Abstract
The vector-borne bacterium Borrelia hermsii, a relapsing fever agent, switches gene expression of a surface protein between different antigenic variants, thereby causing sequential waves of immune escape within hosts and increasing the likelihood of transmission. Analogous programmed systems of antigenic variation occur in African trypanosomes and Plasmodium falciparum. In these examples, switch rates to individual variants differ over a wide range. We studied how B. hermsii determines switch rates in two experimental infections: one where variants were identified by specific antisera and one based on identification by DNA sequence. Unexpressed loci of variant antigens copy into a single expression site at rates determined by extragenic features of silent loci rather than similarity between coding sequences of variants at silent sites and the single expression site. Two elements, in particular, determine switch rates. One set of elements overlaps the 5′ ends of the expressed gene and the silent loci; greater sequence identity between elements was associated with a higher switch rate. The second set of elements flanks the expression site on the 3′ side and occurs at variable distances downstream from silent loci; the nearer an element to a silent locus, the greater the switch rate of that locus into the expression site. In combination, these two features of the genome provide a simple mechanism to modulate switch rate whereby silent loci form a hierarchy of switch rates into the expression site. Although the switching hierarchy causes changes in individual cells that are stochastic, ordering of variants within hosts is semipredictable.
Keywords: antibody, Borrelia, recombination, relapsing fever, vector-borne
Antigenic variation is a common immune evasion strategy among pathogens, particularly bacteria and parasites dependent on arthropods for transmission (1, 2). These pathogens switch gene expression of a dominant surface protein between several different antigenic variants, thus allowing escape of newly appearing variants in the infected animal from prevalent antibodies or other adaptive effectors. Antigenic variation extends the infection, thereby increasing the likelihood of transmission by a tick or insect vector, or in the case of mucosal pathogens by person-to-person contact. In several examples of antigenic variation, there are differences in switch rates between variants. These differences of switch rates play a crucial role in the dynamics of infection and host immune responses (3–6).
The first experimental descriptions of antigenic variation were of relapsing fever (7, 8), an illness with episodes of fever and heavy bacteremia separated by periods of well-being and undetectable bacteria. Early immunologists recognized that the sequential waves of bacteria in the blood during cases of relapsing fever demonstrated the specificity of adaptive immune responses to infection (9). Similar programmed systems of antigenic escape among vector-borne pathogens were also noted in African trypanosomes (10, 11), Plasmodium agents of malaria (12–14), Babesia bovis (15), Anaplasma marginale (16), and Borrelia burgdorferi (17).
Our model system for antigenic variation is Borrelia hermsii, a relapsing fever agent which is transmitted between rodent reservoirs by ticks (18). B. hermsii infections of mice can be initiated with a single organism, and bacterial densities in the blood reach 106 to 107 per ml during peaks. The frequency of new variants in expanding B. hermsii populations either in animals or in culture medium is 10−4 to 10−3 (19). The molecular event underlying the majority of switches in surface proteins is a nonreciprocal recombination involving all or nearly all of a variant gene at a single expression site by one of several variant genes at silent loci on linear plasmids (20–22). We identified most of the silent variants that comprise the antigenic repertoire of B. hermsii, located 59 silent loci on several linear plasmids in the bacterium, and described the 5′ and 3′ flanking sequences (23).
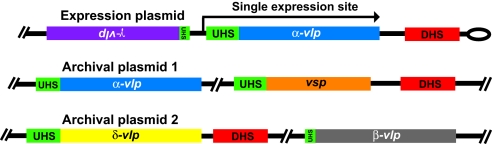
Fig. 1 schematically shows the organization of the expression plasmid and two archival linear plasmids in a cell of B. hermsii. In a switch in phenotype from one variant, or serotype, to another, a variant gene is copied into the expression site. In the example, the “blue” serotype is determined by expression of the blue variant gene. In an infection initiated by the blue serotype, the population of the infection's relapse several days later may be dominated by the “orange” serotype, conferred by replacement of the gene at the expression site by the orange variant. In these and other cases, the silent versions of the variant genes are unaltered.
Fig. 1.
Organization of the expression plasmid and two archival linear plasmids in B. hermsii. The lengths of genes and elements are relative and not to absolute scale. There is a single expression site on one plasmid and silent variants on the same and other plasmids. The direction and extent of transcription of the duplicate gene at the expression site is indicated by the arrow. The colors of genes indicate the family or subfamily of vsp and vlp genes. Most variant genes are arrayed on the same strand in a given plasmid fragment, but some are oppositely oriented, as indicated by inverted text. The UHS element at the expression site comprises 61 nt around the start codon of the variant gene. Silent genes vary in the extent to which their UHS regions are identical to the expression-site UHS, represented here by the relative length of the UHS block. There is a 214-nt noncoding DHS element downstream from the expression site and adjacent to the plasmid telomere and at various locations on the plasmids.
