Abstract
Streptococcus suis is an important pathogen of swine, causing meningitis, arthritis, polyserositis, septicemia, and sudden death in weaning piglets as well as fattening pigs. Recently, 3 molecular tests have been developed in our laboratory: a multiplex polymerase chain reaction (m-PCR) assay for the detection of S. suis species and serotypes 2 and 1/2, and 2 molecular typing methods, pulsed-field gel electrophoresis and an approach based on PCR amplification of a fragment of rRNA genes, including a part of the 16S and 23S genes and the 16S–23S rDNA intergenic spacer region (ISR), followed by restriction fragment length polymorphism (RFLP) analysis (ISR-RFLP). In the present study, we used these tests to analyze tonsil samples from clinically healthy pigs and to identify individual isolates of S. suis during epidemiologic investigations of 8 related herds with a history of septicemia caused by S. suis serotype 2. Capsular typing showed that 58% of the strains were nontypable. Of the 17 serotypes present, serotype 22 was the most prevalent. In the 7 farms without clinical signs on the day of sampling, we detected S. suis serotype 2 or 1/2, or both, in less than 5% of the pigs by m-PCR or by bacteriologic culture. In the 8th farm, on which 2 pigs had clinical signs of septicemia on the day of sampling, we detected S. suis serotype 2 or 1/2, or both, by m-PCR in the tonsils of 40% of fattening pigs (21 wk old) that lacked symptoms. Molecular typing of the serotype 2 strains showed a common origin of contamination in these herds, given that 1 pattern (C1) was detected in the isolates from 6 of the 8 herds. However, up to 4 patterns were associated with septicemia and sudden death. Several patterns of S. suis serotype 2 can be responsible for disease in the same herd. These molecular tools may be useful for confident studies of the transmission of S. suis, thereby contributing to the control of S. suis infection.
Résumé
Streptococcus suis est un agent pathogène important du porc, responsable de pathologies sévères : méningite, polysérosite, septicémie, mortalité brutale. Récemment, trois méthodes moléculaires ont été développées, une réaction d’amplification en chaîne par la polymérase multiplexe (PCR-m) détectant à la fois l’espèce S. suis et les sérotypes 2 et 1/2 ainsi que deux techniques de typage moléculaire, l’électrophorèse en champs pulsés et l’amplification par PCR d’un fragment des gènes de l’ARNr suivie d’une analyse par RFLP (ISR-RFLP). Ces tests ont été appliqués pour analyser des prélèvements d’amygdales de porcs cliniquement sains et pour différencier les isolats de S. suis. Cette étude a été réalisée dans 8 élevages, épidémiologiquement liés et contaminés par S. suis sérotype 2. La sérotypie a montré que 58 % des souches étaient non sérotypables. Parmi les 17 sérotypes détectés, le sérotype 22 était le plus fréquent. Dans 7 élevages, dont les animaux ne présentaient pas de symptôme le jour du prélèvement, les sérotypes 2 et/ou 1/2 ont été détectés, par PCR-m et culture bactérienne, dans moins de 5 % des amygdales. Dans le huitième élevage, hébergeant des porcs souffrant de septicémie le jour du prélèvement, 40 % des animaux contrôlés en fin d’engraissement (21 semaines d’âge) et ne présentant pas de symptôme étaient porteurs de S. suis sérotype 2 et/ou 1/2 au niveau des amygdales. Le typage moléculaire a mis en évidence l’existence d’une origine de contamination commune entre les élevages puisque un profil moléculaire majoritaire (C1) a été détecté dans 6 élevages sur 8. Cependant, quatre profils différents ont été associés à une septicémie et/ou une mortalité subite. Plusieurs profils de S. suis sérotype 2 peuvent être responsables de pathologie dans un même élevage. Ces techniques peuvent être utilisées, avec confiance, lors d’études de transmission de S. suis et peuvent contribuer au contrôle de l’infection.
(Traduit par les auteurs)
Introduction
Streptococcus suis is well recognized worldwide as a swine pathogen of emerging clinical significance in most countries with an intensive swine industry. It is a major pathogen in pigs, causing loss to the swine industry of more than $300 million dollars annually in the United States (1). This pathogen is associated with a range of diseases in pigs, including meningitis, arthritis, pericarditis, polyserositis, septicemia, pneumonia, and sudden death (1). Occasionally, S. suis causes serious zoonotic infections in humans, where it has been associated with septicemia, meningitis, and endocarditis (2–4). Recently, an outbreak in humans of disease due to S. suis serotype 2 related to diseased pigs was reported from China (5,6).
Although 35 capsular types of S. suis (types 1 to 34 and type 1/2) have been described (1), analysis of the sequences of their 16S ribosomal RNA (rRNA) and cpn60 genes has shown that serotypes 32 and 34 are distinct from the other S. suis serotypes and cluster with S. orisratii (7). Virulence differs among and within the serotypes of S. suis. Worldwide, serotype 2 is the S. suis serotype most frequently isolated from diseased pigs, but other serotypes have also been associated with disease in pigs (1). In France, serotype 2 is the most prevalent capsular type, followed by serotypes 1/2, 9, 7, and 3 (8).
