Abstract
IgD has remained a mysterious Ig class and a bane to immunology students since its discovery >40 years ago. Its spotty occurrence in mammals and birds and the discovery of an isotype with similarities to IgD in bony fish are perplexing. We have identified IgD heavy (H) chain (δ) from the amphibian Xenopus tropicalis during examination of the IgH locus. The Xenopus δ gene is in the same position, immediately 3′ of the IgM gene, as in mammals, and it is expressed only in the spleen at low levels, primarily as a transmembrane receptor by surface IgM+ cells. Our data suggest that frog IgD is expressed on mature B cells, like in mouse/human. Unexpectedly, Xenopus IgD is orthologous to IgW, an Ig isotype found only in cartilaginous fish and lungfish, demonstrating that IgD/W, like IgM, was present in the ancestor of all living jawed vertebrates. In striking contrast to IgM, IgD/W is evolutionarily labile, showing many duplications/deletions of domains, the presence of multiple splice forms, existence as predominantly a secretory or transmembrane form, or loss of the entire gene in a species-specific manner. Our study suggests that IgD/W has played varied roles in different vertebrate taxa since the inception of the adaptive immune system, and it may have been preserved as a flexible locus over evolutionary time to complement steadfast IgM.
Keywords: evolution, immune system
Since the pioneering work of comparative immunologists in the 1960s, IgM has been the Ig isotype believed to be primordial and most stable in vertebrate evolution (1). Despite differences in the degree of polymerization of the secretory form in different vertebrate groups (1, 2) and a major splice variant of the transmembrane (TM) form in teleost fish (3), IgM is renowned for its molecular, biochemical, and functional stability. Present in all living jawed vertebrates, IgM is the first isotype to be expressed both in ontogeny and during humoral adaptive immune responses and is found as the major TM receptor on the surface of both conventional and “innate” B cells (4).
By contrast, IgD has remained an enigmatic isotype since its discovery long ago (5). Like IgM, IgD is expressed as a TM receptor on B cells of mouse and human, generated as a result of alternative splicing of pre-mRNA containing the transcribed variable (V) region and the IgM and IgD heavy (H) chain constant (C) regions (μ and δ, respectively). Because of its spotty presence in mammals and absence in birds, IgD was assumed to be a recently evolved isotype (6). However, the discovery of an isotype in ray-finned bony fish with sequence similarity to IgD (7) and its presence in some mammals previously believed to lack IgD (8) have greatly modified our view of Ig isotype evolution and suggested it may have arisen much earlier.
A similarly strange phylogenetic jump surrounds the isotype IgW, which was discovered first in skates (9, 10) and later in sharks (11, 12). The secreted version of the IgW H chain (ω) is present in long and short forms in all elasmobranchs tested to date, and the TM form is also differentially spliced (13). This isotype is believed to be as old as IgM (11, 12) but is also thought to be a dead-end isotype that first evolved in the placoderm or cartilaginous fish lineage. However, recently an isotype orthologous to IgW was reported in lungfish (a lobed-fin bony fish, believed to be related to ancestral amphibians; ref. 14). The discovery of this isotype and the apparent lack of an IgD equivalent in lungfish were unexpected.
Xenopus has three well defined Ig isotypes, IgM, IgY, and IgX (reviewed in ref. 15). Although all of these isotypes are expressed by lymphocytes in the spleen, the only conventional secondary lymphoid tissue in most cold-blooded vertebrates [i.e., no lymph nodes or Peyers patches (16)], there is also expression in other tissues, notably the small intestine (especially for IgX) and the liver (17). “Resting” B cells with surface expression of IgM are found in discrete splenic follicles, which are surrounded by a relatively diffuse T cell zone. Amphibia is the oldest class of vertebrates having the Ig isotype switch (18), and previous work definitively has shown an exchange from one C region to another by means of a deletional mechanism (19, 20). The order of the H-chain genes, however, has not been established. The Xenopus tropicalis genome project web site (www.jgi.doe.gov) has spawned many new possibilities for the phylogenetic study of immune genes. When we inspected genomic scaffolds containing IgH genes in the X. tropicalis database, an array of C1-set Ig superfamily (IgSF) exons was found between the μ and IgX (χ) genes. The location of these exons, their deduced amino acid sequence similarity to δ and ω of other vertebrates, and their exclusive but low expression in spleen suggest that: (i) the Xenopus IgD H-chain equivalent has been uncovered with an expression similar to that in mouse and human; (ii) IgW and IgD H chains are orthologues, and thus both IgD and IgM were present at the inception of the adaptive immune system when gnathostomes emerged; and (iii) whereas IgM is stable over evolutionary time, IgD shows great plasticity.
Results
The Xenopus δ Gene Is Immediately Downstream of μ.
As mentioned, it has been known for 20 years that Xenopus has three H chain isotypes, IgM, IgY, and IgX (15). We examined the genomic scaffold (scaffold 840) containing C domains for these three genes, as well as all of the diversity (D) and junctional (J) and several variable (V)H gene segments. In addition, another unanticipated set of C region exons, as well as one canonical TM exon, was found between the μ and χ genes (Fig. 1). These exons were reconstructed, and the gene was predicted to encode a nine-domain molecule (including the V region) with a TM region like that found in other Ig isotypes (Figs. 2 and 3). A preliminary blastx search with the entire molecule and with individual domains showed similarity to δ chains from other species. However, unlike mammalian IgD H chains, there is no exon encoding a hinge region detected in the genomic scaffold or in the cDNAs (see below). To our surprise, domain by domain blastx searches also selected C domains of lungfish and cartilaginous fish IgW and new antigen receptor (NAR) (21), as well as other Xenopus Ig isotypes (see below).
