Abstract
In soil ecosystems, bacteria must cope with predation activity, which is attributed mainly to protists. The development of antipredation strategies may help bacteria maintain higher populations and persist longer in the soil. We analyzed the interaction between the root-colonizing and biocontrol strain Pseudomonas fluorescens CHA0 and three different protist isolates (an amoeba, a flagellate, and a ciliate). CHA0 produces a set of antibiotics, HCN, and an exoprotease. We observed that protists cannot grow on CHA0 but can multiply on isogenic regulatory mutants that do not produce the extracellular metabolites. The in vitro responses to CHA0 cells and its exoproducts included growth inhibition, encystation, paralysis, and cell lysis. By analyzing the responses of protists to bacterial supernatants obtained from different isogenic mutants whose production of one or more exometabolites was affected and also to culture extracts with antibiotic enrichment, we observed different contributions of the phenolic antifungal compound 2,4-diacetylphloroglucinol (DAPG) and the extracellular protease AprA to CHA0 toxicity for protists and to the encystation-reactivation cycle. The grazing pressure artificially produced by a mixture of the three protists in a microcosm system resulted in reduced colonization of cucumber roots by a regulatory isogenic CHA0 mutant unable to produce toxins. These results suggest that exometabolite production in biocontrol strain CHA0 may contribute to avoidance of protist grazing and help sustain higher populations in the rhizosphere, which may be a desirable and advantageous trait for competition with other bacteria for available resources.
In order to be able to fully express its potential, an inoculated bacterial species has to adapt to soil conditions and to compete for resources with other microorganisms, but it must also overcome predation. In soil ecosystems, predation pressure can be largely attributed to protists. These organisms constitute a key group at the interface between primary consumption and other trophic levels (3). Protists are now recognized as organisms that play a central role in the N and C cycling in soil (10) and through differential grazing strongly influence the structure of bacterial communities (6, 33). As a response to this pressure, bacteria have developed several adaptations, such as morphological changes, increased motility, membrane properties that make them unattractive, and toxin production (28).
Pseudomonas fluorescens strain CHA0, isolated from the rhizosphere of tobacco in Switzerland, is a biocontrol bacterium that protect roots of various plants against several phytopathogens, including the oomycete Pythium ultimum (21), the tomato fungal pathogen Fusarium oxysporum (43), and the nematode Meloydogine incognita (38). One of the salient features of strain CHA0 is that it produces various extracellular compounds, such as the antibiotics 2,4-diacetylphloroglucinol (DAPG), pyoluteorin (PLT), and pyrrolnitrin (PRN), an extracellular protease (AprA), and the volatile compound hydrogen cyanide (HCN) (17). These exoproducts have been demonstrated to contribute to disease suppression (16, 38). In biocontrol pseudomonads, including strain CHA0, the synthesis of extracellular compounds involved in disease control is influenced by nutrient availability, as well as by signals from bacteria, fungi, and plants (16). In strain CHA0, the genes for biosynthesis of each of the extracellular products are subject to specific transcriptional mechanisms (e.g., O2 control of hcnABC expression for HCN production or autoinduction of phl genes for DAPG synthesis [24, 36]). In addition to the transcriptional regulation, CHA0 controls translation of biocontrol genes through the operation of a signal transduction cascade that involves the GacS/GacA two-component system and an unidentified autoinduction signal (5, 43). Activation of the Gac cascade, which occurs in a cell density-dependent manner, results in rapid accumulation of a triad of small regulatory RNAs (RsmX, RsmY, and RsmZ) that counteract the translational repression of the RsmA and RsmE proteins on biocontrol mRNAs (20, 31, 40). Thus, this global posttranscriptional mechanism with quorum-sensing features ensures that there is coordinate production of antibiotics (DAPG, PLT, and PRN), exoprotease, and HCN, which may constitute a potent cocktail that is toxic for many organisms. Production of extracellular compounds by pseudomonads also affects bacterial (19), fungal (15), and microfaunal communities in general. Strain CHA0 is moderately toxic for Tetrahymena pyriformis (34), and growth of the amoeba Hartmanella vermiformis is inhibited by P. fluorescens DR54, which secretes the biosurfactant viscosinamide (1). In soil microcosm experiments, DAPG-producing strains of P. fluorescens have been shown to influence the number of nematodes and protists in soil experiments (7). Despite the accumulated evidence supporting the hypothesis that strain CHA0 has antagonistic activity against various bacteria and eukaryotes, the exact relevance of the exometabolite production for the fitness of this bacterium is still not known.
The purpose of this work was to examine whether the extracellular compounds produced by P. fluorescens CHA0 play a role in the protection against predation by protists. The Gac regulatory pathway is required by CHA0 to persist in nonsterile soil (26) and to colonize roots (43). As part of an effort to better understand the mechanisms permitting P. fluorescens to remain active, we investigated the abilities of model strain CHA0 and various exoproduct-deficient mutants to resist grazing by different soil protists and explored the roles of the different exometabolites in the inhibition of the potential protistan grazers.
MATERIALS AND METHODS
Organisms and cultivation.