Clinically healthy pigs can carry S. suis in their nasal cavities, tonsils, and upper respiratory tract, contributing to the dissemination of this pathogen (9), and S. suis may also colonize the genital and alimentary tracts of pigs (10). Transmission of the infection between herds usually occurs by the movement of healthy carrier pigs (breeding gilts, boars, and weaners). Detection of clinically healthy carrier pigs, which have an essential role in S. suis transmission, requires the use of powerful new molecular techniques. The main difficulty with bacteriologic isolation is to locate S. suis colonies in multi-infected samples, such as tonsils, specimens that are essential for detection of S. suis in live pigs. Several monoplex and multiplex polymerase chain reaction (PCR) tests have been developed that allow the detection of S. suis species and, more specifically, serotypes 2 (and 1/2), 1 (and 14), 7, and 9 (11–15). Other molecular techniques are now available to compare different S. suis strains within the same serotype, including ribotyping (16–21), arbitrarily primed PCR (22–25), multilocus sequence typing (26), and pulsed-field gel electrophoresis (PFGE) (27–29). These genetic tools can be valuable for distinguishing individual isolates of S. suis to establish the origin of the infection in a herd and to monitor the kinetics of an outbreak. Recently, we developed an approach that is based on PCR amplification of a fragment of rRNA genes, including a part of the 16S and 23S genes and the 16S–23S rDNA intergenic spacer region (ISR), followed by restriction fragment length polymorphism (RFLP) analysis with RsaI and MboII endonucleases (30). The ISR-RFLP method is fast and simple, advantages conferred by the PCR procedure, and has a discriminatory power greater than 0.95 and an in vitro reproducibility of 100%.
To improve epidemiologic knowledge of S. suis species and serotype 2 infection on 8 pig farms in France, we conducted the present study, which had 2 objectives: to detect S. suis in tonsils and environmental samples by bacteriologic isolation and by the use of a multiplex PCR (m-PCR) and to differentiate the isolates by means of PFGE and ISR-RFLP.
Materials and methods
Herds
We investigated 8 pig herds (breeders) with 108 to 500 sows, which originated from the same nucleus farm and had a history of clinical signs due to S. suis serotype 2. On 7 farms there had been episodes of S. suis septicemia 6 mo before the visit, and on 1 farm (farm 5) there were 2 cases of S. suis serotype 2 septicemia and sudden death on the day of the visit. There was a particular pattern of expression of S. suis infection in these 8 herds: septicemia and death occurred only during the fattening period. Autogenous bacterins were used routinely on farms 2, 3, 7, and 8. Farm 5 used an autogenous vaccine after our visit.
Samples
During a single visit to each farm, the pigs were examined for clinical signs, and samples were obtained by biopsy and by swabbing the entire surface of the tonsils of 60 animals that did not show clinical signs and were not receiving antibiotic treatment: 15 sows (five 1st- and 2nd-parity sows, five 3rd- and 4th-parity sows, and five older sows with 5 or more parities), 15 pigs in the postweaning section, and 30 pigs in the growing-finishing section (15 pigs aged 14 to 16 wk and 15 aged 20 to 22 wk of age, nearing the end of the finishing phase). Blood samples were collected for bacteriologic analysis. On farm 5, blood samples were collected for S. suis isolation from the 2 pigs that died suddenly (out of 96 pigs in the fattening room). Environmental samples taken in the gestation, postweaning, and growing-finishing sections included samples of feces, feed, drinking water, flies, dust, breeding material, and clothing, as well as drag-swabs rubbed on the air system. All samples were placed in 2 mL of sterile water supplemented with NaCl (8.5 g/L) (initial suspension [IS]), as previously described (15).
Multiplex PCR and bacteriologic analyses
Each sample (or IS) was analyzed by m-PCR without culture and by bacteriologic culture. The m-PCR test is based on amplification of the gene coding for 16S rRNA of S. suis species and on amplification of the cps2J gene, which is involved in the synthesis of the capsules of S. suis serotypes 2 and 1/2. Direct DNA extraction and preparation from biologic and environmental samples and m-PCR conditions were as described previously (15).
For bacteriologic analyses, 10 μL of each sample was placed on selective Columbia medium supplemented with 5% sheep blood, 15 mg/L of nalidixic acid, and 10 mg/L of colistin. Next, the plates were incubated overnight at 37°C in 5% CO2. Four S. suis-like colonies were subcultivated on Columbia medium supplemented with 5% sheep blood, and identification was confirmed by m-PCR. One positive colony per pig was serotyped by the coagglutination test, with the use of 35 different type-specific hyperimmune sera (31).
Bacterial strains
We compared the strains isolated in this study from the tonsils of clinically healthy pigs and from the blood samples of the 2 pigs on farm 5 that died suddenly with 17 field strains of S. suis sero-type 2 that had been isolated in connection with routine diagnostic testing of blood from pigs with septicemia on farms 3 (7 strains), 5 (3 strains), 6 (2 strains), 7 (3 strains), and 8 (2 strains). Of the latter 17 strains, 7 had been used to produce autogenous vaccines.