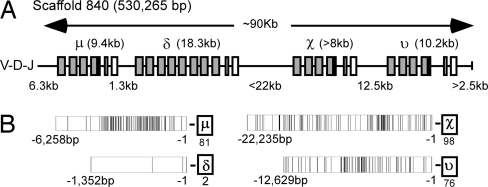
Fig. 1.
The X. tropicalis δ gene is immediately 3′ of the μ gene. (A) X. tropicalis genome scaffold (version 3.0) 840 contains the IgH locus. C, secretory, and TM regions are indicated as gray, black, and open boxes, respectively. The approximate size of the Ig genes and intergenic regions is shown. The χC1 domain is only a prediction, because it has not been assembled into the scaffold. The C domains are located at the edge of the scaffold spanning an ≈90-kb region. (B) AGCT motifs are rich in 5′ upstream region of μ, χ, υ, but not in that of δ. The distance and positions of AGCT motifs are shown as bars. The numbers of AGCT motifs found in these regions are also shown under each Ig isotype. All diagrams are not to scale.
Fig. 2.
Alignment of Xenopus IgD H chain to IgD and IgW H chains from other species. Amino acid sequences were deduced from the δ cDNA sequence (GenBank accession no. DQ387453) isolated from X. tropicalis spleen. Alignments were made by using clustal x with minor manual adjustments. Domain designation was based on exon/intron boundaries. Bars and dots indicate identical amino acids and gaps, respectively. Potential IgSF strands are indicated as A–G with lines over the sequences. Noncanonical cys are marked with an open oval, and canonical cys and trp found in most IgSF domains are shaded. Glycosylation sites are underlined.
Fig. 3.
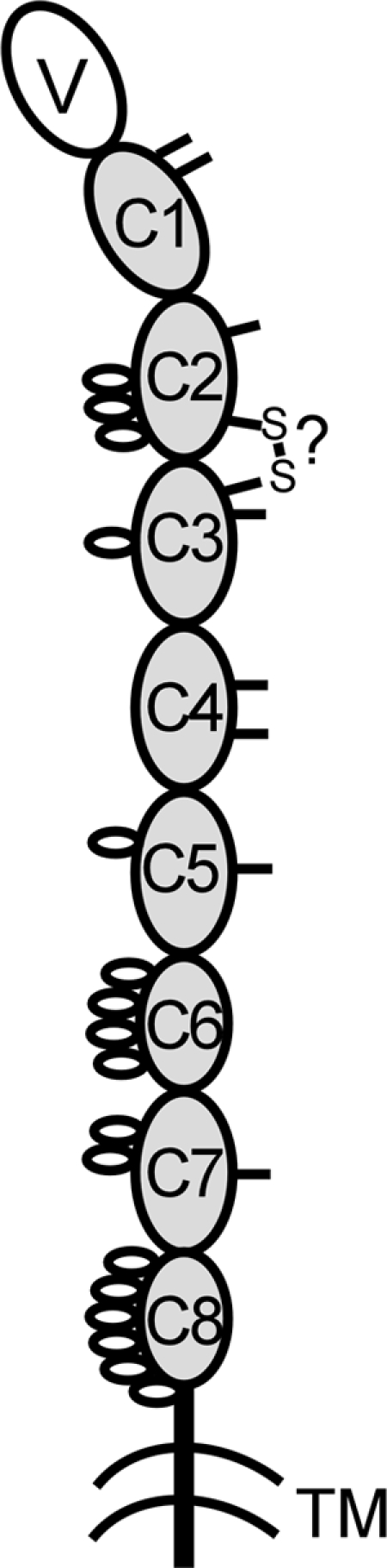
Cartoon of Xenopus IgD H-chain structure. The V and C domain are shown as large ovals, and sugars and noncanonical cys residues are indicated as small open ovals (on the left) and ticks (on the right). GenBank accession nos. are shown in the legend Fig. 6.
“Switch regions,” rich in AGCT motifs that can promote the isotype switch (20), are found upstream of the μ, IgY (υ), and χ genes. Such motifs are found at a much lower number and frequency upstream of the putative Xenopus δ gene (Fig. 1). Furthermore, it is noteworthy that the physical distance between the μ and putative δ genes is very short compared to the distance between δ and χ, χ and υ, and μ and the 3′JH segment (Fig. 1). These data suggest that the makeup of the IgH locus is not conducive to promote isotype switching from μ to the new isotype. Thus, the position of the new gene immediately downstream of μ, the paucity of switch elements in its short upstream region, and preliminary phylogenetic analyses all support the premise that the Xenopus IgD H chain has been revealed.
Expression of Xenopus IgD.
Despite extensive biochemical and molecular analyses of the Ig system in Xenopus (15), there was no hint of another isotype besides the three well studied forms. In retrospect, we think that this gap in knowledge is due to IgD's presence primarily as a TM form and its suspected susceptibility to proteolysis because of its large size (22). Because of this large predicted size and relatively low expression, a complete δ cDNA was not isolated from our libraries, and no EST was found in the public databases. Thus, as described in Materials and Methods, we used PCR to amplify clones from X. tropicalis spleen RNA using primers specific for C8 exon, TM, and 3′-UTR, as well as primers predicted to amplify the majority of Xenopus V gene families (23). Most cDNAs were found to encode nine-domain-long molecules containing TM regions, expected from the genomic scaffold (all coding regions shown in Fig. 2). Some minor variants were detected in which the δC1 and δC2 exons were spliced to small exons leading to premature termination; these cDNAs might encode small secreted forms of IgD (data not shown).