Two gene families comprise the variant antigens that specify the different serotypes: vsp genes of ≈600 nt and vlp genes of ≈1100 nt (24). The vlp genes are further divided into four subfamilies: α, β, γ, and δ (22, 25). Pair-wise differences in amino acid sequences range from 21% to 49% (mean of 37%) for 12 Vsp lipoproteins and 14% to 82% (mean of 65%) for 15 Vlp lipoproteins (23). Most variant genes are arrayed on the same strand in a given plasmid, but some are oppositely oriented. The upstream homology sequence (UHS) element at the expression site comprises 61 nt around the start codon of the variant gene (20, 26). Silent genes vary in the extent to which their UHS regions are identical to the expression site UHS. There is a 214 nt extragenic element, called the downstream homology sequence (DHS), downstream from the expression site and adjacent to the plasmid telomere (20). There are several DHS elements scattered across the plasmids that contain the silent sites; a DHS element does not detectably occur downstream from every silent variant site.
Here, we address the functional significance of these molecular observations. How do the various molecular details combine to determine the relative switch rates of different unexpressed sequences into the expression site? Are rates determined by the variant gene at the expression site or largely by characteristics of silent loci? Could nongenetic factors, such as differences in growth rate, account for the finding that some serotypes are substantially more frequent than others in populations of early relapses? Theoretical studies have shown that programmed antigenic switching provides significant benefits to infectious agents only when the switch rates between variants form a structured hierarchy (4, 5, 27), but the actual connections between the molecular biology and the patterns of switch rates have remained unclear even in intensively studied systems in Trypanosoma (28) and Plasmodium (29, 30).
Results
Serotype Frequencies in the First Relapses.
We inoculated groups of inbred mice with small numbers of one of two serotypes, 7 and 17, which have, respectively, a copy of vlp7 or vlp17 in the expression site (23). That initial serotype proliferated in the blood until day 5 or 6, at which point the host's adaptive immune response rapidly cleared the infecting serotype from the blood (19). When a relapse bacteremia was observed few days later, we used PCR to amplify the expression site and then sequence analysis to identify the predominant variant in the expression site in the relapse population in each mouse. This procedure provided an estimate of the relative rate at which the initial expressed sequence switches to one of the various sequences archived at unexpressed loci (Table 1, which is published as supporting information on the PNAS web site).
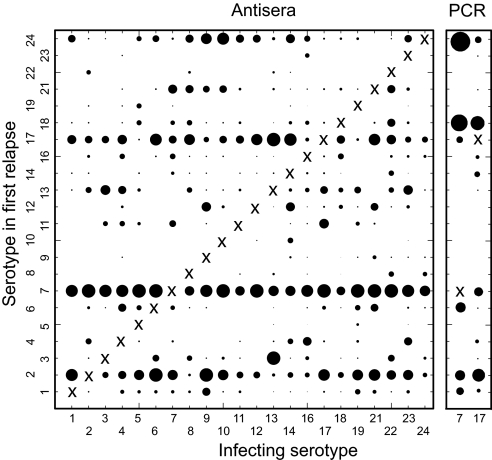
To corroborate our PCR analysis, we compared the estimated frequencies of variants in the relapse populations to the unpublished results from earlier antisera typing experiments that used a battery of variant-specific polyclonal antisera to distinguish between different variants of the same strain of B. hermsii (19). Mice in groups of mean size of 13 were infected with a clonal population of each of 22 variants known at the time. Serotype-specific reactions with antisera were used to identify and get relative counts of each of the typed variants in blood smears under microscopy. Although one serotype predominated in the relapse population, up to 6 serotypes could be detected. We calculated relative switch frequencies between variants by examining mice that started with one variant and had detectable quantities of a second variant in the first relapse (Table 1). The matrix of infecting serotype and first relapse serotypes in Fig. 2 summarizes the analysis of the data. Overall, some serotypes, such as 7 and 17, were much more common than others, such as 19 and 10, in relapse populations.
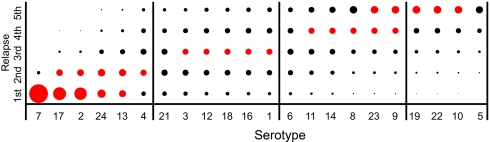
Fig. 2.
Summaries of frequencies of serotypes in first relapse of infections by B. hermsii HS1 in mice, as determined by serotype-specific antisera (Left) or PCR (Right). Data are from Table 1. The typable serotypes that were common to both the antisera analyses were 1–14, 16–19, and 21–24. The infecting serotype (X) was excluded from the denominator of first-relapse serotypes. The area of the circle for each x–y pair is proportional to the probability that a serotype on the y axis was detected in the first relapse of an infection initiated with the serotype on the x axis. The scale for the PCR data was four times that for antisera data to adjust for the greater sensitivity of antisera typing for detecting two or more serotypes in relapse populations (see Results).