Molecular typing by PFGE and ISR-RFLP
Molecular typing of the S. suis isolates was performed by PFGE as previously described (27) and by ISR-RFLP (27,30). Briefly, the mixture used in the ISR-RFLP method contained PCR buffer (67 mM Tris-HCl, 16 mM (NH4)2SO4, 0.01% Tween 20, and 2.5 mM MgCl2 [pH 8.8]), 1.5 mM of each deoxyribonucleoside triphosphate (Eurobio, Les Ulis, France), 400 nM of each primer, 2 units of Taq DNA polymerase (Eurobio), and 5 μL of the DNA template in a total volume of 50 μL. Amplification was performed in a GeneAmp PCR system 9600 (Applied Biosystems, Courtaboeuf, France). The reaction procedure consisted of 40 cycles of amplification at 94°C for 30 s, 60°C for 30 s, and 72°C for 5 min and a postelongation at 72°C for 5 min. Next, 15 μL of the products (approximately 1714 pb) was digested with either RsaI or MboII as described by the manufacturers (Amersham France, Les Ulis, France, and Roche Diagnostics, Meylan, France). Products digested with RsaI were separated in a 2.5% low-melting-point agarose gel in TBE buffer (90 mM Tris, 90 mM borate, and 2.5 mM ethylenediamine tetraacetic acid [pH 8]) for 2.5 h at a constant voltage of 125 V. Products digested with MboII were separated in a 2% standard agarose gel in TBE buffer for 2 h at a constant voltage of 125 V. Patterns were detected by ultraviolet transillumination after ethidium bromide staining. A “ladder” of 50 base pairs (Pfizer, Paris, France) was used as a molecular size standard. The patterns were digitized and analyzed with use of the Biogene package (Vilber-Lourmat, Marne la Vallée, France) as previously described (27). The strains were compared according to “combined” patterns (named C1 to C13) based on the results of both PFGE and ISR-RFLP.
Statistical analysis
Laboratory results obtained for each farm were compared by the Fisher exact test (n ≤ 5) or the chi-squared test (n > 5) of independence in 2 × 2 tables. Statistical tests were performed with Systat 9.0 for Windows (Systat Software GmbH, Erkrath, Germany), and differences were considered significant when the probability (P-value) was less than 0.05.
Results
Detection of S. suis in blood
All blood samples collected from clinically healthy pigs on the 8 farms tested negative by m-PCR and by bacteriologic analysis. Strains 331 and 332 of S. suis serotype 2 were isolated from the 2 pigs on farm 5 that died suddenly on the day of the visit.
Detection of S. suis in tonsils of clinically healthy pigs
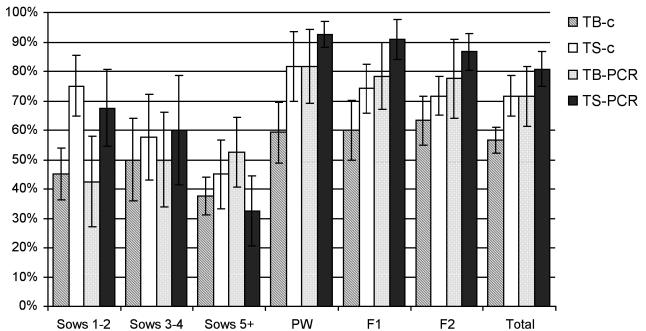
Both m-PCR and bacteriologic culture detected S. suis on all 8 farms, but with variable frequency (Figure 1). On average, 71% of the tonsil biopsy samples and 81% of the tonsil swab samples from clinically healthy pigs were m-PCR positive, whereas 57% of the tonsil biopsy samples and 72% of the tonsil swab samples yielded S. suis on culture. The difference between the 2 tests and between the 2 methods of sampling was significant (P < 0.05). The prevalence of S. suis positivity was significantly higher (P < 0.05) in the pigs or young sows than in the older sows (parity ≥ 5).
Figure 1.
Proportions of clinically healthy pigs positive for Streptococcus suis in tonsil biopsy (TB) and tonsil swab (TS) samples by culture (c) and multiplex polymerase chain reaction (PCR) assay. Sows 1–2 — 1st- and 2nd-parity sows (n = 40); Sows 3–4 — 3rd- and 4th-parity sows (n = 40); Sows 5+ — sows with 5 or more parities (n = 40); PW — pigs in the postweaning section (n = 120); F1 — fattening pigs aged 14 to 16 wk (n = 120); F2 — fattening pigs aged 20 to 22 wk (n = 120).
Of the 406 S. suis isolates recovered from the 8 herds, 90 were from sows, 111 from weaned pigs, and 205 from fattening pigs (102 aged 14 to 16 wk and 103 aged 20 to 22 wk). Nontypable strains accounted for 237 (58%) of the 406 strains. Of the 17 serotypes of S. suis identified in the tonsil samples (Table I), serotype 22 was the most frequent, accounting for 14%, followed by serotypes 5, 11, 2, and 27, at approximately 3% each.
Table I.
