused on the appended domains of the ARSs, and the crosslinking experiments dealt with the whole ARS structures. Because of the intrinsic differences between the two approaches and the structural complexity of the multi-ARS complex, the results of this work may not be consistent with those obtained by crosslinking.
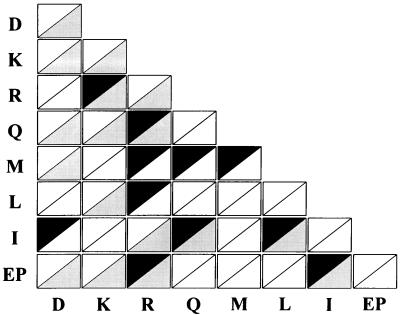
We thought it would be useful to compare the results of this work with those of the crosslinking methods (Fig. 6), because the two experiments could be complementary. Of the 11 interacting pairs identified in this work, 6 (R–K, Q–R, EP–R, I–Q, I–L, and EP–I) were detected by genetic and crosslinking methods. The R–K and EP–I pairs also were suggested by other methods (4, 13). These results suggest that many interactions between ARSs would be mediated by their appendices, although additional interactions may take place via their core domains. The remaining five pairs (I–D, M–R, L–R, M–Q, and M–M) were found only by the two-hybrid analyses, and the homologous (R–R, K–K, and D–D) and heterologous (K–D, Q–D, M–D, EP–D, Q–K, L–K, EP–K, and I–R) pairs were found only by the crosslinking method. The fact that some pairs were not detected by the crosslinking methods may have been a result of crosslinkers that did not reach the interacting regions or peptides that did not have chemical groups reactive to the crosslinkers. On the contrary, the pairs that were observed only by the crosslinking methods were likely in close proximity, but theses pairs do not make specific interactions via their respective appendices.
Figure 6.
ARS pairs identified by two-hybrid and crosslinking methods. The 28 heterologous and 8 homologous pairs of the complex-forming ARSs are displayed as rectangles consisting of two triangles. The interacting pairs between the appendices of ARSs determined by the two-hybrid analyses (dark triangles) were compared with all of the neighboring pairs suggested by the crosslinking methods (gray triangles). White triangles represent the pairs detected by neither of the two methods.
DISCUSSION
Proteins are adapted to different environments by amino acid replacements and recruitment of additional domains to their core bodies. Modular arrangement is prominent in the primary and tertiary structures of ARSs (36–39). Their core bodies essentially consist of two modules responsible for catalysis and tRNA recognition. Some ARSs like Escherichia coli methionyl-tRNA synthetase (40) and alanyl-tRNA synthetase (41) have dispensable domains that are required for oligomerization. In this paper, we focused on such extra domains of ARS, especially higher eukaryote-specific domains that must have been acquired in the course of evolution (42). Recent advances in genome sequencing enable us to perform systematic comparison of ARS sequences from various organisms and to infer these extra domains from eight ARSs (Fig. 1). Using these defined peptides, we systematically examined the interaction abilities of these extra domains.
It has been thought that the acquired domains are related to the occurrence of the multi-ARS complex. Indeed, there is some experimental evidence supporting this relationship (21, 22, 43). The work presented here was designed to address this possibility in all eight human ARSs that form the complex. Interactions of the peptide appendices were determined by using a yeast two-hybrid system. With 8 different ARS peptides, 28 heterologous and 8 homologous combinations are possible. Because we fused each peptide with both LexA and B42 in the two-hybrid system, the 28 heterologous pairs were analyzed in two directions, and thus a total of 64 combinations were searched exhaustively (Fig. 2). In some cases, the interaction was sensitive to the polarity of the fused proteins. For example, LRS-C and KRS-N fused to LexA and QRS-N fused to B42 did not show any interaction, whereas the same peptides fused to the counterpart did (Fig. 2). The polarity effect of the fused proteins has been observed frequently in the two-hybrid assay. The negative interaction of these hybrid proteins may have resulted from a conformational change or steric hindrance that interfered with the interaction, and thus we took only the positive results. Because we used single-repeated motifs in IRS and EPRS in the exhaustive search (Figs. 1 and 2), we separately tested the interaction between the complete repeats of IRS and EPRS as well as another six domains (Fig. 4). From this combinatorial analysis, we observed one additional interaction (I–D) and confirmed the interaction previously reported (EP–R; ref. 19). Together, a total of 10 heterologous and 1 homologous interaction pairs were found.