Although antisera typing identified variants at prevalences as low as 2%, the PCR typing with expression site primers was limited to the most abundant one or two. Even so, the results of the PCR analysis, in terms of rankings of serotype frequencies, correlated closely with the earlier antibody-based study (Spearman correlation coefficient 0.72; 95% confidence interval 0.55–0.89; P = 0.0004). The five most prevalent serotypes in relapses in descending order were 7, 2, 17, 24, and 13 for the antibody-typing study and 18, 24, 2, 7, and 17 for the PCR-typing study (Fig. 2). These sets, which differed by single serotypes, accounted for 753 (59%) of 1272 and 63 (66%) of 95 of the relapse serotypes identified in each study, respectively (χ2 1.86; P = 0.2). Although the set compositions were similar, we noted that serotype 7, which was the most prevalent serotype in relapses by antibody-typing, was only the fourth most common by PCR-typing. Serotype 7 numbers may be underestimated by PCR for the complete expression site of all mouse serotypes, because the amplification product in this case would be a lengthy 2.3 kb instead of the 1.0–1.5 kb for almost all other serotypes (22, 31). To circumvent this possible bias, we developed PCR assays that were specific for serotype 7 and, as a control, serotype 17. In 19 mice infected with serotype 17, 18 (95%) had detectable serotype 7 in the relapse. This finding was similar to the 239 (82%) of 291 of mice that had serotype 7 in the relapse populations by antibody typing (χ2 1.21; P = 0.22). In comparison to serotype 7, serotype 17 was detected less commonly in relapses by serotype-specific PCR: 10 (42%) of 24 mice undergoing relapses after infection with serotype 7 (χ2 13.2; P = 0.0003). Again, this value was similar to the 164 (56%) of 291 of mice with detectable serotype 17 in relapses by antibody typing (χ2 1.39; P = 0.2; Table 1).
Variants Differ in Frequency but Not in Growth Rate in Unselected Heterologous Populations.
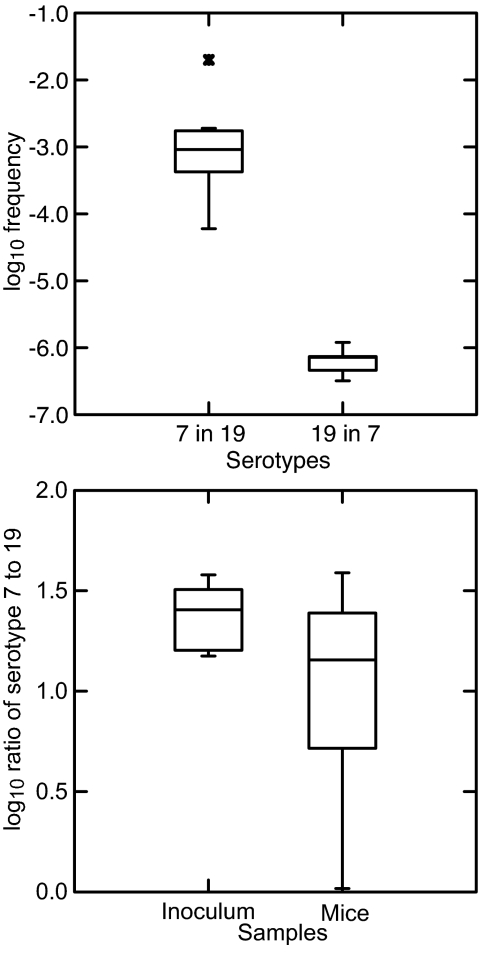
Differences in serotype frequencies in the relapse populations may be attributable to causes other than varied switch rates at the genetic level. For instance, cross-reactivity between variant antigens could be the primary cause of differences in the frequencies of these types, as reported by Recker et al. (32) for Plasmodium falciparum. Accordingly, we validated our use of frequency in the relapse population as a measure of relative switch rate. We infected groups of 9 immunodeficient mice with either serotype 7 or 19. In mice, serotype 7 was frequently found in the relapse population in both the antisera and PCR typing studies, whereas 19 was infrequent in the antisera study and not detected at all in the relapses in the PCR experiment (Table 1). The infection of the mice was monitored as the spirochete populations expanded, and blood was collected for extraction when the heterologous serotype (e.g., serotype 7 in an infection population derived from serotype 19) was expected, on the basis of pilot studies, to appear. We then used serotype-specific quantitative PCR to measure the ratios of these two types. Fig. 3Upper summarizes the results for five mice in which serotype 19 was detectable in serotype 7 populations and eight mice in which serotype 7 was found in serotype 19 populations (Table 2, which is published as supporting information on the PNAS web site). The medians for the ratios of serotypes 7 to 19 for all mice in a group were 6.0 × 10−4 (n = 9) in infection populations initiated by serotype 19, and 3.2 × 10−7 (n = 9) for serotypes 19 to 7 in populations initiated by serotype 7 (Wilcoxon–Mann–Whitney P = 0.001). These results match our observations on the differences in relative switch rates in immunocompetent mice measured by antisera or PCR typing.
Fig. 3.
Quantitative PCR assays for serotypes 7 and 19 in mouse infections. (Upper) Box-and-whisker plots, which give the median, first, and third quartiles, and 1.5 times interquartile range, of log10 frequencies of serotype 19 in serotype 7 populations (n = 5) and of serotype 7 (n = 8) in serotype 19 populations in the blood of immunodeficient mice that had detectable amounts of both serotypes (Table 2). (Lower) Box-and-whisker plots of log10 ratios of serotypes 7–19 in five samples of the plasma inoculum and in blood samples recovered from each of 10 immunocompetent mice infected with the mixture.
