Abstract
Immunohistochemical (IHC) testing and electron microscopy have implicated Papillomavirus (PV) as the etiologic agent for equine papillomas and aural plaques, but Equine papillomavirus (EPV) DNA has yet to be demonstrated in these lesions by polymerase chain reaction (PCR). The purpose of this study was to evaluate formalin-fixed, paraffin-embedded tissues from naturally occurring cases of equine papillomas, aural plaques, and sarcoids for the presence of EPV DNA by means of PCR and for the presence of PV antigen by means of IHC testing. We used EPV-specific primers that amplified a region of 384 base pairs (bp) spanning the E4 and L2 genes of the EPV genome and consensus PV primers that amplified a 102-bp region of the L1 gene. Group-specific PV structural antigens were detected with the use of a streptavidin–biotin–alkaline phosphatase IHC stain. With IHC testing, 23 of 38 papillomas, 4 of 9 aural plaques, and 0 of 10 sarcoids were positive for PV antigen; EPV DNA was found in 20 of the 38 papillomas and 1 of the 10 sarcoids but 0 of the 9 aural plaques. The consensus primers did not amplify novel PV DNA in any of the tissues. Nucleotide sequencing of viral DNA from 7 papillomas amplified with EPV-specific primers revealed DNA fragments that were 96% to 99% identical to known EPV sequences. Some samples had nucleotide substitutions in common, which suggests infection with related strains. Together, EPV DNA or PV antigen (or both) was demonstrated in 26 (68%) of the 38 equine papillomas. Although aural plaques contained PV antigen, they were negative for EPV DNA; therefore, we hypothesize that aural plaques contain a PV distinct from EPV.
Résumé
Les épreuves immunohistochimiques (IHC) et la microscopie électronique ont mis en cause le Papillomavirus (PV) comme agent étiologique des papillomes équins et des plaques auriculaires (papillomes de l’oreille), mais l’ADN du papillomavirus équin (EPV) n’a pas encore été mis en évidence par réaction d’amplification en chaîne par la polymérase (PCR) dans ces lésions. Le but de l’étude était d’évaluer la présence d’ADN de l’EPV par épreuve PCR et la présence d’antigène du PV par épreuve IHC dans des tissus fixés à la formaline et paraffinés provenant de cas cliniques de papillomes équins, de plaques auriculaires et de sarcoïdes. Des amorces spécifiques à EPV amplifiant une région de 384 paires de bases (bp) couvrant les gènes E4 et L2 du génome d’EPV et des amorces consensus du PV amplifiant une région de 102 bp du gène L1 ont été utilisées. Les antigènes structuraux spécifiques de groupe du PV ont été détectés en IHC par coloration à la streptavidine–biotine–phosphatase alcaline. Par épreuve IHC, 23 des 38 papillomes, 4 des 9 plaques auriculaires et 0 des 10 sarcoïdes étaient positifs pour la présence d’antigène du PV; l’ADN d’EPV a été trouvé dans 20 des 38 papillomes, 1 des 10 sarcoïdes et 0 des 9 plaques auriculaires. Les amorces consensus n’ont pas amplifiées d’ADN nouveaux de PV dans aucun des tissus. Le séquençage nucléotidique de l’ADN viral amplifié provenant de 7 papillomes avec les amorces spécifiques à EPV a révélé des fragments d’ADN de 96 % à 99 % identiques à des séquences connues d’EPV. Quelques cas avaient des substitutions nucléotidiques en commun, ce qui suggère une infection par des souches reliées. Dans l’ensemble, l’ADN d’EPV ou l’antigène de PV (ou les deux) ont été démontrés dans 26 (68 %) des 38 papillomes équins. Bien que les plaques auriculaires contenaient de l’antigène de PV, elles se sont avérées négatives pour la présence d’ADN d’EPV; ainsi, l’hypothèse est émise que les plaques auriculaires contiennent un PV distinct de l’EPV.
(Traduit par Docteur Serge Messier)
Introduction
Papillomaviruses (PVs) are nonenveloped double-stranded DNA viruses that induce benign proliferative epithelial tumors (papillomas) in a variety of species (1,2). Many PVs that infect animal species, including humans and equines, have been characterized (3). Recently, the complete nucleotide sequence of Equus caballus papillomavirus type 1 (EcPV-1), obtained from an equine cutaneous papilloma, was published (4). Evidence for a causal link between PV and disease is limited. However, it is hypothesized that 2 different types of equine PVs (EPVs) cause viral papillomatosis (warts) and ear papillomas (aural plaques) (5,6), and a 3rd EPV may be associated with penile papillomas (3).