Distribution of Streptococcus suis serotypes isolated from tonsils in 8 herds of pigs
| Fattening pigs; age (wk)
|
|||||
|---|---|---|---|---|---|
| Serotype | Sows (n = 90) | Weaned pigs (n = 111) | 14 to 16 (n = 102) | 20 to 22 (n = 103) | All pigs tested (n = 406) |
| 2 | 0 | 0 | 3 | 9 | 12 |
| 1/2 | 2 | 0 | 4 | 0 | 6 |
| 3 | 0 | 0 | 1 | 0 | 1 |
| 4 | 1 | 2 | 0 | 1 | 4 |
| 5 | 4 | 4 | 6 | 0 | 14 |
| 7 | 0 | 0 | 1 | 1 | 2 |
| 8 | 2 | 5 | 0 | 0 | 7 |
| 11 | 4 | 3 | 4 | 2 | 13 |
| 16 | 1 | 5 | 3 | 0 | 9 |
| 18 | 1 | 0 | 3 | 2 | 6 |
| 22 | 6 | 14 | 19 | 18 | 57 |
| 27 | 10 | 0 | 1 | 1 | 12 |
| 28 | 0 | 5 | 0 | 2 | 7 |
| 29 | 3 | 4 | 1 | 1 | 9 |
| 30 | 1 | 1 | 0 | 0 | 2 |
| 31 | 0 | 0 | 1 | 0 | 1 |
| 34 | 0 | 0 | 1 | 0 | 1 |
| Autoagglutinable | 1 | 4 | 0 | 1 | 6 |
| Nontypable | 54 | 64 | 54 | 65 | 237 |
Detection of S. suis serotypes 2 and 1/2 in tonsils of clinically healthy pigs
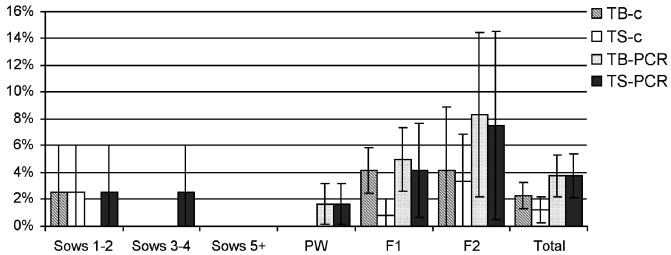
The proportions of clinically healthy pigs on the 8 farms whose tonsil samples tested positive for S. suis serotype 2 or 1/2, or both, by m-PCR and bacteriologic analysis are shown in Figure 2. In total, 15 strains were isolated (on farms 1, 2, 3, 4, 5, and 7), serotype 2 or 1/2, or both, being detected in 2.3% of the tonsil biopsy samples and in 1.25% of the tonsil swab samples by bacteriologic analysis and in less than 4% of the pigs by m-PCR. The difference in results obtained with the 2 techniques was significant (P < 0.05), but there were no significant differences among the sampling sites.
Figure 2.
Percentages of clinically healthy pigs positive for S. suis serotypes 2 and 1/2. Abbreviations as for Figure 1.
Weaned pigs had significantly higher rates of infection with S. suis serotype 2 or 1/2, or both, than did sows (P < 0.05), and older pigs in the fattening section had significantly higher rates of infection with 1 or both of these serotypes than did younger pigs (P < 0.05). On the 7 farms with chronic S. suis infection (all but farm 5), less than 5% of the tonsil samples were positive for S. suis serotype 2 or 1/2, or both. However, on farm 5, where pigs showed clinical signs on the day of the visit, the carrier prevalence of the 2 serotypes together was 40% in the pigs 20 to 22 wk old.
Detection of S. suis in environmental samples
Neither PCR nor culture demonstrated S. suis serotypes 2 and 1/2 in the environment of the pigs. However, 2 nontypable S. suis strains were recovered from 2 drinking water samples in the postweaning section of farm 1. These 2 isolates had an identical biochemical profile (in the Api20 Strep system) that was different from that classically obtained from S. suis strains isolated in disease cases; however, 16S rDNA sequencing confirmed that they were members of the species S. suis (99% identity; data not shown).
Using m-PCR, we recovered S. suis DNA (different from that of serotypes 2 and 1/2) from at least 1 environmental sample from each farm (Table II). In total, 58 of the 354 samples tested positive; S. suis DNA was recovered from feed, drinking water, feces, litter, and dust, as well as some flies (Musca domestica) captured on farm 5. On farm 5, S. suis DNA was present in 14 of 45 samples. The PCR results for this farm were significantly different from the PCR results for farms 1, 7, and 8 (P < 0.05).
Table II.
Results of multiplex polymerase chain reaction (m-PCR) testing for S. suis species in the environmental samples collected at the 8 farms
| Farm number | Pig group | n | PCR+ | Sample source (and number) |
|---|---|---|---|---|
| 1 | S | 12 | 0 | |
| WP | 15 | 2 | Drinking water (2) | |
| FP | 17 | 0 | ||
| Total | 44 | 2 | ||
| 2 | S | 16 | 2 | Feeding trough (2) |
| WP | 19 | 7 | Feed (1), drinking water (2), feces (1), dust (1), feeding trough (2) | |
| FP | 18 | 2 | Feeding trough (2) | |
| Total | 53 | 11 | ||
| 3 | S | 12 | 3 | Dust (1), feeding trough (2) |
| WP | 12 | 5 | Feed (2), feces (1), feeding trough (2) | |
| FP | 18 | 1 | Feeding trough (1) | |
| Total | 42 | 9 | ||
| 4 | S | 12 | 2 | Feed (2) |
| WP | 19 | 1 | Drinking water (1) | |
| FP | 13 | 5 | Feed (1), litter (2), dust (1), clothes drag-swab (1) | |
| Total | 44 | 8 | ||
| 5 | S | 16 | 6 | Feed (1), feeding trough (2), fly (3) |
| WP | 15 | 8 | Drinking water (3), dust (3), feeding trough (2) | |
| FP | 14 | 0 | ||
| Total | 45 | 14 | ||
| 6 | S | 12 | 6 | Feed (2), feces (1), feeding trough (2), drag-swab on sow (1) |
| WP | 12 | 0 | ||
| FP | 21 | 3 | Drinking water (3) | |
| Total | 45 | 9 | ||
| 7 | S | 12 | 0 | |
| WP | 13 | 1 | Feed (1) | |
| FP | 15 | 0 | ||
| Total | 40 | 1 | ||
| 8 | S | 14 | 4 | Feed (1), feeding trough (3) |
| WP | 13 | 0 | ||
| FP | 14 | 0 | ||
| Total | 41 | 4 | ||
| Total | 354 | 58 |
S — sows; WP — weaned pigs; FP — fattening pigs; n — number of samples analyzed; PCR+ — number of samples positive for S. suis species by m-PCR.