Both the δC1 and δTM probes revealed a ≈3.8-kb mRNA present only in spleen (Fig. 4). As mentioned above, the H chains for the other isotypes (IgM, IgY, and IgX) are found predominantly as secretory forms and are expressed not only in the spleen but also in other tissues (IgM shown in Fig. 4). There was a weakly hybridizing δ band of ≈1.8 kb; this mRNA has been detected neither in our cDNA libraries nor by PCR. Thus, the preliminary sequence and expression analyses suggest that Xenopus IgD is predominantly a TM receptor, found exclusively in the distinct secondary lymphoid tissue. That μ is more highly expressed in other tissues (colon, small intestine, liver, and oviduct) than in spleen strongly suggests that δ expression is extinguished upon B cell activation. We hypothesized, therefore, that IgD would be expressed in IgM+ B cells, like its orthologue in mouse and human.
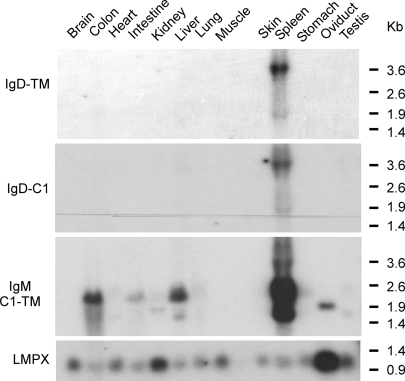
Fig. 4.
Xenopus IgD is expressed predominantly in the spleen. Twenty micrograms of total RNA from various tissues of an X. tropicalis frog was loaded on the gel, and the Northern blot was hybridized with X. tropicalis δC1 and TM probes separately. An X. tropicalis μ C1-TM probe was also used to compare the level of expression and tissue distribution. Two bands detected with the μ probe account for the different 3′-UTR lengths between secretory and TM forms or unknown splice variants specific to this species. Ubiquitously expressed LMPX was used as the loading control. Although the loading amounts are not equal on this blot, IgD is found only in the spleen, despite the fact that less spleen RNA was loaded. RNA size markers are shown on the right.
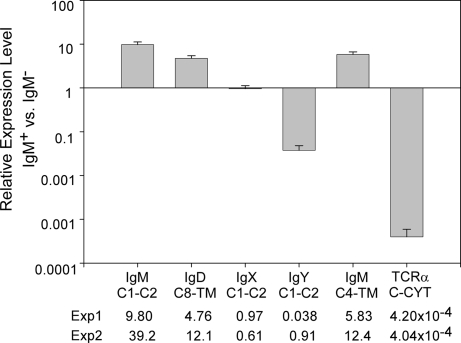
To test this assertion, we performed real-time quantitative PCR (qPCR) analysis, using RNA from splenic IgM+ cells purified with a mAb specific for IgM (10A9) (Fig. 5 and Table 1, which is published as supporting information on the PNAS web site). In two sets of RNAs derived from different animal pools, we detected increased δ expression in IgM+ cells relative to the IgM− cells, with a ratio almost identical to μTM mRNA in the same cell populations (4.76 vs. 5.83 and 12.1 vs. 12.4); thus, the majority of δ mRNA was found in the IgM+ cells. We did not anticipate that expression detected with the μC1–C2 primers, which amplify both the μ secretory and TM forms, would be higher than the expression of μTM mRNA in the IgM+ cells. Perhaps in Xenopus, lymphocytes that secrete IgM remain surface IgM+ (as well as surface IgD+). As predicted, the negative control T cell receptor α (TCRα) mRNA was highly enriched in the IgM− cells. In these experiments, υ expression was not consistent, either being enriched in the IgM− cells (Experiment 1) or equally represented in the surface IgM+ and IgM− cells (Experiment 2). Previous work has suggested that, despite the Ig class switch, IgY and IgM could be coexpressed in B cells, perhaps because of a long lifetime of the μ mRNA or protein (24). We suspect that the uneven expression of υ is related to the immune status or age of the animals, and this problem certainly warrants further study. The animal's immune status may also affect μ and δ expression, because both were somewhat elevated in the 10A9+ cells in Experiment 2. χ was expressed in much lower amounts in all experiments, which was expected because of reports of relatively low levels of IgX in spleen relative to the small intestine (17). Most important for this investigation, δ mRNA is highly enriched in IgM+ cells, again suggesting that Xenopus IgD is principally a TM receptor encoded by differentially spliced mRNA, as in mouse and human (25).
Fig. 5.
Relative expression level of four Ig isotypes in IgM+ vs. IgM− splenocytes. qPCR analysis was done by using SYBR green, and each run was performed in triplicate. The relative expression level was calculated by using 2−ΔΔCt method as 10A9+ vs. 10A9− cells and plotted on a log scale in sigma plot. Primer sets are shown underneath. The figure represents the mean value of Experiment 1 (three separate runs) with standard deviations shown in bars. Mean values of two separate experiments are also shown underneath. Each threshold cycle (Ct) was normalized to the Ct of LMPX. The raw threshold cycle (Ct) values are presented in Table 1.