Warts are considered the most common tumor in horses between 1 and 3 y of age (5,7). They affect various cutaneous sites, as well as oral, ocular, and genital mucous membranes (8), and can cause problems because of physical location and esthetics. Spontaneous remission is common within 1 to 9 mo (8). Aural plaques occur in horses of all ages, manifesting as well-demarcated raised, depigmented, hyperkeratotic plaques 1 to 3 cm in diameter on the inner surface of the ear pinnae (9). They rarely, if ever, spontaneously resolve and are susceptible to fly bites and secondary infection.
In warts, PV virions have been observed in nuclei of cells of the stratum granulosum, corneum, and spinosum by electron microscopy (EM) (7), and PV antigen has been identified with immunohistochemical (IHC) techniques (3,10); however, to our knowledge, EPV DNA has not been demonstrated in formalin-fixed, paraffin-embedded (FFPE) tissue with the use of polymerase chain reaction (PCR). Similarly, although viral particles have been observed with EM and PV antigen has been identified by IHC study in aural plaques (11), these lesions have not been investigated for the presence of EPV DNA for definitive identification of the virus.
Sarcoids are common, locally aggressive fibroblastic skin tumors with a high rate of recurrence after surgical removal (12). They may be single or multiple and can occur at any location, although they have a predilection for the head, neck, legs, and ventral body surfaces. They frequently occur at sites of previous injury and scarring (5,13). Some sarcoids resemble equine papillomas; however, the spontaneous regression that is common with equine papillomas is rarely seen with sarcoids (14,15).
Most equine sarcoids examined with the use of molecular hybridization, restriction enzyme analysis, PCR, and in situ hybridization (ISH) have demonstrated Bovine papillomavirus (BPV) DNA sequences (16–18). Equine sarcoids are therefore considered to be the result of a nonproductive BPV infection, the viral DNA persisting episomally (16). However, a direct causal relationship has not been established (19), and involvement of other PVs has not been investigated. Fibromatous lesions exhibiting histologic characteristics similar to those of dermal sarcoids and bovine fibropapillomas were identified in the dermis of 10 spontaneous and experimentally induced equine warts, suggesting that fibropapillomas in horses may be induced by EPV (20).
The purpose of this study was to confirm the etiologic link of EPV with equine papillomas and aural plaques by demonstrating EPV DNA and PV antigen in these lesions with the use of PCR and IHC techniques, respectively. In addition, we wanted to compare the effectiveness of PCR and IHC testing of equine papillomas. Since PV is difficult to culture, definitive identification of PV infection generally relies on techniques that identify viral DNA, such as PCR, rather than on virus isolation. Confirmation of the diagnosis in clinical cases is primarily based on histologic study (5); we therefore used this method as the gold standard. Depending on the quality of biopsy specimens, it can be difficult to differentiate histopathologically between sarcoid, papilloma, fibroma, fibrosarcoma, and other inflammatory lesions (5,21). Demonstration of EPV DNA or PV antigen would help distinguish between lesion types.
Materials and methods
Tissue specimens
We selected for study 58 archived FFPE tissue blocks of equine biopsy specimens with accompanying slides stained with hematoxylin and eosin submitted between 1985 and 2002 from 2 veterinary diagnostic laboratories: Prairie Diagnostic Services (PDS), Saskatoon, Saskatchewan, and Central Laboratory for Veterinarians, Langley, British Columbia. The specimens included 38 papillomas from various anatomic locations (including 2 congenital and 6 genital papillomas), 10 aural plaques, and 10 sarcoids. Sample selection was based on previously described histologic characteristics for papillomas, aural plaques, and sarcoids (9,12,20). As well, 9 normal samples, 5 of skin and 4 of noncutaneous tissues, were randomly selected as controls from specimens from 6 horses with no visible evidence of skin lesions that had been submitted to the PDS postmortem diagnostic service during the time of the study.