ARSs seem to have developed idiosyncratic ways to interact with other molecules. RRS-N showed the ability to interact with several different peptides. EPRS and IRS increased the strength and versatility of their interactions by sequence repetition (Figs. 2 and 4). MRS contains the N-terminal extension that can make a homologous interaction (Fig. 2) and an additional C-terminal motif homologous to the EPRS repeats (Fig. 1) that may be involved in extra protein–protein interaction (18). The interactions occurring at both extremes of MRS may explain its flexible stoichiometry in the multi-ARS complex (44). The multivalency of the interactions or the multiplicity of the sequence units may help to stabilize the multi-ARS complex. Alternatively, different peptides may compete for binding to a particular ARS in vivo, and this competition may facilitate the dissociation of the component from the complex.
The N-terminal truncated DRS did not associate to the multi-ARS complex, suggesting the role of the N-terminal region in the complex-formation. However, its N-terminal extension did not take the connected heterologous domain to the complex (34). These results imply that an additional interaction in the core domain is also important in the complex. Analyses of the interactions with the N- and C-terminal peptides of DRS showed that the two regions interact with IRS and EPRS, respectively (Fig. 5). Our conclusions concerning different interactions between the appendices and core domains of the ARS components are supported further by comparing the pairs determined in this work with those suggested by the crosslinking method. A portion of the crosslinked pairs were confirmed by the two-hybrid results, suggesting that many contacts may take place other than the peptide appendices.
The dissociation constant between the repeated sequences of EPRS and IRS was determined to be 2.9 μM (19). We expect that the other pairs would interact at similar affinity, because the induction levels of the reporter genes in the two-hybrid assay were similar between the interacting pairs (Fig. 2). If the ARS components were maintained only by the interactions of their peptide appendices, they would be dissociated easily from the complex and thus the complex would be fragile. However, the components of the multi-ARS complex remained associated during various purification procedures (8, 42, 45). Interactions of the core bodies of ARSs and other components such as p43 (10), p38 (11), and p18 (9) also may contribute to the stability of the complex. In this regard, it is worthwhile to note a recent report showing that p38 plays an important role in the assembly of the multi-ARS complex (11).
Although our results showed that many peptide appendices are involved in protein–protein interactions between the components of the multi-ARS complex, we do not rule out the possibility that the appended domains have other functions. Other types of heterologous oligomerization are also apparent in the eukaryotic tRNA synthetases. VRS is associated with elongation factor complex (46, 47). MRS and ERS of Saccharomyces cerevisiae are bound to the Arc1p that facilitate the delivery of tRNA (48, 49). The possible function as a targeting signal also has been proposed for the N-terminal extension of DRS (35). Other functional possibilities of these peptide appendices are supported further by the fact that these extra peptide appendices are also present among the non-complex-forming ARSs (50–53). Therefore, the general function of these peptides may not be to maintain the multi-ARS complex, although they may contribute to its assembly. Perhaps they structurally and functionally link these enzymes to other biological systems, such as protein synthesis machinery and the cytoskeleton, or facilitate the transfer or association of tRNA (19, 54–56).
This work was supported in part by Biotech 2,000 Grant 97-N1-06-01-A-06, by a grant from the National Creative Research Initiatives of the Ministry of Science and Technology of Korea (to S.K.), and by a grant from the Ministry of Education, Science, and Culture, Japan (to K.S.).