An apparent scarcity of serotype 19 in serotype 7 populations could be accounted for by a slower growth rate or other disadvantage for serotype 19 in serotype 7's presence. We evaluated this hypothesis by first mixing clonal populations of serotype 7 or 19 in freshly obtained plasma from immunodeficient mice and then by using this mixture to inoculate ≈100 cells into each of 10 immunocompetent mice. Blood was sampled before the peak of infection and the appearance of the neutralizing immune response. We determined numbers of serotypes 7 and 19 in the samples of the inoculum and of blood of the mice by variant-specific quantitative PCR. Fig. 3 Lower shows the ratios of serotype 7 to 19 before inoculation and after growth of ≈13 generations in the 10 mice. The mean log10 ratio (95% confidence limits) of serotype 7 to 19 was 1.4 (1.3–1.5) in the inoculum and 1.0 (0.7–1.3) in the mouse samples (P > 0.1). Thus, there was no evidence that serotype 7 grew faster than or otherwise dominated serotype 19 in the blood of mice.
Homology Between Coding Regions at Expression and Silent Sites Does Not Affect Switching Rates.
We carried out three tests of another hypothesis: greater sequence identity between variant coding regions at the expression site and a silent site fosters recombination and manifests as a higher frequency switch between that pair of variants. We first examined the correlation between the pair-wise nucleotide distance between variants and pair-wise frequencies of switches by using the Mantel test. Serotypes with vsp variant genes were examined separately from vlp gene serotypes, because of the lack of homology between these types of genes (Fig. 1). Table 3, which is published as supporting information on the PNAS web site, gives the pair-wise nucleotide distances, and Table 1 shows the frequencies of switches from the antisera typing set. Because switches between pairs of serotypes had different values for different directions, e.g., serotype 5 to 14 and serotype 14 to 5, left-to-right and right-to-left switches were done separately.
The correlation coefficients between sequence identity and switch frequency for left-to-right switches were −0.006 (P = 0.51) for 9 vsp genes and their corresponding serotypes and −0.057 (P = 0.70) for 13 vlp genes and serotypes. For right-to-left switches the coefficients were 0.057 (P = 0.37) for vsp serotypes and −0.146 (P = 0.90) for vlp serotypes.
Another test of directionality in switch frequency is whether there is an association between switches in each direction. If sequence similarity between variants is important, then switches between similar variants should be high in both directions, that is, no matter which variant of a pair is in the expression site and which is at a silent locus. The antisera typing data in Table 1 can be rewritten in pairs corresponding to left-to-right and right-to-left switches. By Kendall's rank correlation the association between the frequency of switching in each direction is close to zero (z = 0.58; P = 0.56; t = 0.03), suggesting that sequence homology does not affect switch frequency.
If the degree of sequence identity between variant genes was a major determinant of switch rates, then the relapse serotypes would be biased toward the same gene family (vsp, α-vlp, β-vlp, γ-vlp, and δ-vlp) as the infecting serotype. For instance, if the infecting serotype expressed one of the vsp genes, then there might be a higher proportion of relapse serotypes expressing one of the other vsp genes than if the infecting serotype expressed a β-vlp. We used the PCR typing data set for this study, because this method would capture all possible variants at the expression site and would not be limited to the serotypes represented by the antisera battery. From the 32 mice infected with serotype 7 (α-vlp), 42 serotypes were identified in relapses, and from the 41 mice infected with serotype 17 (δ-vlp), 53 serotypes were identified. The distributions of relapse serotypes by gene family were as follows: 28 vsp, 10 α-vlp, 0 β-vlp, 0 γ-vlp, and 4 δ-vlp from serotype 7-infected mice, and 24 vsp, 18 α-vlp, 5 β-vlp, 0 γ-vlp, and 6 δ-vlp for serotype 17-infected mice. Relapse serotypes expressing genes of the same family, e.g., a δ-vlp gene in the case of serotype 17 infections, were not significantly more or less common than expected for serotype 7 infections (odds ratio 0.6, 95% confidence interval of 0.2–1.7, P = 0.37) or for serotype 17 infections (1.2, 0.3–6.3, P = 0.78).
In summary, we found little or no correlation between pair-wise sequence similarity and frequencies in relapses. This independence of the coding regions allows the switching patterns to be modulated by mechanisms unconstrained by natural selection of the surface protein. The three studies above also address a hypothesis about frequency differences that invokes low levels of cross-reactivity between variant antigens as an important determinant (5). Weak cross-reactivity might cause the relapse populations to have fewer than expected serotypes of the same variant family as the infecting serotype, but this was not observed.
Genome Determinants of Switching Rates.
These findings led us next to ask whether noncoding features of the silent loci were determinants of relative switch rates. As Fig. 1 illustrates, variant genes comprise five sequence families of different sizes, are located on different plasmids including the expression plasmid, and occur in different orientations with respect to each other and the DHS element. The UHS elements of genes at silent sites vary in their identities with the expression site UHS, and archival vsp and vlp genes vary in their distance to DHS element on their downstream sides. Each of these sequence features of the genome could plausibly affect switch rates.