Extraction of DNA
We cut 10 sections 10 μm thick from each tissue block. The microtome blade was cleaned with a DNA degradation solution (Dnase Away; Molecular Biologic Products, San Diego, California, USA) between samples to prevent cross-contamination. The DNA was extracted with the use of the Qiagen DNeasy tissue kit (Qiagen, Mississauga, Ontario) in accordance with the manufacturer’s instructions, except that we replaced the suggested 1200 μL of xylene with 1000 μL of xylene and absolute ethanol and repeated the xylene step once. The concentration of extracted DNA was determined by means of a spectrophotometer (Hewlett-Packard [Canada], Edmonton, Alberta) at a wavelength of 260 nm. A negative-extraction control that did not contain tissue was included for every 10 samples extracted to monitor for contamination.
Detection of PV DNA by PCR
To detect PV DNA, we used 2 sets of primers in a PCR assay. One primer set (EPV-F, 5′ -TGC GTT CGC CCC AAT AGT CAT CTT-3′ , at nucleotide positions 3441 to 3464 in the EPV genome; and EPV-R, 5′ -ACC GCC CGC CTC ACC CTT GTC-3′ , at nucleotide positions 3804 to 3824) was designed from EcPV-1 (GenBank accession number NC_003748) with the use of Primer Select 3.10 (DNASTAR, Madison, Wisconsin, USA) and synthesized by a commercial laboratory (Invitrogen Life Technologies, Burlington, Ontario). To ensure the specificity of the primer set for EPV, we compared the primer sequences with the sequences of PVs of other species by means of GenBank’s automated alignment-search program, BLAST (www.ncbi.nlm.nih.gov/BLAST). We found no significant matches, whereas the sequences of the consensus primers matched a variety of PV sequences. To demonstrate that the EPV-F and EPV-R primers were specific for the EPV nucleotide sequence, we performed PCR assays using DNA extracted from a bovine, a feline, and a canine cutaneous papilloma, each positive by IHC testing for PV antigen: no amplifiable DNA was identified in the nonequine papillomas with use of the EPV primer set. The primer set amplified a 384 base pair (bp) fragment of the E4 and L2 genes of the EPV genome. We used DNA extracted from an equine cutaneous papilloma that generated a PCR product having a nucleotide sequence identical to EPV as the positive control. Samples of skin, spleen, liver, kidney, intestine, and lung from horses with no clinically evident skin lesions were used as negative controls. A negative-amplification control containing no DNA was also included with each assay to monitor for contamination.
The amplification reaction mixture consisted of 28.35 μL of sterile ultrapure water, 5.0 μL of 10X PCR buffer without ammonium sulfate (MBI Fermentas, Burlington, Ontario), 7.0 μL of 25-mM magnesium chloride (MBI Fermentas), 0.4 μL of 25-mM deoxyribonucleotide triphosphate (dNTP; MBI Fermentas), 2.0 μL of each primer (25 pmol/μL), 0.25 μL of Taq polymerase (5 U/μL; Invitrogen, Burlington, Ontario), and 5 μL of template DNA, for a final volume of 50 μL. Amplification was carried out in a thermocycler (PTC-200 Peltier Thermal Cycler; MJ Research, Waltham, Massachusetts, USA) with the following program: 94°C for 3 min, then 35 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min, and a final extension step at 72°C for 7 min.
A 2nd primer set (IFNR-2 and IDNT-2) was used to amplify a 102-bp fragment of the L1 gene of animal and human PVs (consensus), as previously described (22). The amplification reaction mixture consisted of 34.83 μL of sterile ultrapure water, 5.0 μL of 10X PCR buffer with ammonium sulfate, 3.0 μL of 25-mM magnesium chloride, 1.0 μL of 10-mM dNTP, 2.0 μL of each primer (25 pmol/μL), 0.17 μL of Taq polymerase (5 U/μL), and 2 μL of template DNA, for a final volume of 50 μL. Amplification was performed by a “touchdown” method, as follows: 94°C for 5 min, 16 cycles of 94°C for 1 min, the annealing temperature being decreased by 1°C each cycle from 50°C to 35°C, and 72°C for 1 min. This was followed by 24 cycles of 94°C for 1 min, 50°C for 2 min, and 72°C for 1 min; the final step was a 6-min extension at 72°C.