Genotyping of S. suis serotype 2 and 1/2
Using PFGE with SmaI endonuclease and the ISR-RFLP method with RsaI and MboII endonucleases, we characterized 34 strains of S. suis serotypes 2 and 1/2 and the 2 nontypable strains isolated from the environment (Table III). Among the 34 strains of serotypes 2 and 1/2, 3 PFGE patterns were identified. With the ISR-RFLP method, 8 and 5 patterns were detected with the use of RsaI and MboII, respectively.
Table III.
Genetic diversity of 36 S. suis strains isolated from the 8 farms
| Farm number | Strain | Serotypea | Origin | Used in autogenous vaccine? | PFGE pattern | RsaI pattern | MboII pattern | Combined patternb |
|---|---|---|---|---|---|---|---|---|
| 1 | 347 | 2 | T | No | P3 | R31 | M15 | C2 |
| 348 | 1/2 | T | No | P3 | R31 | M15 | C2 | |
| 349 | 2 | T | No | P3 | R31 | M15 | C2 | |
| 350 | 2 | T | No | P3 | R31 | M15 | C2 | |
| 351 | NT | E | No | P5 | R43 | M6 | C3 | |
| 352 | NT | E | No | P5 | R43 | M6 | C3 | |
| 2 | 353 | 1/2 | T | No | P1 | R24 | M8 | C1 |
| 3 | 311 | 2 | S | No | P2 | R27 | M10 | C4 |
| 330 | 2 | S | No | P1 | R24 | M8 | C1 | |
| 336 | 2 | S | No | P1 | R28 | M8 | C5 | |
| 337 | 2 | S | No | P1 | R24 | M7 | C6 | |
| 338 | 2 | S | No | P1 | R24 | M8 | C1 | |
| 340 | 2 | S | Yes | P1 | R24 | M8 | C1 | |
| 341 | 2 | S | Yes | P1 | R24 | M8 | C1 | |
| 354 | 2 | T | No | P1 | R24 | M8 | C1 | |
| 4 | 355 | 1/2 | T | No | P1 | R10 | M8 | C7 |
| 5 | 289 | 2 | S | No | P1 | R24 | M8 | C1 |
| 326 | 2 | S | No | P1 | R24 | M8 | C1 | |
| 331c | 2 | S | No | P1 | R24 | M8 | C1 | |
| 332c | 2 | S | No | P1 | R24 | M8 | C1 | |
| 342 | 2 | S | Yes | P1 | R38 | M10 | C8 | |
| 356 | 2 | T | No | P1 | R24 | M8 | C1 | |
| 357 | 2 | T | No | P1 | R5 | M8 | C9 | |
| 358 | 2 | T | No | P1 | R16 | M8 | C10 | |
| 359 | 2 | T | No | P1 | R24 | M8 | C1 | |
| 360 | 2 | T | No | P1 | R24 | M8 | C1 | |
| 361 | 2 | T | No | P1 | R24 | M8 | C1 | |
| 362 | 2 | T | No | P1 | R24 | M8 | C1 | |
| 6 | 320 | 2 | S | No | P1 | R24 | M8 | C1 |
| 345 | 2 | S | Yes | P1 | R24 | M8 | C1 | |
| 7 | 288 | 2 | S | No | P2 | R38 | M10 | C11 |
| 343 | 2 | S | Yes | P2 | R10 | M11 | C12 | |
| 344 | 2 | S | Yes | P1 | R24 | M8 | C1 | |
| 363 | 2 | T | No | P1 | R24 | M8 | C1 | |
| 8 | 297 | 2 | S | No | P1 | R24 | M8 | C1 |
| 346 | 2 | S | Yes | P1 | R24 | M8 | C1 |
PFGE — pulsed-field gel electrophoresis; T — tonsil; NT — nontypable; E — environment; S — septicemia.
Strains 331 and 332, as well as the tonsil and environment isolates, were collected in the current study; the others strains were collected from earlier outbreaks.
Obtained from the combined results with PFGE and ISR-RFLP (an approach based on PCR amplification of a fragment of rRNA genes, including a part of the 16S and 23S genes and the 16S–23S rDNA intergenic spacer region [ISR], followed by restriction fragment length polymorphism).
These strains were isolated from blood samples of 2 pigs that died suddenly on the day of the visit to farm 5.
Ten “combined” patterns were identified among the 31 S. suis isolates of serotype 2. The most common pattern, C1, was detected on 6 farms (2, 3, 5, 6, 7, and 8) in 13 strains isolated from pigs with septicemia and 7 tonsil samples from clinically healthy pigs. The C1 pattern was also detected in 1 S. suis strain of serotype 1/2. The 2 nontypable strains, isolated from environmental samples, had a singular pattern (C3). Two strains from farms 5 and 7 used to produce autogenous vaccines did not have the C1 pattern.
Discussion
In this study of S. suis typing among 8 herds, m-PCR was more sensitive than bacteriologic culture. With multi-infected samples, such as tonsil specimens, PCR facilitates detection of S. suis. The PCR assay that we used detected and identified S. suis serotypes 2 and 1/2 among α-hemolytic colonies on blood agar medium (15). The high sensitivity and specificity of m-PCR, as well as the presence of an internal control, allow direct analysis of samples without a culture step. The use of molecular biology techniques is essential for identification of carrier pigs, which play an important epidemiologic role in S. suis infection.