ABBREVIATIONS
- ARS
aminoacyl-tRNA synthetase
- XRS
ARS for amino acid X
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Schimmel P. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- 2.Mirande M. Prog Nucleic Acid Res Mol Biol. 1991;40:95–142. doi: 10.1016/s0079-6603(08)60840-5. [DOI] [PubMed] [Google Scholar]
- 3.Kisselev L L, Wolfson A D. Prog Nucleic Acid Res Mol Biol. 1994;48:83–142. doi: 10.1016/s0079-6603(08)60854-5. [DOI] [PubMed] [Google Scholar]
- 4.Yang D C H. Curr Top Cell Regul. 1996;34:101–136. doi: 10.1016/s0070-2137(96)80004-5. [DOI] [PubMed] [Google Scholar]
- 5.Mirande M, Kellermann O, Waller J P. J Biol Chem. 1982;257:11049–11055. [PubMed] [Google Scholar]
- 6.Cirakoglu B, Waller J-P. Eur J Biochem. 1985;151:101–110. doi: 10.1111/j.1432-1033.1985.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 7.Mirande M, Le Corre D, Waller J-P. Eur J Biochem. 1985;147:281–289. doi: 10.1111/j.1432-1033.1985.tb08748.x. [DOI] [PubMed] [Google Scholar]
- 8.Norcum M T. J Biol Chem. 1989;264:15043–15051. [PubMed] [Google Scholar]
- 9.Quevillon S, Mirande M. FEBS Lett. 1996;395:63–67. doi: 10.1016/0014-5793(96)01005-8. [DOI] [PubMed] [Google Scholar]
- 10.Quevillon S, Agou F, Robinson J-C, Mirande M. J Biol Chem. 1997;272:32573–32579. doi: 10.1074/jbc.272.51.32573. [DOI] [PubMed] [Google Scholar]
- 11.Quevillon S, Robinson J-C, Berthonneau E, Siatecka M, Mirande M. J Mol Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 12.Sihag R K, Deutscher M P. J Biol Chem. 1983;258:11846–11850. [PubMed] [Google Scholar]
- 13.Norcum M T. J Biol Chem. 1991;266:15398–15405. [PubMed] [Google Scholar]
- 14.Dang C V, Yang D C H. J Biol Chem. 1979;254:5350–5356. [PubMed] [Google Scholar]
- 15.Filonenko V V, Deutscher M P. J Biol Chem. 1994;269:17375–17378. [PubMed] [Google Scholar]
- 16.Norcum M T, Warrington J A. Protein Sci. 1998;7:79–87. doi: 10.1002/pro.5560070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields S, Sternglanz R. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 18.Rho S B, Lee K H, Kim J W, Shiba K, Jo Y J, Kim S. Proc Natl Acad Sci USA. 1996;93:10128–10133. doi: 10.1073/pnas.93.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rho S B, Lee J S, Jeong E-J, Kim K-S, Kim Y G, Kim S. J Biol Chem. 1998;273:11267–11273. doi: 10.1074/jbc.273.18.11267. [DOI] [PubMed] [Google Scholar]
- 20.Jacobo-Molina A, Peterson R, Yang D C H. J Biol Chem. 1989;264:16608–16612. [PubMed] [Google Scholar]
- 21.Escalante C, Yang D C. J Biol Chem. 1993;268:6014–6023. [PubMed] [Google Scholar]
- 22.Girjes A A, Hobson K, Philip C, Lavin M F. Gene. 1995;164:347–350. doi: 10.1016/0378-1119(95)00502-w. [DOI] [PubMed] [Google Scholar]
- 23.Vellekamp G, Sihag R K, Deutscher M P. J Biol Chem. 1985;260:9843–9847. [PubMed] [Google Scholar]
- 24.Schuler G D, Altschul S F, Lipman D J. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 25.Geourjon C, Deleage G, Roux B. J Mol Graphics. 1991;9:188–190. doi: 10.1016/0263-7855(91)80008-n. [DOI] [PubMed] [Google Scholar]
- 26.Lupas A, Dyke M V, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 27.Lamour V, Quevillon S, Diriong S, N’Guyen V C, Lipinski M, Mirande M. Proc Natl Acad Sci USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiba K, Suzuki N, Shigesada K, Namba Y, Schimmel P, Noda T. Proc Natl Acad Sci USA. 1994;91:7435–7439. doi: 10.1073/pnas.91.16.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fett R, Knippers R. J Biol Chem. 1991;266:1448–1455. [PubMed] [Google Scholar]