To ensure that the extracted DNA contained amplifiable DNA, primers that amplify a 108-bp segment of the feline p53 tumor suppressor gene sequence (GenBank accession number D26608) were used in a PCR assay as previously described (22). Equine p53 (GenBank accession number X91793) is nearly identical in nucleotide sequence to feline p53 in the region of the primers, with only 1 nucleotide difference, in the forward primer. The reaction mixture consisted of 34.75 μL of sterile ultrapure water, 5.0 μL of 10X buffer solution with ammonium sulfate, 3.0 μL of 25-mM magnesium chloride, 1.0 μL of 10-mM dNTP, 2.0 μL of each primer (10 pmol/μL), 0.25 μL of Taq polymerase (5 U/μL), and 2.0 μL of template DNA, for a final volume of 50 μL. Amplification was performed as follows: 94°C for 3 min, then 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 2 min.
The PCR products (10 μL) were separated by electrophoresis on a 2.5% agarose gel (Topogen, Columbus, Ohio, USA), stained with ethidium bromide, and photographed under ultraviolet transillumination (Fisher Scientific Company, Edmonton, Alberta). Images were captured with the use of a documentation and analysis system (Alpha Imager 2000; Alpha Innotech Corporation, San Leandro, California, USA).
Nucleotide sequencing of PCR products
Products resulting from amplification of DNA from equine tissues with the EPV and consensus primers were purified by phenol/chloroform extraction according to standard procedures. The purified PCR products were submitted for bidirectional sequencing with the same primers as used for amplification to the National Research Council of Canada, in Saskatoon. Sequence data from the specimens were compared with nucleotide sequences of other PVs in GenBank (M20219, J02044, and NC_003748) by means of the BLAST algorithm.
Detection of PV antigen
To test for PV antigen, we used a streptavidin–biotin–alkaline phosphatase IHC staining method (23) on 5-μm-thick sections cut from the tissue blocks. The sections were stained with an automated stainer (Ventana Medical Systems, Tucson, Arizona) and a 1:2000 dilution of rabbit type-1 antibody against BPV (Dako Corporation, Santa Barbara, California, USA). Tissue from a bovine cutaneous papilloma was included as a positive control in each IHC assay.
Statistical analysis
We compared the EPV-PCR, IHC, and histologic results using the McNemar chi-squared test for paired samples of papillomas (24). We tested agreement between the EPV-PCR and IHC results using Cohen’s kappa coefficient with the program WinEpiscope 2.0 and a 95% confidence interval; agreement is poor at κ ≤ 0.20, fair at 0.21 ≤ κ ≤ 0.40, moderate at 0.41 ≤ κ ≤ 0.60, substantial at 0.61 ≤ κ ≤ 0.80, and good at > 0.80 (25).
Results
The feline p53 tumor suppressor gene PCR used to determine whether specimens contained sufficient amplifiable DNA yielded a single band at the expected location for all but 2 of the tissue specimens (normal skin and an aural plaque); results from these samples were therefore eliminated from the data analysis.
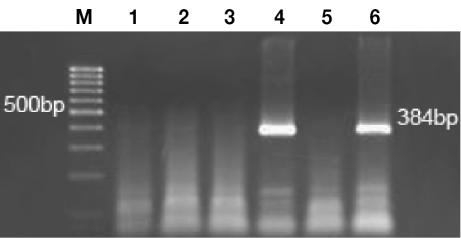
The EPV-PCR amplified a product of the expected size (384 bp) (Figure 1) in 20 (53%) of the 38 papilloma samples (Table I); of these 20 samples, 17 (85%) were also positive for PV antigen by IHC testing. The consensus primers amplified a 102-bp product from 2 (5%) of the 38 papillomas; the same 2 samples were positive for PV antigen. Of the 38 papillomas, 23 (60%) were positive by IHC staining of keratinocyte nuclei within the superficial layers; 6 (26%) of the 23 were negative by PCR. Overall, 26 (68%) of the 38 papillomas were positive for EPV by either EPV-PCR or IHC.
Figure 1.
Photomicrograph of an ethidium-bromide-stained agarose gel of polymerase chain reaction products obtained with use of an Equine papillomavirus primer set. Lane M is a 100-base-pair (bp) DNA ladder; lanes 1 to 4 represent DNA from normal equine skin, sarcoid, aural plaque, and papilloma, respectively; lane 5 is a negative control (no DNA); and lane 6 is amplified DNA from the positive control.
Table I.