We also found that tonsil swabs were more sensitive than tonsil biopsy for the detection of S. suis in live sows and growing pigs. Tonsil swabbing is easier to perform and is not traumatic for the pig. These results confirm those observed in experimentally infected pigs (15). Recently, tonsil swabs were used to detect S. suis serotype 2 in live sows (32).
In the 8 herds, S. suis was detected by m-PCR in 81% of the tonsil swabs on average; 58% of the strains were nontypable. The high prevalence of nontypable S. suis strains isolated from tonsil samples was previously reported by Han and colleagues (33), who found 26 strains to be nontypable among 55 isolates recovered from slaughter pigs in Korea. In Canada, Brisebois and associated (34) showed that 79% of strains isolated from nasal cavities on 49 farms were nontypable. These results suggest the existence of nonencapsulated S. suis or more than 35 serotypes. In our study, 17 S. suis serotypes were detected; serotype 22 was the most prevalent, and serotypes 5, 11, 2, and 27 next most prevalent. Serotype 2 is generally associated with severe lesions, although some strains, belonging to less common capsular types, such as serotypes 22 and 5, have been associated with septicemia, meningitis, and sudden death (10,25,35–37). Serotype 34 (or S. orisratti) was recovered from 1 pig (Table I).
In the 7 herds with a history of S. suis serotype 2 infection but no clinical signs on the day of investigation, we detected serotype 2 or 1/2, or both, in less than 5% of the tonsil samples. In the 8th herd, in which pigs had clinical signs of disease on the day of sampling, the carriage rate of these serotypes was 40% in the older fattening pigs (20 to 22 wk old). In the 8 herds, disease was observed in the fattening section. However, S. suis serotypes 2 and 1/2 were also detected in tonsil samples from weaned pigs in 4 herds and from sows in 2 herds. These results are in agreement with previous findings that sows presumably infect their piglets during farrowing or via the respiratory route after farrowing, or both (38). Since S. suis is also found in the genital and digestive tracts, piglets may be exposed during the birth process and suckling (1,10,38–40).
Several serotypes of S. suis (though not serotypes 2 and 1/2) were detected in the environment in 58 of 354 samples in this study. The presence of S. suis in the environment is probably transitory (10). Nevertheless, S. suis serotype 2 has been shown to survive in feces and dust at 25°C for 8 d and 24 h, respectively, and for 10 and 25 d at 9°C (41). Moreover, S. suis serotype 2 can be isolated from a variety of animal species, especially mammalians, birds, and flies, which may play an epidemiologic role in S. suis infection. Growing pigs are generally susceptible. Depending on different factors, such as the virulence of the strain, coexisting infection, environmental factors affecting bacterial survival, and moving and mixing of animals, transmission of S. suis may occur. It is also known that S. suis serotype 2 can be transmitted from diseased pigs to healthy pigs through direct or indirect contact (1,10,42).
Our results confirm that serotyping is not sufficiently discriminating to differentiate S. suis strains (27,30). The PFGE and ISR-RFLP methods have been shown to be more useful in genotyping S. suis serotype 2 strains. Moreover, using these molecular methods, we found that 56% of S. suis serotype 2 isolates had a common pattern (C1), which was detected in 6 of the 8 farms. This finding suggests that many isolates had a common origin of contamination. Isolates with the C1 pattern were also detected in the nucleus herd, suggesting transmission via the gilts (data not shown). In addition, molecular typing of 17 strains, isolated from 13 herds of commercial farms that were related to the 8 farms studied here, revealed that S. suis with the C1 pattern was the most prevalent strain (data not shown). Therefore, vertical transmission (nucleus herd to multiplier herds to commercial farms) can be considered. Nevertheless, 10 patterns were also detected in serotype 2, suggesting a multifactorial origin of contamination.
Surprisingly, up to 4 genotypic patterns of serotype 2 were observed for each farm, and up to 4 profiles were associated with septicemia. In this regard, the molecular epidemiologic features of the infection appear to be different from those described classically for serotype 2. Indeed, it was previously reported that in closed infected herds a single clone of S. suis was responsible for disease (23,35).
Therefore, a common autogenous vaccine prepared with a C1-pattern strain should be used with caution in a vaccination program. Two strains isolated from diseased pigs on 2 farms were different from the strain used to prepare the autogenous vaccine used on those farms. Although a clear correlation between different genotypic patterns and antigens has not been demonstrated, the presence of multiple strains of S. suis serotype 2 in a single herd might result in vaccination failures.
In conclusion, different strains of S. suis serotype 2 may be present in a herd and responsible for clinical signs. Therefore, genotypic characterization (with the use of PFGE, ISR-RFLP, or other molecular typing methods) is essential to better understand S. suis infection by identifying individual isolates of S. suis, establishing the origin of the infection, and monitoring the kinetics of an outbreak.
Acknowledgments
We thank Thierry Ogel for skilled technical assistance, the farmers and the production companies for their cooperation in this project, and Christelle Fablet and Anne Gautier-Bouchardon for critical reading of the manuscript.