Structure and Phylogenetic Position of Xenopus IgD.
Xenopus IgD has eight C domains, all prototypic members of the C1-set IgSF (ref. 26; Figs. 2 and 3). The N-terminal C1 domain has two cysteines (cys) in the A strand, one of which is predicted to associate with light (L) chains. This double-cys feature is also found in the lungfish IgW C1 domain and in Xenopus IgY (Fig. 2 and not shown for IgY). The Xenopus C1 domain has a large gap in the D and E strands, making it unique among the C1 domains. The C2 domain has two cys in the A-B strands, suggesting a hinge of sorts; this domain is also potentially heavily glycosylated (underlined in Fig. 2 and open ovals in Fig. 3), likely for protection from proteolysis. In general, the C2 domain in all Ig isotypes evolves rapidly (27), to the point of being deleted from mouse IgD (Fig. 7); it was therefore difficult to align the Xenopus sequence with any of the other isotypes. The C3 domain, as in lungfish IgW and catfish IgD, does not have the canonical tryptophan found in practically all IgSF domains, and a noncanonical cys in the F-G strand loop is conserved among the three domains; it is likely, based upon modeling, that this cys makes a disulfide bond with the aforementioned cys in the C2 domain A-B loop (Fig. 3). Other noncanonical cys in the C1–C4 domains might be involved in intradomain disulfide bonds, whereas the impaired cys in the C5 and C7 domains might be involved in interchain bonding. The C5 and C7 domains, and the C6 and C8 domains, are quite similar, suggesting a recent tandem duplication, similar to that described by our group for elasmobranch IgW (12) and others for certain ray-finned fish IgD (ref. 28; Fig. 2). The duplicated C6 and C8 domains seem to lack the IgSF A strand and are generally smaller (more compact?), suggesting that these domains will have an unusual domain structure. The C-terminal domains and the connecting piece have many potential sites for glycosylation, again likely functioning to protect from proteolysis (Figs. 2 and 3).
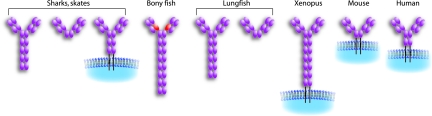
Fig. 7.
Plasticity of IgD/W in different vertebrate species. TM and secretory forms are obviously displayed. TM forms are the major type in frog, mouse, and human. The TM and secretory forms are shown for IgW to emphasize the alternative splicing. No TM forms have been described in lungfish, and the TM/secretory forms in bony fish are the same size. L-chain association is predicted only in bony fish, Xenopus, and lungfish. Only one form of ray-finned bony fish IgD (the original catfish model) among several other forms is shown here, and the light-red domain is the C1 domain donated from the μC1 domain. The hinge regions are not shown for mouse and human δ chains.
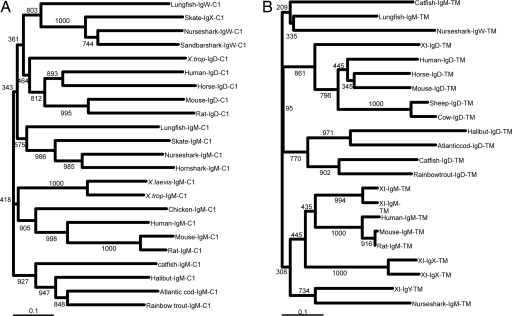
The entire Xenopus IgD sequence was most similar in blastx analysis to the lungfish IgW H chain (which has seven C domains) followed by δ chains from other species and the other Xenopus isotypes, but all with low (26–38%) identity at the amino acid level. As mentioned above, rapid evolution is typical of Ig isotypes and immune genes in general, making phylogenetic analysis difficult (1, 27, 29). Because the C1 and TM domains tend to evolve more slowly than the rest of the molecule (see Discussion), we used these regions for a more rigorous phylogenetic analysis. For the C1 domain, Xenopus δ clusters with δC1 from mammals, and this cluster is most closely associated with the IgW H-chain C1 clade (Fig. 6A). The TM region tree showed a strong association of the Xenopus molecule with the same regions of mammalian δ (Fig. 6B). Unfortunately, the lungfish IgW H-chain TM exon/cDNA has not been isolated, which might allow a more insightful analysis of the fish δ and ω sequences, similar to the C1 tree. Taken together, the data show there is no doubt that the orthologue of the mammalian IgD H chain has been uncovered in Xenopus. The data further support the argument that the IgD and IgW H chains are in the same clade, and thus IgD is as ancient as IgM.
Fig. 6.
Phylogenetic relationship of Xenopus IgD to IgD and IgW from other species. Neighbor-joining bootstrapping trees (1,000 runs) of C1 (A) and TM (B) domains were made. Gaps were included, and multiple substitutions were not taken into account. The scales show the genetic distances. The scale shown as a bar represents the genetic distance (i.e., number of amino acid changes in the given scale). GenBank accession nos. for each sequence are as follows: Human-IgD (AAH21276), Mouse-IgD (AAB59654), Rat-IgD (AAO19643), Horse-IgD (AAU09793; ref. 44), Lungfish-IgW (AAO52811), Sandbar shark-IgW (AAB03680), Nurse shark-IgW (AAB08972), Skate-IgX (AAA49546), Catfish-IgD (AAC60133), Halibut-IgD (BAB41204), Atlantic Cod-IgD (AAF72566), Rainbow trout-IgD (AAW66976), Human-IgM (AAH11857), Mouse-IgM (AAH18315), Rat-IgM (AAH92582), Chicken-IgM (P01875), X. laevis-IgM (AAA49774), X. tropicalis-IgM (AAH89679), Lungfish-IgM (AAO52808), Nurse shark-IgM (AAA50817), Horn shark-IgM (P23085), and Skate-IgM (AAA49547).