Results of polymerase chain reaction (PCR) and immunohistochemical (IHC) testing for Equine papillomavirus (EPV) DNA and antigen, respectively, in equine papillomas, aural plaques, and sarcoids
Amplified DNA from 7 of the EPV-PCR-positive cutaneous papillomas was sequenced and found to be 96% to 99% identical in sequence to NC_003748. Three samples shared a nucleotide substitution of cytosine for adenosine at site 3502 of the EPV genome. The same samples had a substitution of guanosine for adenosine at site 3589. These samples and 2 additional samples demonstrated a substitution of cytosine for thymidine at site 3549.
Of the 6 genital papillomas, 3 produced a weak band at 384 bp with the EPV-PCR primers, but nucleotide sequencing of the PCR products and a BLAST search failed to yield any significant matches. Two of the genital papillomas were positive for PV antigen with IHC staining, but only 1 of these was PCR-positive. Neither congenital papilloma was positive by EPV-PCR or IHC testing. The nucleotide sequence of the PCR products obtained from 2 papillomas with use of the consensus primers failed to yield any significant BLAST matches with other PVs.
Of the 38 samples identified histologically as papillomas, significantly fewer were positive by EPV-PCR (20/38) or by IHC testing (23/38) (P < 0.001). There was no significant difference between the proportions identified as positive by EPV-PCR as compared with IHC testing (P = 0.51). There was moderate agreement between the 2 tests (κ = 0.52).
Only 1 of the 10 sarcoids yielded a 384-bp band with the use of EPV-specific primers; sequencing demonstrated 94% identity to the nucleotide sequence of EPV within a 150-bp region. A 102-bp band was present with the use of consensus PV primers in 7 of the 10 sarcoids; nucleotide sequencing, performed on 2 of these samples, revealed 90% identity to BPV type 2 for 1 sample and 90% identity to BPV type 1 for the other sample.
All the negative-control equine tissues had negative results with both EPV-PCR and IHC testing. The consensus primers demonstrated DNA amplification in 1 negative-control skin sample; sequencing showed 94% identity to BPV type 2.
Discussion
Both PCR and IHC gave positive results for more than 50% of the equine papillomas studied. The proportion of positive samples was not significantly different with either test, although it must be remembered that IHC identifies only generic PV antigen, whereas PCR confirms the presence of EPV. Cohen’s kappa statistic indicated moderate agreement between the results of the 2 tests. All the negative- control samples were negative by both tests.
Amplification and identification of EPV DNA in equine cutaneous papillomas suggest that EPV is involved in the development of these lesions. Nucleotide substitutions may reflect polymerase-copying errors or inherited substitutions, indicating descent of these organisms from a common ancestor. The high degree of identity in the 384-bp nucleotide sequence suggests that the product amplified is EPV DNA, although our conclusions are based on a small segment of a 7610-bp genome. The amount of natural variation in genetic sequence between isolates of EPV is unknown.
Although the technique of DNA extraction from FFPE tissues is considered robust and most DNA can be extracted and amplified, preparations from fixed tissues always exhibit certain limitations for PCR. Formalin fixation can induce degradation of DNA, resulting in a reduction in test sensitivity. The PCR primers were designed to amplify a relatively small (384-bp) region to minimize the loss of sensitivity due to sheared DNA (26). Fixation time has been shown to reduce amplification efficiency, especially when tissue is fixed in buffered 10% formalin for 1 to 4 wk; there is little or no effect when tissue is fixed for 48 h or less (27). Although tissues are usually fixed for 48 h or less, the precise fixation time was unknown for the tissues in our study.
The presence of paraffin-embedded material or other contaminants in extracted DNA can inhibit DNA amplification, causing false-negative results (28,29). Detection of amplifiable p53 DNA in our study confirmed that the DNA extraction protocol was adequate and ruled out the possibility of interference in the PCR assay, particularly inhibition of the Taq polymerase, by contaminants in the DNA samples (30). Age of the sample is also an important consideration: successful amplification has been reported in 40-y-old FFPE tissues, but researchers have reported a decline in amplification from fixed tissues that are 5 y old or older (30). In our study, tissue blocks were up to 17 y old, although amplification of p53 DNA was demonstrated in tissue of various ages.