References
- 1.Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21:381–407. doi: 10.1023/a:1005870317757. [DOI] [PubMed] [Google Scholar]
- 2.Kopic J, Paradzik MT, Pandak N. Streptococcus suis infection, a zoonosis we should have in mind — 2 case reports. Lijec Vjesn. 2003;125:134–137. [PubMed] [Google Scholar]
- 3.Lopreto C, Lopardo HA, Bardi MC, Gottschalk M. [Primary Streptococcus suis meningitis: first case in humans described in Latin America. ] Enferm Infecc Microbiol Clin. 2005;23:110. doi: 10.1157/13071618. [DOI] [PubMed] [Google Scholar]
- 4.Mazokopakis EE, Kofteridis DP, Papadakis JA, Gikas AH, Samonis GJ. First case report of Streptococcus suis septicemia and meningitis from Greece. Eur J Neurol. 2005;12:487–489. doi: 10.1111/j.1468-1331.2005.00998.x. [DOI] [PubMed] [Google Scholar]
- 5.Huang YT, Teng LJ, Ho SW, Hsueh PR. Streptococcus suis infection. J Microbiol Immunol Infect. 2005;38:306–313. [PubMed] [Google Scholar]
- 6.Normile D. Infectious diseases. WHO probes deadliness of China’s pig-borne disease. Science. 2005;309:1308–1309. doi: 10.1126/science.309.5739.1308a. [DOI] [PubMed] [Google Scholar]
- 7.Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Han Goh S. Biochemical analysis, cpn60 and 16S rRNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet Microbiol. 2005;107:63–69. doi: 10.1016/j.vetmic.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Berthelot-Hérault F, Morvan H, Kéribin AM, Gottschalk M, Kobisch M. Production of muramidase released protein (MRP), extracellular factor (EF) and haemolysin by field isolates of Streptococcus suis capsular type 2, 1/2, 9, 7 and 3 isolated from swine in France. Vet Res. 2000;31:473–479. doi: 10.1051/vetres:2000133. [DOI] [PubMed] [Google Scholar]
- 9.Mwaniki CG, Robertson ID, Trott DJ, Atyeo RF, Lee BJ, Hampson DJ. Clonal analysis and virulence of Australian isolates of Streptococcus suis type 2. Epidemiol Infect. 1994;113:321–334. doi: 10.1017/s095026880005175x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins R, Gottschalk M. Streptococcal diseases. In: Straw BE, D’Allaire S, Mengeling WL, Taylor DJ, eds. Diseases of Swine. 8th ed. Ames, Iowa: Iowa State University Press, 1999:563–578.
- 11.Smith HE, van Bruijnsvoort L, Buijs H, Wisselink HJ, Smits MA. Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol Lett. 1999;178:265–270. doi: 10.1111/j.1574-6968.1999.tb08686.x. [DOI] [PubMed] [Google Scholar]
- 12.Smith HE, Veenbergen V, van der Velde J, Damman M, Wisselink HJ, Smits MA. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J Clin Microbiol. 1999;37:3146–3152. doi: 10.1128/jcm.37.10.3146-3152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wisselink HJ, Joosten JJ, Smith HE. Multiplex PCR assays for simultaneous detection of 6 major serotypes and 2 virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J Clin Microbiol. 2002;40:2922–2929. doi: 10.1128/JCM.40.8.2922-2929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okwumabua O, O’Connor M, Shull E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol Lett. 2003;218:79–84. doi: 10.1111/j.1574-6968.2003.tb11501.x. [DOI] [PubMed] [Google Scholar]
- 15.Marois C, Bougeard S, Gottschalk M, Kobisch M. Multiplex PCR assay for detection of Streptococcus suis species and sero-types 2 and 1/2 in tonsils of live and dead pigs. J Clin Microbiol. 2004;42:3169–3175. doi: 10.1128/JCM.42.7.3169-3175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaudoin M, Harel J, Higgins R, Gottschalk M, Frenette M, MacInnes JI. Molecular analysis of isolates of Streptococcus suis type 2 by restriction-endonuclease-digested DNA separated on SDS-PAGE and by hybridization with an rDNA probe. J Gen Microbiol. 1992;138:2639–2645. doi: 10.1099/00221287-138-12-2639. [DOI] [PubMed] [Google Scholar]
- 17.Harel J, Higgins R, Gottschalk M, Bigras-Poulin M. Genomic relatedness among reference strains of different Streptococcus suis serotypes. Can J Vet Res. 1994;58:259–262. [PMC free article] [PubMed] [Google Scholar]
- 18.Smith HE, Rijnsburger M, Stockhofe-Zurwieden N, Wisselink HJ, Vecht U, Smits MA. Virulent strains of Streptococcus suis sero-type 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J Clin Microbiol. 1997;35:1049–1053. doi: 10.1128/jcm.35.5.1049-1053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen SR, Aarestrup FM, Jensen NE, Jorsal SE. Associations of Streptococcus suis serotype 2 ribotype profiles with clinical disease and antimicrobial resistance. J Clin Microbiol. 1999;37:404–408. doi: 10.1128/jcm.37.2.404-408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okwumabua O, Staats J, Chengappa MM. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping) J Clin Microbiol. 1995;33:968–972. doi: 10.1128/jcm.33.4.968-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarradas C, Luque I, de Andres D, et al. Epidemiological relationship of human and swine Streptococcus suis isolates. J Vet Med B Infect Dis Vet Public Health. 2001;48:347–355. doi: 10.1046/j.1439-0450.