We used logistic regression to evaluate the association between these different variables and an ordinal classification scheme for serotype frequency in the first relapse. We based our logistic analysis on the combined data from PCR typing, antisera typing, and genome sequencing. We classified each of the 57 variants in the study as being switched to at high, intermediate, or low frequency, and included in the latter the 30 that were not observably expressed (Table 4, which is published as supporting information on the PNAS web site; we excluded two silent variants, see Materials and Methods). Here, our measure of switch frequency describes the relative rate at which a silent locus converts the expression site aggregated over all variants in the expression site. We aggregated over all candidates in the expression site because we found no evidence that the variant at the expression site influenced the subsequent switch process.
The odds ratios (95% confidence limits) for explanatory information about switch frequency in various predictor variables were as follows: number of genes in a variant family, 1.00 (0.92–1.08; P = 0.98); rank of the family size, 0.91 (0.68–1.23; P = 0.58); DNA strand the variant gene is located on, 0.39 (0.04–1.51; P = 0.25); and plasmid or sequence fragment the silent variant is located on, 0.99 (0.84–1.19; P = 0.61). In contrast, the odds ratios for fraction of UHS identity was 16.5 (2.15–190; P = 0.004) and for log10 DHS distance was 3.44 (1.63–7.28; P = 10−5) by the same analysis. There was little covariation between a silent site's UHS sequence and its distance to the nearest downstream DHS (Spearman coefficient = 0.10; P = 0.47).
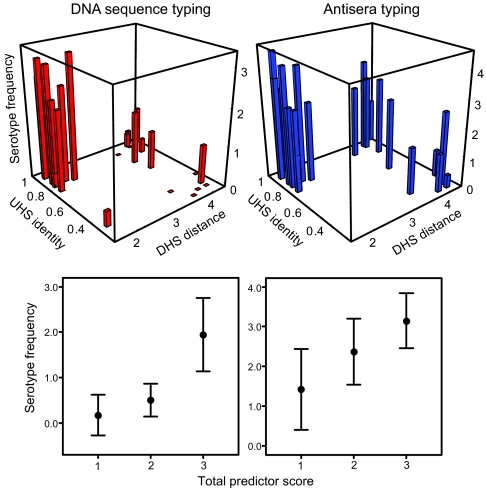
Fig. 4 shows that UHS and DHS together provide a good prediction for the relative rate at which an unexpressed sequence switches into the expression site. This result held for both the DNA sequence and antisera typing data sets, which were independent. If a silent variant both has high UHS sequence identity and is adjacent to a DHS, this defines a total predictor score of 3, and that variant is associated with the highest switch rates. If an unexpressed sequence has either a high degree of UHS identity or a close DHS element, it has a score of 2 and an intermediate switch rate. Those sequences that lack both a high UHS and a nearby DHS have a score of 1 and switch least frequently.
Fig. 4.
UHS and DHS elements are determinants of serotype switch rates in B. hermsii. (Upper) More frequent serotypes in relapses have high UHS identity and are close to a DHS element. The height (z axis) shows loge of frequencies per 100 immunocompetent mice for each variant; frequency depends on the fraction of UHS nucleotide identity and log10 of the minimum distance to the nearest downstream DHS element. Results shown separately for the DNA sequencing experiment (red; 27 variants; Table 4) and the antisera typing experiment (blue; 22 variants). The general linear model correlation coefficients for variant frequency regressed on UHS identity and DHS distance were 0.64 (F2 = 8.41; P = 0.002) for the DNA set and 0.66 (F2 = 7.23; P = 0.005) for the antisera set. (Lower) Loge of the relapse frequency per 100 mice vs. the total predictor score, defined as follows: 1, UHS identity <0.6 and log10 DHS distance ≥2.5; 2, UHS identity ≥0.6 or DHS distance <2.5; and 3, UHS identity ≥0.6 and DHS distance <2.5 (Table 4). We chose this scoring system to separate the clearly defined groups (Upper). Spearman correlation coefficients (95% confidence intervals) between total predictor score and loge of variant frequency per 100 mice were 0.67 (0.43–0.92; P = 0.0002) for DNA sequence typing and 0.62 (0.36–0.89; P = 0.002) for antisera typing.
The 27 variants that were known to be expressed (Table 2) had a mean predictor score of 2.2 (1.9–2.5). By contrast, the remaining 30 variants, which have not been observably expressed in this or previous studies, had a significantly lower mean predictor score of 1.5 (1.3–1.7; Wilcoxon–Mann–Whitney P = 0.001). One or more of these silent variants with low scores may be expressed at a low rate, perhaps contributing to relapse bacteremias that develop late in the infection.
Discussion
We have used relative frequency of variants in the relapse population as a measure of switch rate and argue that molecular properties of UHS and DHS explain much of the variation in switch rate. Two lines of evidence support our interpretation. First, molecular properties of UHS and DHS are the only correlates of the frequency of variants in the relapse population among several alternatives that we have tested. Second, we have shown that UHS and DHS provide the 5′ and 3′ sites for recombination between the expression site and a silent locus (23), thus linking our observations on serotype frequencies to specific features of the genome. The demonstration here that homology between the protein coding regions at the expression and silent sites has little effect on switching suggests to us the action of site-specific recombination or an illegitimate recombination process, alone or in combination with general homologous recombination, as may be the case in Trypanosoma brucei (33).