Discussion
Discovery of Xenopus IgD.
From the gene position, expression, and phylogenetic analysis, it is unequivocal that the IgD H chain equivalent in Xenopus has been detected. Although this gene is highly plastic throughout evolution (see below), the C1 and TM regions are orthologous to their mammalian counterparts (Fig. 6), and the δ mRNA is highly enriched in the surface IgM+ population. It is not surprising that the C1 domain and TM region were most informative in generating the trees, because these parts of IgH chains have been shown previously to evolve at a slower rate than the other domains (1, 14); this is most likely because of the functional constraints placed upon these portions: association with the Ig L chain for the C1 domain and interactions with signaling molecules and general TM stability for the TM region. As described in more detail below, the entire phylogenetic history of this bewildering molecule should become clearer when sequences from other vertebrates become available.
IgD/W Is as Old as IgM.
Our analysis strongly supports the hypothesis that IgD and IgM are present in all extant vertebrate classes (except perhaps Avians), and that both μ and δ arose near the time that the adaptive immune system emerged ≈500 million years ago (1). The Xenopus sequence helps to unite mammalian δ with the reported δ chain in ray-finned bony fish as well as with the IgW H chain in cartilaginous fish and lungfish. Indeed, at the time of the discovery of catfish IgD (7), it was thought strange that this molecule would be found in ray-finned bony fish and placental mammals but not in all other animal groups. It was also unexpected that lungfish (14), like cartilaginous fish, would have IgW and not IgD (although this analysis was hampered by the fact that a ray-finned fish δ C1 domain does not exist, its L chain-binding domain being encoded by the μ mRNA by means of alternative splicing; Fig. 7). Our interpretation is that lungfish, like sharks, have retained the ancestral characteristic of having a high level of secretory IgD/W, which apparently was lost in the ray-finned fish. The common ancestor of lungfish and ray-finned fish is as old as that of lungfish and amphibians, and yet we detected higher similarity between the lungfish IgW and Xenopus IgD sequences; for many immune genes, we commonly find that ray-finned fish sequences make clusters in phylogenetic trees among themselves that are out of the mainstream, i.e., often appear more divergent than expected based on the fossil record (e.g., ref. 30). We think it will be informative to examine IgD/W from chondrosteans such as the sturgeon, which are derived from ancestors intermediate to those of extant ray-finned and cartilaginous fish (16). Sequences from other divergent amphibians, like ranid frogs and urodeles, and from reptiles will also be useful in testing this hypothesis. It should be pointed out that Hordvik et al. (28) already suggested a phylogenetic connection between IgW and IgD when the first teleost δ sequences were reported.
Another question we must ask is, if our hypothesis is correct and the δ and ω genes are derived from a common ancestor, how did these genes become physically associated in the genome? The IgH gene organization is not known in lungfish, but at least from teleosts through mammals the δ gene is downstream of μ, and the δ mRNA is generated by alternative splicing from a large pre-mRNA [this has not been shown definitively in ray-finned fish but must be true (7)]. Because cartilaginous fish have discrete clusters of μ and ω genes (10, 13, 31), each cluster with its own set of V exons, how did the genes for the two isotypes become closely linked? It is difficult to even speculate on this problem, and it may never be solved, but the “functional clustering” of these genes must have occurred before the advent of isotype switching in amphibians. Perhaps we will obtain some clues to this puzzle from a thorough analysis of elasmobranch genomes.
IgD/W: The Adaptive Immune System's Plaything.
An interesting feature of this IgD/W H chain locus is that it is highly plastic in evolution, both in terms of the number of C domains and the plethora of splice variants found, especially in fish (Fig. 7). In cartilaginous fish, two of the C domains were derived ≈250 million years ago by a tandem duplication event (12), in Xenopus a recent two-domain tandem duplication event occurred, and in some bony fish there was a three-domain duplication (e.g., ref. 28). Within ray-finned teleost fish, the number of C exons for this isotype is different in various species (7, 28, 32–34), and the secreted and TM forms seem to be encoded by different loci in the catfish (34). In mammals, there are different numbers of C domains in different species, even between mouse and human (27). In artiodactyl species (pig, sheep, and cow), the δ1 domain-encoding exon has been deleted and replaced with a duplicated μ1 exon (8, 35, 36), and the δ gene has been dispensed with altogether in chicken and rabbit (ref. 37 and unpublished observations). In addition to the modifications in the gene over evolutionary time, the types of splice variants reported for the different fish taxa are remarkable. As mentioned above, in cartilaginous fish and lungfish, there are two major secretory forms either with five to six C domains or two C domains, and the major TM form in sharks has four C domains. In all ray-finned fish, the μ1 domain exon is spliced into the IgD transcript, encoding the domain predicted to associate with L chains (Fig. 7). Thus, it is our impression that this is the locus that evolution “tinkers with,” perhaps using IgD/W for multiple functions at different stages of phylogeny, the most obvious feature being the differences in levels of the secretory and TM forms but also the widely varying number of domains. We are obviously at the beginning of our analysis of this system, but perhaps an evolutionary perspective of IgD/W will eventually lead to an understanding of the function of this elusive isotype. Because Xenopus is a “high-connectivity model” in immunology (e.g., ref. 15), which has been rather well defined and with existing mAbs to all hematopoietic cell types, future experiments with IgD-specific mAbs will permit analyses of the biochemistry, cellular distribution, and perhaps function of IgD.