Depending on the developmental stage of the papilloma, EPV DNA may have been present at levels less than detectable by PCR. Studies of bovine and equine papillomas have shown that in the early infection and growth phase the lower layers of the squamous epithelium contain little PV DNA (1,10,20), viral-protein expression and virus assembly being in the upper epidermal layers (1). Another possible explanation for false-negative results is deletion or mutation within the region of EPV targeted by the PCR, resulting in failure of the primers to recognize the gene sequence (28). It is possible that nested PCR would have increased the ability to amplify very low amounts of viral genome, thus reducing the risk of false-negative results. However, a greatly increased risk of contamination is associated with nested PCR, limiting its usefulness as a diagnostic tool.
Congenital papillomas have occasionally been reported in newborn foals (31,32). Our study included 2 congenital papillomas; the absence of amplifiable EPV DNA and PV antigen is consistent with previous reports that showed no evidence of inclusion bodies and negative IHC results for PV (10,32).
When the PCR products from the genital (penile and vulvar) papillomas in our study were sequenced, matches with EPV DNA nucleotide sequences were not found. These results are consistent with a previous report demonstrating that PV from 2 penile papillomas failed to hybridize with an EPV probe, suggesting the existence of a novel equine PV type (3).
The primary antibody used in this study for IHC testing reacts with PV-specific common structural capsid antigens that are well conserved across species (2,10,11). Since PV virions are assembled in the superficial keratinocytes (3,7,10,20), IHC staining is visualized in the superficial layers of the squamous epithelium when replicating PV is present. Lack of detection of viral antigen in some of our equine papillomas may indicate that there was no replicating PV or that antigenicity was inhibited by fixation (23,33). Nonproductive PV lesions, such as equine sarcoids, are not expected to contain viral antigen (2,34); therefore, negative IHC results for the equine sarcoids in our study were expected. Since papillomas initially have basal cell hyperplasia without virion assembly, some papillomas may contain PV DNA but not antigen (10), as found in 3 of the papillomas in our study.
Both the EPV-PCR and the consensus primers failed to amplify DNA in aural plaques, but PV antigen was detected in some, suggesting the possibility of a novel PV that differs sufficiently in nucleotide sequence to prevent the primers from binding. This finding is in agreement with a previous report (5) that introduced the possibility of different types of EPV as etiologic agents for cutaneous papillomas and aural plaques. Differences between clinical manifestations of warts and aural plaques may be attributed to genetic variance between strains of EPV: it has been recognized that, within a given host species, different PV strains appear to preferentially affect different epithelial locations (35).
The consensus primers allow the detection in FFPE tissues of DNA from PVs related to BPV1, BPV2, BPV5, ovine PV, European elk PV, deer PV, BPV3, BPV4, BPV6, canine oral PV, cottontail rabbit PV, and Human papillomavirus (HPV) types 1, 41, and 63 (22). Consensus primers amplified DNA in 2 of the papilloma tissues in our study. In the absence of successful sequencing, the DNA amplification was most likely nonspecific. Amplification with the consensus primers was also demonstrated in most of the equine sarcoids, and this was expected on the basis of reports that types 1 and 2 BPV are present in most sarcoids (16,17). Given the large amount of connective tissue in certain types of sarcoids, it is possible that a particular section may contain little or no viral DNA and yield a false-negative result (36). The presence of a PCR-positive result with the use of EPV primers in 1 sarcoid is difficult to interpret, as the sequencing results indicated homology within only a 150-bp region of the 384-bp sequence. The presence of a latent virus is a possibility, since the PV genome can often be found in normal epithelium, and normal epithelium is the accepted site of latent infection (34). Although the sample size for sarcoids was small, if EPV is etiologically involved, we might have expected more positive results with the EPV primers.
In conclusion, we demonstrated EPV DNA in a high proportion of equine papillomas but not in aural plaques or sarcoids, which suggests that EPV may have a direct involvement in the pathogenesis of cutaneous papillomas. Nucleotide differences in EPV strains could explain the inability to detect EPV DNA in aural plaques; continued research is required to determine the cause of these lesions. Since PV demonstrates a higher degree of conservation within the E2 (3), E1, and L1 genes (21) than in the E4 gene (1), additional primers designed to amplify these regions may allow amplification of EPV DNA from a greater proportion of equine papillomas and aural plaques. In conjunction with IHC testing, PCR may be useful in confirming the presence of EPV in some equine papillomas, especially those in which histologic differentiation from other lesions is problematic.
Acknowledgments
We thank Dr. Sally Lester, Central Laboratory for Veterinarians, for the tissue blocks, Dr. Sarah Parker for assistance with the statistical analysis, and Ms. Brenda Trask and the staff of the PCR and IHC sections of PDS for technical assistance. This study was supported by an Interprovincial Graduate Student Fellowship from the Western College of Veterinary Medicine, awarded to Dr. Postey.