Several other bacterial pathogens vary a surface antigen through recombination between an expression site locus and complete or partial variant sequences at silent loci (18). But the closest analogy to antigenic variation in Borrelia is found in the protozoa that cause African trypanosomiasis (28, 34). In T. brucei, Morrison et al. (35) found different switch rates associated with different genomic locations for silent variants. However, the overall controls for the semipredictable ordering of variants in Trypanosoma, as well as in Plasmodium and other parasites, remain undefined. We have shown in B. hermsii that two elements flanking the expression site have homologs on the silent archival plasmids, and those two elements together explain much of the tendency for the switch rate of a silent site to be relatively high or low.
The key puzzle in such systems of programmed antigenic variation is how a stochastic process of switching leads to repeated relapses caused by a partially ordered sequence of variant surface antigens (10, 19, 36). Previous theory emphasized the need for switch rates to be structured in such a way that the rise and fall of variants flow through a connected pathway of types with tendencies to peak at different time (4, 5, 27). In those theories, the probability of switching to a particular new type depends on the type currently expressed. By contrast, we found that the switch rates of B. hermsii can be grouped into a simple hierarchy. This simple hierarchy defines a loose ordering in which switches to certain variants occur at relatively high frequencies, switches to other variants occur at relatively low frequencies, and the currently expressed variant has little effect on the switch frequencies.
Our observations raise the question: Is a simple hierarchy of switch rates sufficient to drive sequential relapses in which new variants escape host immunity? We studied this question with a quantitative model, illustrated in Fig. 5. That figure shows cumulatively along its x-axis the descending frequencies of the different serotypes in first relapses in the antisera typing study (Fig. 2 and Table 1). Using a model described in the caption, we show how the observed switching hierarchy favors some serotypes to appear in early waves of bacterial population growth and other serotypes to occur in later waves. That separation between early and late types causes a sequence of relapses.
Fig. 5.
Model of relapses of B. hermsii infection when there is a hierarchy of switch rates. The bottom row of circles shows the frequencies, pi, at which each serotype is detectable in the first-relapse bacteremia from the antisera-typing data (frequencies by serotype among the total of mice in Table 1), where the subscript i is the serotype number. Moving upwards, each row of circles shows the predicted frequency of detection in subsequent-relapse bacteremias. The total area of the circles across a row sums to a constant, so the circle areas represent the relative frequency at which a serotype is detectable in a particular bacteremia. In each row, the five red circles show the serotypes with the highest frequency of detection. We assumed that the current variant did not influence the switch rate (see Results) and that the abundance level that stimulated immunity, y, was 10-fold lower than the level at which it was detectable by antisera typing, x (see Supporting Methods, which is published as supporting information on the PNAS web site). In the absence of immunity, let the probability that the abundance of a type stimulates protective immunity be qi = piy/x. Then fi = pi(1 − qi)n−1, where the value (1 − qi)n−1 is the probability that, in the nth relapse, a serotype has not previously generated an immune response by appearing during one of the first n − 1 relapses. We used y/x = 0.1 to generate this figure; lower values of y/x, such as 0.01, cause only small qualitative changes in the patterns of dominance in successive relapses.
The simple switching hierarchy of B. hermsii, independent of the currently expressed type, forms an interesting contrast with more complex systems of antigenic variation. For example, an in vitro study of P. falciparum indicated that the currently expressed type significantly affects the rates at which the parasite switches to different variants (37). Why might B. hermsii have a homogeneous switch process, entirely or largely independent of the currently expressed type, whereas P. falciparum apparently has a heterogeneous switch process, dependent on the current type? We believe that the homogeneous process in B. hermsii represents a simpler form of antigenic variation associated with a few relapses after initial infection, whereas the heterogeneous process in P. falciparum represents a more complex form of switching associated with an extended sequence of relapses. The heterogeneity in P. falciparum may be influenced by more complex switch mechanisms and by immune selection (3, 32). As more information accumulates on different systems, we will be able to understand how the complexity of the switch process matches the patterns of recurring parasitemias and the degree to which antigenic variation extends the time of infection.
Materials and Methods
Mouse Infections.
The origin of B. hermsii isolate HS1 was described by Stoenner et al. (19). Serotypes 7 and 19 were cloned by limiting dilution and then propagated in adult female CB17 scid mice (Charles River Laboratories, Wilmington, MA). Serotype identities were confirmed by sequencing of the expression site as described (22). Cells were counted in a Petroff–Hausser counting chamber by phase-contrast microscopy. For studies of relapses and serotype frequencies 4- to 6-week-old female BALB/c mice (Charles River Laboratories) were inoculated i.p. with spirochetes in 0.1 ml of PBS or in 0.1 ml of plasma from infected CB17 scid mice. The mice were monitored daily for the presence and density of spirochetes by phase-contrast microscopy of tail-vein blood.