Materials and Methods
Screening of the Genomic Scaffold.
We started data mining the X. tropicalis genome scaffold version 3.0 (www.jgi.doe.gov) for the IgH locus. We performed a blastn search with Xenopus laevis μ cDNA sequence and found that only scaffold 840 (530,265 bp) contained a full-length μ gene. Scaffold 840 was then retrieved from the Joint Genome Institute browser window, and all “fgenesh” entries were examined manually for Ig domains. Each Ig domain was manually crosschecked blastx in the National Center for Biotechnology Information vertebrate database using the blosum 45 matrix.
Isolation of cDNAs by PCR.
Because there are no EST entries for Xenopus δ in the X. laevis or X. tropicalis databases, we isolated it by PCR. First-strand cDNA was made by using the Superscript III First-Strand synthesis system for RT-PCR (Invitrogen) using an oligo(dT) adaptor primer for 3′-RACE from ≈1 μg of X. tropicalis spleen total RNA. We selected nested primer sets, one for the V to TM regions and the other for the C8 domain to the 3′-UTR. Primer sets were 5′-AAC TAC CCT TCA ACT GAC ATG-3′ and 5′-ACG GAG ACT ATT ACA GTA CC-3′, 5′-AGC ACA GTT AGC ATT GAC CTT G-3′ and 5′-CTC GAG AAG CTT GAA TTC GGA TCC-3′ [artificial adaptor sequence flanked a the 3′-end of in-house oligo(dT) primer]. PCR fragments were subsequently cloned into TA cloning pCR2.1 vector (Invitrogen) and sequenced. Two overlapping fragments were joined manually and submitted to GenBank (accession no. DQ387453).
Northern Blotting.
An X. tropicalis δ C1 domain probe was made from X. tropicalis genomic DNA by PCR using the C1 primer set 5′-TCA GAT ACA GGT ACA TCT GC-3′ and 5′-AAT CTT CTT TGT AGG TCG AAC-3′. The TM probe was also made from X. tropicalis genomic DNA by PCR using the TM primer set 5′-ACT ACG CAG CCT ACC GAT AC-3′ and 5′-ACG GAG ACT ATT ACA GTA CC-3′. Probes were subcloned into the TA cloning vector pCR2.1 (Invitrogen), and the sequence was confirmed before use. The μC1-TM probe was made from X. tropicalis spleen cDNA using primer set 5′-ACC TGT GAT GCA AGC TTC CG-3′ and 5′-AAT GAA GGT TGA GGC AGT GG-3′. RNA was isolated from various tissues of an X. tropicalis frog using the TRIzol reagent (Invitrogen). Total RNA (20 μg) was electrophoresed in 1% agarose gels by using 4-morpholinepropanesulfonic acid as running buffer (Quality Biological, Gaithersburg, MD) and then transferred to an Optitran nitrocellulose membrane (Schleicher & Schuell). The membrane was hybridized with the δC1 probe under high-stringency conditions (38). We waited until the radioactivity was not detectable on the blots and then reprobed with δ TM, μ C1-TM, and ubiquitously expressed 20S proteasome β-subunit, LMPX (ref. 39; loading control).
Phylogenetic Trees.
The deduced Xenopus δ C1 and TM domains were aligned by using clustal x, and neighbor-joining bootstrapping trees (1,000 trial runs) were made and viewed in the treeview 1.6.6 program (40). For both trees, gaps were included, and multiple substitutions were not taken into account.
Separation of Membrane-Bound IgM-Positive Splenocytes.
X. laevis spleen cell suspensions were obtained from the spleens of five frogs in Experiment 1 and four frogs in Experiment 2 by gentle dissociation with blunt-end forceps and suspended in amphibian PBS (APBS, mammalian PBS with 25% water) supplemented with 5% heat-inactivated FCS. The splenocyte suspension was depleted of red blood cells (≈30%) by layering cells over 52% Percoll (Amersham Pharmacia Biosciences) in APBS (41). After centrifugation at 900 × g for 20 min, the leukocytes found at the interface were collected and washed several times with APBS supplemented with 5% FCS. Finally, the cells were pelleted and resuspended in medium containing the mAb 10A9, which is specific for X. laevis IgM, and incubated for 1 h at 4°C. The cells were then centrifuged, washed several times with APBS supplemented with 5% FCS, and resuspended in modified MACS buffer (0.5% BSA/2 mM EDTA in APBS). Goat anti-mouse IgG microbeads (Miltenyi Biotec, Auburn, CA) were added and incubated for 1 h at 4°C. The cells were then spun down and washed several times with modified MACS buffer. 10A9− cells were separated with two rounds of the MACS LS separation column (Miltenyi Biotec). 10A9− cells were collected as “flowthrough” from first round of separation. Total RNA was isolated from cells by using TRIzol (Invitrogen).
qPCR.