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 22.Chatellier S, Gottschalk M, Higgins R, Brousseau R, Harel J. Relatedness of Streptococcus suis serotype 2 isolates from different geographic origins as evaluated by molecular fingerprinting and phenotyping. J Clin Microbiol. 1999;37:362–366. doi: 10.1128/jcm.37.2.362-366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez G, Harel J, Lacouture S, Gottschalk M. Genetic diversity of Streptococcus suis serotypes 2 and 1/2 isolates recovered from carrier pigs in closed herds. Can J Vet Res. 2002;66:240–248. [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez G, Pestana de Castro AF, Ribeiro Pagnani KJ, Nakazato G, Dias da Silveira W, Gottschalk M. Clonal distribution of an atypical MRP+, EF*, and suilysin+ phenotype of virulent Streptococcus suis serotype 2 strains in Brazil. Can J Vet Res. 2003;67:52–55. [PMC free article] [PubMed] [Google Scholar]
- 25.Cloutier G, D’Allaire S, Martinez G, Surprenant C, Lacouture S, Gottschalk M. Epidemiology of Streptococcus suis serotype 5 infection in a pig herd with and without clinical disease. Vet Microbiol. 2003;97:135–151. doi: 10.1016/j.vetmic.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 26.King SJ, Leigh JA, Heath PJ, et al. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulence clones and potential capsular serotype exchange. J Clin Microbiol. 2002;40:3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berthelot-Hérault F, Marois C, Gottschalk M, Kobisch M. Genetic diversity of Streptococcus suis strains isolated from pigs and humans as revealed by pulsed-field gel electrophoresis. J Clin Microbiol. 2002;40:615–619. doi: 10.1128/JCM.40.2.615-619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allgaier A, Goethe R, Wisselink HJ, Smith HE, Valentin-Weigand P. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J Clin Microbiol. 2001;39:445–453. doi: 10.1128/JCM.39.2.445-453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vela AI, Goyache J, Tarradas C, et al. Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J Clin Microbiol. 2003;41:2498–2502. doi: 10.1128/JCM.41.6.2498-2502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marois C, Le Devendec L, Gottschalk M, Kobisch M. Molecular characterization of Streptococcus suis strains by 16S–23S intergenic spacer polymerase chain reaction and restriction fragment length polymorphism analysis. Can J Vet Res. 2006;70:94–104. [PMC free article] [PubMed] [Google Scholar]
- 31.Gottschalk M, Higgins R, Jacques M, Mittal KR, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–2636. doi: 10.1128/jcm.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swildens B, Wisselink HJ, Engel B, et al. Detection of extracellular factor-positive Streptococcus suis serotype 2 strains in tonsillar swabs of live sows by PCR. Vet Microbiol. 2005;109:223–228. doi: 10.1016/j.vetmic.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Han DU, Choi C, Ham HJ, et al. Prevalence, capsular type and antimicrobial susceptibility of Streptococcus suis isolated from slaughter pigs in Korea. Can J Vet Res. 2001;65:151–155. [PMC free article] [PubMed] [Google Scholar]
- 34.Brisebois LM, Charlebois R, Higgins R, Nadeau M. Prevalence of Streptococcus suis in four to eight week old clinically healthy piglets. Can J Vet Res. 1990;54:174–177. [PMC free article] [PubMed] [Google Scholar]
- 35.Reams RY, Harrington DD, Glickman LT, Thacker HL, Bowersock TL. Multiple serotypes and strains of Streptococcus suis in naturally infected swine herds. J Vet Diagn Invest. 1996;8:119–121. doi: 10.1177/104063879600800121. [DOI] [PubMed] [Google Scholar]
- 36.Higgins R, Gottschalk M. Distribution of Streptococcus suis capsular types in 2000. Can Vet J. 2001;42:223. [PMC free article] [PubMed] [Google Scholar]
- 37.Salasia SI, Lammler C. Distribution of serotype, virulence markers and further characteristics of Streptococcus suis isolates from pigs. Zentralbl Veterinarmed B. 1995;42:78–83. doi: 10.1111/j.1439-0450.1995.tb00685.x. [DOI] [PubMed] [Google Scholar]
- 38.Clifton-Hadley FA, Alexander TJ, Enright MR. Monitoring herds for Streptococcus suis type 2: chance contamination of slaughter pigs. Vet Rec. 1986;118:274. doi: 10.1136/vr.118.10.274. [DOI] [PubMed] [Google Scholar]
- 39.Robertson ID, Blackmore DK. Prevalence of Streptococcus suis types 1 and 2 in domestic pigs in Australia and New Zealand. Vet Rec. 1989;124:391–394. doi: 10.1136/vr.124.15.391. [DOI] [PubMed] [Google Scholar]
- 40.Amass SF, SanMiguel P, Clark LK. Demonstration of vertical transmission of Streptococcus suis in swine by genomic fingerprinting. J Clin Microbiol. 1997;35:1595–1596. doi: 10.1128/jcm.35.6.1595-1596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clifton-Hadley FA, Enright MR. Factors affecting the survival of Streptococcus suis type 2. Vet Rec. 1984;114:584–586. doi: 10.1136/vr.114.24.584. [DOI] [PubMed] [Google Scholar]
- 42.Berthelot-Hérault F, Cariolet R, Labbé A, Gottschalk M, Cardinal JY, Kobisch M. Experimental infection of specific pathogen free piglets with French strains of Streptococcus suis capsular type 2. Can J Vet Res. 2001;65:196–200. [PMC free article] [PubMed] [Google Scholar]