The DNA sequence-based study of relapses is described in Dai et al. (23). In brief, we infected mice with 1–10 spirochetes of serotype 7 or 17. At the time of relapse, blood was subjected to PCR with primers for the expression locus, and the resultant fragments were sequenced (31). Each expressed variant identified in a relapse was accounted for by an archived silent version. For the present study, 13 additional mice were included, giving a total of 32 infected with serotype 7 and 41 infected with serotype 17.
The methods for the antisera typing experiment with strain HS1 and a battery of serotype-specific, fluorescein-labeled, polyclonal mouse antisera have been described (19). Twenty-two of 24 serotypes in the original stocks were subsequently confirmed by DNA sequence (22, 23). Only serotype-specific reactions, that is, with fluorescence scores of 5 of 5, as defined by Stoenner et al. (19), were used. Original laboratory records for experiments carried out between 1/5/1980 and 4/11/1981 were examined, and the results of infections of 291 outbred, 18- to 19-day-old Swiss mice with these 22 serotypes were used to calculate pair-wise relapse frequencies (Table 1). Overall, 95% of the bacteria in blood smears were typable.
PCR.
DNA was extracted from 20- to 50-μl samples of whole blood or plasma with the QIAamp DNA Micro Kit (Qiagen, Valencia, CA). For serotype-specific PCR, the forward primer was 5′-TAAACTTTGAAAGTTGAGGTATAATGC-3′), and the reverse primers (and annealing conditions) were 5′-CTCTCCTGATTTAACTATTGAGCT-3′ (54°C for 1 min) for serotype 7 and 5′-TATCAGCTCTATTCGCATCAGTGG-3′ (60°C for 1 min) for serotype 17. Reaction conditions were as follows: 94°C for 5 min and then 40 cycles of 1 min at 94°C, followed by 1 min at 72°C. Samples were subjected to an initial denaturation at 94°C for 5 min and a final extension at 72°C for 7 min. For each of the 40 cycles, the denaturation step was 94°C for 1 min, and extension was 72°C for 1 min. For serotype-specific quantitative PCR, the forward primer was 5′-TGTAAACTTTGAAAGTTGAGGTATAATGC-3′. For serotype 7, the FAM-labeled probe was 5′-AGGTAAGACCGGAGTATCAGGAGGAGTAAATGGA-3′, and the reverse primer was 5′-TCCTACCTAATTCCATTAGTGAATTGC-3′. For serotype 19, the probe was 5′-TGCGGAAGACCCTCAGAGTAAATTTTTAAAGTCA-3′, and the reverse primer was 5′-GAAACTATATCTCCAAATGATGTAAACACA-3′. Assays were performed in a Rotor-Gene RG-3000 apparatus (Corbett Research, Sydney, Australia). The PCR conditions were as follows: 94°C for 10 min and then 50 cycles of 10 s at 94°C, followed by 1 min at 60°C. The standards for the assay were recombinant plasmids with cloned expressed vlp7 or vlp19 genes (20, 22), and the units of measurement were gene copies. The slopes of the standard curves were same in the presence or absence of excess heterologous DNA.
DNA Sequences.
Dai et al. (23) describe the locations of 59 silent variant genes and 14 DHS elements on plasmids of B. hermsii strain HS1 (accession nos. DQ218042, DQ166207, DQ172919, AY840995, AY838879, DQ173930, and CP000273–CP000281). We included 57 silent variants with these characteristics: location on a fragment that had at least one DHS element or was ≥15 kb in length and potential activation through gene replacement rather than a deletion at the expression site (38). The fraction of identity for UHS sequences was determined for a region beginning 35 nt upstream of the start codon and extending downstream for a total of 61 nt (23). DHS distance in nt was measured from the stop codon to the next DHS element on the same strand. If the fragment ended before a DHS was noted, we recorded the length to the end. The values for UHS and DHS factors are given in Table 4.
Statistical Analysis.
Standard statistical analyses were carried out with SYSTAT v. 11 or StatXact v. 6.3 (Cytel Software, Cambridge, MA), and significance tests were two-sided. We examined the correlation between the pair-wise nucleotide distance between variants and pair-wise frequencies of switches using the Mantel test (39) and Arlequin 2.0 software (http://anthro.unige.ch/arlequin) with 10,000 permutations. We used exact polytomous logistic regression (LogXact v. 6.3, Cytel Software) to evaluate the association between scalar predictor variables and discrete outcomes.
Supplementary Material
Acknowledgments
We thank Tom Schwan and Steve Porcella for access to unpublished sequence data, Chris Crowder for data analysis, and Carol Carter and Hany Mattaous for technical assistance. This work was supported by National Institutes of Health Grants AI24424 (to A.G.B.) and GM076499 (to S.A.F.) and National Science Foundation Grant DEB-0089741 (to S.A.F.).