qPCR was done by using the Superscript III Platinum SYBR green Two-Step qRT-PCR kit with ROX (Invitrogen), following the manufacturer's recommendation. Approximately 1 μg of total RNA was used for the first-strand cDNA synthesis, and 1/21 of the cDNA were used for each reaction. qPCR was done in three experiments of triplicates in each run (MX3000P instrument; Stratagene). PCR conditions were 50°C for 2 min (to activate Uracil DNA Glycosylase), followed by 95°C for 4 min, 40 cycles of 95°C for 30 sec, 58°C for 30 sec, and 72°C for 30 sec, linked to the cycle for dissociation curve. PCR primers were designed interexonically so that amplification from genomic DNA contamination could be excluded. All primers were tested to determine whether only a single PCR product could be detected on agarose gels as well as only a single peak on the dissociation curve. Primers used for PCR are: IgM C1-C2, 5′-AAC GTT GCC TCT GCA GTC TG-3′, 5′-TTC TTC AAC TCT GAC ACC TTC-3′; IgX C1-C2, 5′-GCT CAG CGA TGT TGA TGG AC-3′, 5′-AAT ACG CAG TTG GCT GCT GG-3′; IgY C1-C2, 5′-CCT TCC TGC ACC AGT AGA TG-3′, 5′-GTG TTC CTT TGG AAC AGA CAC-3′; IgD C8-TM, 5′-AAA AGC ACA GTT AGC ATC GGC-3′, 5′-GAT GTT GTC CAC ACR CTA CTA G-3′; IgM C4-TM, 5′-AAA GGA CAG AAG AGT GGA AAG-3′, 5′-ACG GAG ACT ATT ACA GTA CC-3′; TCRα C-CYT, 5′-GAG GTC CCT GAA TAT GTG TG-3′, 5′-TAT GCT GAC CAG AGT CGT AG-3′; and LMPX exon1-exon2, 5′-AGA TCT GTC TTG GAA CCG TC-3′, 5′-TAT GAG CCA GCT GTA GCA CG-3′. As for the Northern blot, LMPX was used to normalize for RNA amounts. Relative gene expressions of 10A9-positive cells vs. 10A9-negative cells were calculated by using the 2−ΔΔCt method (42, 43).
Supplementary Material
Acknowledgments
We thank Helen Dooley and Mike Criscitiello for critical reading and suggestions. This work is supported by National Institutes of Health Grants RR06603 (to M.F.) and AI27877 (to M.F. and Y.O.).
Abbreviations
- TM
transmembrane
- H
heavy
- C
constant
- L
light
- IgSF
Ig superfamily
- qPCR
quantitative PCR
- cys
cysteines
- APBS
amphibian PBS
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition footnote: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ387453).
References
- 1.Flajnik M. F., Miller K., Du Pasquier L. In: Fundamental Immunology, 5th Ed. Paul W. E., editor. Philadelphia: Lippincott Williams & Wilkins; 2003. pp. 519–570. [Google Scholar]
- 2.Kaattari S., Evans D., Klemer J. Immunol. Rev. 1998;166:133–142. doi: 10.1111/j.1600-065x.1998.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilson M. R., Marcuz A., van Ginkel F., Miller N. W., Clem L. W., Middleton D., Warr G. W. Nucleic Acids Res. 1990;18:5227–5233. doi: 10.1093/nar/18.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preud’homme J. L., Petit I., Barra A., Morel F., Lecron J. C., Lelièvre E. Mol. Immunol. 2000;37:871–887. doi: 10.1016/s0161-5890(01)00006-2. [DOI] [PubMed] [Google Scholar]
- 5.Rowe D. S., Fahey J. L. J. Exp. Med. 1965;121:171–184. doi: 10.1084/jem.121.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler J. E., Sun J., Navarro P. Int. Immunol. 1996;8:1897–1904. doi: 10.1093/intimm/8.12.1897. [DOI] [PubMed] [Google Scholar]
- 7.Wilson M., Bengten E., Miller N. W., Clem L. W., Du Pasquier L., Warr G. W. Proc. Natl. Acad. Sci. USA. 1997;94:4593–4597. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y., Kacskovics I., Pan Q., Liberles D. A., Geli J., Davis S. K., Rabbani H., Hammarström L. J. Immunol. 2002;169:4408–4416. doi: 10.4049/jimmunol.169.8.4408. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi K., Tomonaga S., Kajii T. Mol. Immunol. 1984;21:397–404. doi: 10.1016/0161-5890(84)90037-3. [DOI] [PubMed] [Google Scholar]
- 10.Harding F. A., Amemiya C. T., Litman R. T., Cohen N., Litman G. W. Nucleic Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berstein R. M., Schluter S. F., Shen S., Marchalonis J. J. Proc. Natl. Acad. Sci. USA. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg A. S., Hughes A. L., Guo J., Avila D., McKinney E. C., Flajnik M. F. Eur. J. Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 13.Rumfelt L. L., Diaz M., Lohr R. L., Mochon E., Flajnik M. F. J. Immunol. 2004;173:1129–1139. doi: 10.4049/jimmunol.173.2.1129. [DOI] [PubMed] [Google Scholar]