- 30.Shiba K, Stello T, Motegi H, Noda T, Musier-Forsyth K, Schimmel P. J Biol Chem. 1997;272:22809–22816. doi: 10.1074/jbc.272.36.22809. [DOI] [PubMed] [Google Scholar]
- 31.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 32.Lazard M, Mirande M. Gene. 1993;132:237–245. doi: 10.1016/0378-1119(93)90201-d. [DOI] [PubMed] [Google Scholar]
- 33.Lage H, Dietel M. Gene. 1996;178:187–189. doi: 10.1016/0378-1119(96)00313-7. [DOI] [PubMed] [Google Scholar]
- 34.Mirande M, Lazard M, Martinez R, Latreille M T. Eur J Biochem. 1992;203:459–466. doi: 10.1111/j.1432-1033.1992.tb16570.x. [DOI] [PubMed] [Google Scholar]
- 35.Agou F, Mirande M. Eur J Biochem. 1997;243:259–267. doi: 10.1111/j.1432-1033.1997.0259a.x. [DOI] [PubMed] [Google Scholar]
- 36.Burbaum J J, Schimmel P. J Biol Chem. 1991;266:16965–16968. [PubMed] [Google Scholar]
- 37.Delarue M, Moras D. Bioessays. 1993;15:675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- 38.Schimmel P, Ripmaster T. Trends Biochem Sci. 1995;20:333–334. doi: 10.1016/s0968-0004(00)89067-2. [DOI] [PubMed] [Google Scholar]
- 39.Martinis S A. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 887–901. [Google Scholar]
- 40.Cassio D, Waller J-P. Eur J Biochem. 1971;20:283–300. doi: 10.1111/j.1432-1033.1971.tb01393.x. [DOI] [PubMed] [Google Scholar]
- 41.Jasin M, Regan L, Schimmel P. Nature (London) 1983;306:441–447. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- 42.Kerjan P, Cerini C, Semeriva M, Mirande M. Biochim Biophys Acta. 1994;1199:293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Deutscher M P. Biochem Biophys Res Comm. 1991;180:702–708. doi: 10.1016/s0006-291x(05)81122-2. [DOI] [PubMed] [Google Scholar]
- 44.Lazard M, Mirande M, Waller J-P. J Biol Chem. 1987;262:3982–3987. [PubMed] [Google Scholar]
- 45.Kellermann O, Brevet A, Tonetti H, Waller J-P. Eur J Biochem. 1979;99:541–550. doi: 10.1111/j.1432-1033.1979.tb13286.x. [DOI] [PubMed] [Google Scholar]
- 46.Bec G, Kerjan P, Zha X D, Waller J P. J Biol Chem. 1989;264:21131–21137. [PubMed] [Google Scholar]
- 47.Motorin Y A, Wolfson A D, Orlovsky A F, Gladilin K L. FEBS Lett. 1988;238:262–264. doi: 10.1016/0014-5793(88)80492-7. [DOI] [PubMed] [Google Scholar]
- 48.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt E C. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 49.Simos G, Sauer A, Fasiolo F, Hurt E C. Mol Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 50.Frolova L Y, Sudomoina M A, Grigorieva A Y, Zinovieva O L, Kisselev L L. Gene. 1991;109:291–296. doi: 10.1016/0378-1119(91)90624-k. [DOI] [PubMed] [Google Scholar]
- 51.Shiba K, Schimmel P, Motegi H, Noda T. J Biol Chem. 1994;269:1–7. [PubMed] [Google Scholar]
- 52.Shiba K, Motegi H, Yoshida M, Noda T. Nucleic Acids Res. 1998;26:5045–5051. doi: 10.1093/nar/26.22.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsui F W, Siminovith L. Nucleic Acids Res. 1987;15:3349–3367. doi: 10.1093/nar/15.8.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reed V S, Yang D C H. J Biol Chem. 1994;269:32937–32941. [PubMed] [Google Scholar]
- 55.Reed V S, Wasteny M E, Yang D C H. J Biol Chem. 1994;269:32932–32936. [PubMed] [Google Scholar]
- 56.Whelihan E F, Schimmel P. EMBO J. 1997;16:2968– 2974. doi: 10.1093/emboj/16.10.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]