References
- 1.Lowy DR, Howley PM. Papillomavirus and their replication. In: Knipe DM, Howley PM, eds. Fields Virology. 4th ed. Philadelphia, Pennsylvania: Lippincott Williams & Wilkins, 2001:2197–2257.
- 2.Lancaster WD, Olson C. Animal papillomaviruses. Microbiol Rev. 1982;46:191–207. doi: 10.1128/mr.46.2.191-207.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Banion MK, Reichmann ME, Sundberg JP. Cloning and characterization of an equine cutaneous papillomavirus. Virology. 1986;152:100–109. doi: 10.1016/0042-6822(86)90375-2. [DOI] [PubMed] [Google Scholar]
- 4.Ghim S, Rector A, Delius H, Sundberg JP, Bennett Jenson A, VanRanst M. Equine papillomavirus type 1: complete nucleotide sequence and characterization of recombinant virus-like particles composed of the EcPV-1 L1 major capsid protein. Biochem Biophys Res Commun. 2004;324:1108–1115. doi: 10.1016/j.bbrc.2004.09.154. [DOI] [PubMed] [Google Scholar]
- 5.Scott DW, Miller WH, eds. Epithelial neoplasms. In: Equine Dermatology. St. Louis: Saunders, 2003:700–731.
- 6.Williams MA. Papillomatosis: warts and aural plaques. In: Robinson NE, ed. Current Therapy in Equine Medicine. 4th ed. Philadelphia: WB Saunders, 1997:389–399.
- 7.Fulton RE, Doane FW, Macpherson LW. The fine structure of equine papillomas and the equine papillomavirus. J Ultrastruct Res. 1970;30:328–343. doi: 10.1016/s0022-5320(70)80066-1. [DOI] [PubMed] [Google Scholar]
- 8.Cook RH, Olson C., Jr Experimental transmission of cutaneous papilloma of the horse. Am J Pathol. 1951;27:1087–1097. [PMC free article] [PubMed] [Google Scholar]
- 9.Binninger CE, Piper RC. Hyperplastic dermatitis of the equine ear. J Am Vet Med Assoc. 1968;153:69–75. [PubMed] [Google Scholar]
- 10.Sundberg JP, Junge RE, Lancaster WD. Immunoperoxidase localization of papillomaviruses in hyperplastic and neoplastic lesions of animals. Am J Vet Res. 1984;45:1441–1446. [PubMed] [Google Scholar]
- 11.Fairley RA, Haines DM. The electron microscopic and immunohistochemical demonstration of a papillomavirus in equine aural plaques. Vet Pathol. 1992;29:79–81. doi: 10.1177/030098589202900110. [DOI] [PubMed] [Google Scholar]
- 12.Ragland WL, Keown GH, Spencer GR. Equine sarcoid. Equine Vet J. 1970;2:2–11. [Google Scholar]
- 13.Marti E, Lazary S, Antczak DF, Gerber H. Report of the first international workshop on equine sarcoid. Equine Vet J. 1993;25:397–407. doi: 10.1111/j.2042-3306.1993.tb02981.x. [DOI] [PubMed] [Google Scholar]
- 14.Ragland WL, Spencer GR. Attempts to relate bovine papillomavirus to the cause of equine sarcoid: equidae inoculated intradermally with bovine papillomavirus. Am J Vet Res. 1969;30:743–752. [PubMed] [Google Scholar]
- 15.Brostrom H. Equine sarcoids, a clinical epidemiological study in relation to equine leucocyte antigens (ELA) Acta Vet Scand. 1995;36:223–236. doi: 10.1186/BF03547691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amtmann E, Muller H, Sauer G. Equine connective tissue tumors contain unintegrated bovine papillomavirus DNA. J Virol. 1980;35:962–964. doi: 10.1128/jvi.35.3.962-964.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angelos JA, Marti E, Lazary S, Carmichael LE. Characterization of BPV-like DNA in equine sarcoids. Arch Virol. 1991;119:95–109. doi: 10.1007/BF01314326. [DOI] [PubMed] [Google Scholar]