- 14.Ota T., Rast J. P., Litman G. W., Amemiya C. T. Proc. Natl. Acad. Sci. USA. 2003;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du Pasquier L., Robert J., Courtet M., Mussmann R. Immunol. Rev. 2000;175:201–213. doi: 10.1111/j.1600-065x.2000.imr017501.x. [DOI] [PubMed] [Google Scholar]
- 16.Zapata A, Amemiya C. T. Curr. Top. Microbiol. Immunol. 2000;248:67–107. doi: 10.1007/978-3-642-59674-2_5. [DOI] [PubMed] [Google Scholar]
- 17.Mussmann R., Du Pasquier L., Hsu E. Eur. J. Immunol. 1996;26:2823–2830. doi: 10.1002/eji.1830261205. [DOI] [PubMed] [Google Scholar]
- 18.Stavnezer J., Amemiya C. T. Semin. Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Mussmann R., Courtet M., Schwager J., Du Pasquier L. Eur. J. Immunol. 1997;27:2610–2619. doi: 10.1002/eji.1830271021. [DOI] [PubMed] [Google Scholar]
- 20.Zarrin A. A., Alt F. W., Chaudhuri J., Stokes N., Kaushal D., Du Pasquier L., Tian M. Nat. Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg A. S., Avila D., Hughes M., Hughes A., McKinney E. C., Flajnik M. F. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 22.Dooley H., Flajnik M. F. Eur. J. Immunol. 2005;35:936–945. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 23.Hsu E., Schwager J., Alt F. W. Proc. Natl. Acad. Sci. USA. 1989;86:8010–8014. doi: 10.1073/pnas.86.20.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadji-Azimi I., Parrinello N. Cell. Immunol. 1978;39:316–324. doi: 10.1016/0008-8749(78)90107-7. [DOI] [PubMed] [Google Scholar]
- 25.Liu C. P., Tucker P. W., Mushinski J. F., Blattner F. R. Science. 1980;209:1348–1353. doi: 10.1126/science.6774414. [DOI] [PubMed] [Google Scholar]
- 26.Williams A. F., Barclay A. N. Annu. Rev. Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 27.Kolar G. R., Capra J. D. In: Fundamental Immunology, 5th Ed. Paul W. E., editor. Philadelphia: Lippincott Williams & Wilkins; 2003. pp. 47–68. [Google Scholar]
- 28.Hordvik I., Thevarajan J., Samdal I., Bastani N., Krossøy B. Scand. J. Immunol. 1999;50:202–210. doi: 10.1046/j.1365-3083.1999.00583.x. [DOI] [PubMed] [Google Scholar]
- 29.Hughes A. L. Immunogenetics. 1998;47:283–296. doi: 10.1007/s002510050360. [DOI] [PubMed] [Google Scholar]
- 30.Greenberg A. S., Steiner L., Kasahara M., Flajnik M. F. Proc. Natl. Acad. Sci. USA. 1993;90:10603–10607. doi: 10.1073/pnas.90.22.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson M., Amemiya C., Luer C., Litman R., Rast J., Niimura Y., Litman G. Int. Immunol. 1994;6:1661–1670. doi: 10.1093/intimm/6.11.1661. [DOI] [PubMed] [Google Scholar]
- 32.Stenvik J., Jørgensen T. O. Immunogenetics. 2000;51:452–461. doi: 10.1007/s002510050644. [DOI] [PubMed] [Google Scholar]
- 33.Saha N. R., Suetake H., Kikuchi K., Suzuki Y. Immunogenetics. 2000;56:438–447. doi: 10.1007/s00251-004-0693-y. [DOI] [PubMed] [Google Scholar]
- 34.Bengten E., Quiniou S. M., Stuge T. B., Katagiri T., Miller N. W., Clem L. W., Warr G. W., Wilson M. J. Immunol. 2002;169:2488–2497. doi: 10.4049/jimmunol.169.5.2488. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y., Pan-Hammarström Q., Kacskovics I., Hammarström L. J. Immunol. 2003;171:1312–1318. doi: 10.4049/jimmunol.171.3.1312. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y., Kacskovics I., Rabbani H., Hammarström L. J. Biol. Chem. 2003;278:35024–35032. doi: 10.1074/jbc.M301337200. [DOI] [PubMed] [Google Scholar]
- 37.Lanning D. K., Zhai S. K., Knight K. L. Gene. 2003;309:135–144. doi: 10.1016/s0378-1119(03)00500-6. [DOI] [PubMed] [Google Scholar]
- 38.Bartl S., Baish M. A., Flajnik M. F., Ohta Y. J. Immunol. 1997;159:6097–6104. [PubMed] [Google Scholar]
- 39.Salter-Cid L., Nonaka M., Flajnik M. F. J. Immunol. 1998;160:2853–2861. [PubMed] [Google Scholar]
- 40.Page R. D. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 41.Flajnik M. F., Du Pasquier L. Dev. Biol. 1988;128:198–206. doi: 10.1016/0012-1606(88)90282-5. [DOI] [PubMed] [Google Scholar]
- 42.Fairman J., Roche L., Pieslak I., Lay M., Corson S., Fox E., Luong C., Koe G., Lemos B., Grove R., et al. BioTechniques. 1999;27:566–574. doi: 10.2144/99273rr04. [DOI] [PubMed] [Google Scholar]
- 43.Fang Y., Wu W. H., Pepper J. L., Larsen J. L., Marras S. A., Nelson E. A., Epperson W. B., Christopher-Hennings J. J. Clin. Microbiol. 2002;40:287–291. doi: 10.1128/JCM.40.1.287-291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner B., Miller D. C., Lear T. L., Antczak D. F. J. Immunol. 2004;173:3230–3242. doi: 10.4049/jimmunol.173.5.3230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.