Abbreviations
- DHS
downstream homology sequence
- UHS
upstream homology sequence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Barbour AG, Restrepo BI. Emerg Infect Dis. 2000;6:449–457. doi: 10.3201/eid0605.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank SA. Immunology and Evolution of Infectious Disease. Princeton: Princeton Univ Press; 2002. [PubMed] [Google Scholar]
- 3.Agur Z, Abiri D, van der Ploeg LHT. Proc Natl Acad Sci USA. 1989;86:9626–9630. doi: 10.1073/pnas.86.23.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank SA. Proc R Soc London Ser B; 1999. pp. 1397–1401. [Google Scholar]
- 5.Gupta S. Curr Opin Microbiol. 2005;8:428–433. doi: 10.1016/j.mib.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Turner CMR. Parasitology. 2002;125:S17–S23. doi: 10.1017/s0031182002002470. [DOI] [PubMed] [Google Scholar]
- 7.Jançso N. Zentbl Bakt Orig. 1918;81:457–474. [Google Scholar]
- 8.Meleney HE. J Exp Med. 1928;48:65–82. doi: 10.1084/jem.48.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell H. Trans R Soc Trop Med Hyg. 1936;30:179–190. [Google Scholar]
- 10.Vickerman K. Parasitology. 1989;99:S37–S47. doi: 10.1017/s0031182000083402. [DOI] [PubMed] [Google Scholar]
- 11.Barry JD, McCulloch R. Adv Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- 12.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su X, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JV, Peterson DS, Ravetch JVW. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 14.Deitsch KW, Pinal AD, Wellems TE. Mol Biochem Parasitol. 1999;101:107–116. doi: 10.1016/s0166-6851(99)00062-6. [DOI] [PubMed] [Google Scholar]
- 15.Al-Khedery B, Allred DR. Mol Microbiol. 2006;59:402–414. doi: 10.1111/j.1365-2958.2005.04993.x. [DOI] [PubMed] [Google Scholar]
- 16.Futse JE, Brayton KA, Knowles DP, Jr, Palmer GH. Mol Microbiol. 2005;57:212–221. doi: 10.1111/j.1365-2958.2005.04670.x. [DOI] [PubMed] [Google Scholar]
- 17.Lawrenz MB, Wooten RM, Norris SJ. Infect Immun. 2004;72:6577–6585. doi: 10.1128/IAI.72.11.6577-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbour AG. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: Am Soc Microbiol; 2002. pp. 972–994. [Google Scholar]
- 19.Stoenner HG, Dodd T, Larsen C. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitten T, Barbour AG. Proc Natl Acad Sci USA. 1990;87:6077–6081. doi: 10.1073/pnas.87.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plasterk RH, Simon MI, Barbour AG. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- 22.Restrepo BI, Kitten T, Carter CJ, Infante D, Barbour AG. Mol Microbiol. 1992;6:3299–3311. doi: 10.1111/j.1365-2958.1992.tb02198.x. [DOI] [PubMed] [Google Scholar]
- 23.Dai Q, Restrepo BI, Porcella SF, Raffel SJ, Schwan TG, Barbour AG. Mol Microbiol. 2006;60:1329–1343. doi: 10.1111/j.1365-2958.2006.05177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbour AG. In: Antigenic Variation. Craig A, Scherf A, editors. London: Academic; 2003. pp. 319–356. [Google Scholar]
- 25.Hinnebusch BJ, Barbour AG, Restrepo BI, Schwan TG. Infect Immun. 1998;66:432–440. doi: 10.1128/iai.66.2.432-440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbour AG, Burman N, Carter CJ, Kitten T, Bergström S. Mol Microbiol. 1991;5:489–493. doi: 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 27.Frank SA, Barbour AG. Infect Genet Evol. 2006;6:141–146. doi: 10.1016/j.meegid.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Barry JD, McCulloch R. In: Antigenic Variation. Craig A, Scherf A, editors. London: Academic; 2003. pp. 224–242. [Google Scholar]
- 29.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 30.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 31.Restrepo BI, Barbour AG. Cell. 1994;78:867–876. doi: 10.1016/s0092-8674(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 32.Recker M, Nee S, Bull PC, Kinyanjui S, Marsh K, Newbold C, Gupta S. Nature. 2004;429:555–558. doi: 10.1038/nature02486. [DOI] [PubMed] [Google Scholar]
- 33.Conway C, Proudfoot C, Burton P, Barry JD, McCulloch R. Mol Microbiol. 2002;45:1687–1700. doi: 10.1046/j.1365-2958.2002.03122.x. [DOI] [PubMed] [Google Scholar]
- 34.Borst P. In: Mobile DNA II. Craig N, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: Am Soc Microbiol; 2002. pp. 953–971. [Google Scholar]
- 35.Morrison LJ, Majiwa P, Read AF, Barry JD. Int J Parasitol. 2005;35:961–972. doi: 10.1016/j.ijpara.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Coffey EM, Eveland WC. J Infect Dis. 1967;117:29–34. doi: 10.1093/infdis/117.1.29. [DOI] [PubMed] [Google Scholar]
- 37.Horrocks P, Kyes S, Pinches R, Christodoulou Z, Newbold C. Mol Biochem Parasitol. 2004;134:193–199. doi: 10.1016/j.molbiopara.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Restrepo BI, Carter CJ, Barbour AG. Mol Microbiol. 1994;13:287–299. doi: 10.1111/j.1365-2958.1994.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 39.Mantel N. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.