- 18.Teifke JP, Hardt M, Weiss E. Detection of bovine papillomavirus DNA in formalin-fixed and paraffin-embedded equine sarcoids by polymerase chain reaction and nonradioactive in situ hybridization. Eur J Vet Pathol. 1994;1:5–10. [Google Scholar]
- 19.Chambers G, Ellsmore VA, O’Brien PM, et al. Association of bovine papillomavirus with the equine sarcoid. J Gen Virol. 2003;84:1055–1062. doi: 10.1099/vir.0.18947-0. [DOI] [PubMed] [Google Scholar]
- 20.Hamada M, Oyamada T, Yoshikawa H, Yoshikawa T, Itakura C. Histopathological development of equine cutaneous papillomas. J Comp Pathol. 1990;102:393–403. doi: 10.1016/s0021-9975(08)80161-2. [DOI] [PubMed] [Google Scholar]
- 21.Goodrich L, Gerber H, Marti E, Antczak DF. Neoplasia: equine sarcoids. Vet Clin North Am Equine Pract. 1998;14:607–623. doi: 10.1016/s0749-0739(17)30189-x. [DOI] [PubMed] [Google Scholar]
- 22.Kidney BA, Haines DM, Ellis JA, et al. Evaluation of formalin-fixed paraffin-embedded tissues from vaccine site-associated sarcomas of cats for papillomavirus DNA and antigen. Am J Vet Res. 2001;62:833–839. doi: 10.2460/ajvr.2001.62.833. [DOI] [PubMed] [Google Scholar]
- 23.Haines DM, Chelack BJ. Technical considerations for developing enzyme immunohistochemical staining procedures on formalin-fixed paraffin-embedded tissues for diagnostic pathology. J Vet Diagn Invest. 1991;3:101–112. doi: 10.1177/104063879100300128. [DOI] [PubMed] [Google Scholar]
- 24.Norman GR, Streiner DL. In: Biostatistics. The Bare Essentials. St. Louis: Mosby, 1994:155–156.
- 25.Petrie A, Watson P. In: Statistics for Veterinary and Animal Science. London: Blackwell Science, 1999:168–181.
- 26.Greer CE, Peterson SL, Kiviat NB, et al. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am J Clin Pathol. 1991;95:117–124. doi: 10.1093/ajcp/95.2.117. [DOI] [PubMed] [Google Scholar]
- 27.Rogers BB, Alpert LC, Hines EAS, et al. Analysis of DNA in fresh and fixed tissue by the polymerase chain reaction. Am J Pathol. 1990;136:541–548. [PMC free article] [PubMed] [Google Scholar]
- 28.Quinn ED. Sample preparation. In: Lee H, Morse S, Olsvik O, eds. Nucleic Acid Amplification Technologies. Natick, Massachusetts: Eaton Publishing, 1997:49–60.
- 29.Eeles RA, Stamps AC. Polymerase Chain Reaction (PCR). The Technique and Its Applications. Austin, Texas: R.G. Landes Company, 1993:1–32.
- 30.Shibata D. Preparation of nucleic acids for archival material. In: Mullis KB, Ferre F, Gibbs RA, eds. The Polymerase Chain Reaction. Boston: Birkhauser, 1994:47–54.
- 31.Avina-Garma A. Equine congenital cutaneous papillomatosis: a report of 5 cases. Equine Vet J. 1981;13:59–61. doi: 10.1111/j.2042-3306.1981.tb03455.x. [DOI] [PubMed] [Google Scholar]
- 32.Schueler RL. Congenital equine papillomatosis. J Am Vet Med Assoc. 1973;162:640. [PubMed] [Google Scholar]
- 33.Rickert RR, Maliniak RM. Intralaboratory quality assurance of immunohistochemical procedures. Arch Pathol Lab Med. 1989;113:673–679. [PubMed] [Google Scholar]
- 34.Campo SM. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- 35.Johnson PJ. Dermatologic tumors (excluding sarcoids) Vet Clin North Am Equine Pract. 1998;14:636–658. doi: 10.1016/s0749-0739(17)30190-6. [DOI] [PubMed] [Google Scholar]
- 36.Carr EA, Theon AP, Madewell BR, et al. Bovine papillomavirus DNA in neoplastic and nonneoplastic tissues obtained from horses with and without sarcoids in the western United States. Am J Vet Res. 2001;62:741–744. doi: 10.2460/ajvr.2001.62.741. [DOI] [PubMed] [Google Scholar]