The soil protists and P. fluorescens strains used in this study are listed in Table 1. Protist culture stocks were maintained on a suspension of P. fluorescens CHA19 at a density of 108 CFU ml−1 in modified amoeba saline (AS) (30). To reduce the number of CHA19 cells before experiments, precultures were allowed to encyst, air dried, resuspended in AS, and reactivated in a fresh suspension containing 107 CFU ml−1 of CHA19. P. fluorescens CHA0 and isogenic mutants of this strain were maintained on plates containing nutrient agar (NA) (40 g liter−1 blood agar base, 5 g liter−1 yeast extract). Unless otherwise indicated, bacterial suspensions were prepared from overnight cultures in nutrient yeast broth (25 g liter−1 nutrient broth, 5 g liter−1 yeast extract) grown at 30°C with agitation (200 rpm). One-milliliter aliquots were washed twice in AS, and suspensions were normalized on the basis of the optical density at 600 nm. Bacterial suspensions were plated on NA to check the number of CFU ml−1.
TABLE 1.
Organisms and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Protist isolates | ||
| And12 | Unidentified lobose amoeba morphologically identified as Vahlkampfia sp.; isolated from a pristine soil in Jaén, Spain; GenBank accession no. AY965862 | 23 |
| And31 | Neobodo designis; flagellate isolated from a pristine soil in Jaén, Spain; GenBank accession no. AY965872 | 23 |
| Sp1 | Colpoda steinii; ciliate isolated from a polycyclic aromatic hydrocarbon-contaminated soil in Jaén, Spain; GenBank accession no. DQ388599 | E. Lara (EPFL, Switzerland) |
| Strains of P. fluorescens | ||
| ARQ1 | miniTn7-gfp1 at attTn7 site of CHA0; Kmr | This study |
| ARQ2 | miniTn7-gfp1 at attTn7 site of CHA19; Kmr | This study |
| ARQ3 | miniTn7-gfp1 at attTn7 site of CHA1144; Kmr | This study |
| CHA0 | Wild type; DAPG+ PLT+ PRN+ HCN+ AprA+ | 41 |
| CHA19 | ΔgacS DAPG− PLT− PRN− HCN− AprA− | 43 |
| CHA207 | hcnA′-′lacZ DAPG+ PLT+ PRN+ HCN− AprA+ | 5 |
| CHA631 | ΔphlA DAPG− PLT++ PRN+ HCN+ AprA+ | 36 |
| CHA805 | aprA′-′lacZ DAPG+ PLT+ PRN+ HCN+ AprA− | 38 |
| CHA1012 | pltB::Tn5 Kmr DAPG++ PLT− PRN+ HCN+ AprA+ | 2 |
| CHA1018 | ΔphlA derivative of CHA1012; DAPG− PLT− PRN+ HCN+ AprA+ | 2 |
| CHA1144 | ΔrsmX ΔrsmY ΔrsmZ DAPG− PLT− PRN− HCN− AprA− | 20 |
| Plasmids | ||
| pBK-miniTn7-gfp1 | pUC19-based delivery plasmid for miniTn7-gfp1; Kmr Cmr Aprmob+ | 22 |
| pUX-BF13 | Helper plasmid for Tn7-based transposon mutagenesis containing the transposition functions; R6K replicon; Apr | 4 |
PLT++ and DAPG++, higher levels of PLT and DAPG, respectively, than the levels in CHA0.
GFP tagging of P. fluorescens strains.
A single copy of the gene encoding green fluorescent protein (GFP) was introduced at a neutral chromosomal site into P. fluorescens strains CHA0, CHA19, and CHA1144 using a mini-Tn7 delivery system. Briefly, P. fluorescens cells were electrotransformed with ca. 1 μg of the gfp shuttle vector pBK-miniTn7-gfp1 (22) and ca. 1 μg of the Tn7 helper vector pUX-BF13 (4). Transformants were selected on NA plates containing 25 μg ml−1 of kanamycin, and expression of gfp was confirmed by using an epifluorescence microscope (Nikon Alphaphot-2). Introduction of gfp at the attTn7 chromosomal locus did not modify the observed effects of P. fluorescents strains on the protists used in this study.
Cocultivation of protists and bacteria.
Cocultivation experiments were performed using a factorial design with three protist isolates and seven P. fluorescens strains in 96-well microtiter plates, with three replicates per treatment. In each well, 100 μl of a 107-CFU ml−1 P. fluorescens suspension prepared in AS was mixed with a suspension of active protists. The plates were covered with lids and incubated at 17°C with gentle agitation (50 rpm). For each well, four to eight fields were analyzed daily for 5 days. Before each measurement, the suspensions were gently homogenized with micropipette tips. In all cases, protists were counted using an inverted microscope (Nikon MTS; magnification, ×200). Encysted, dead, and active organisms were counted separately.
Short-term responses of active Vahlkampfia sp. and Colpoda steinii to bacterial supernatants.
Suspensions of P. fluorescens strains (108 CFU ml−1 in AS) were incubated for 72 h at room temperature and then centrifuged (14,000 rpm, 10 min) to remove most of the cells. Ten microliters of each supernatant was added to a 90-μl suspension of active Vahlkampfia sp. or C. steinii cells. The numbers of active and encysted organisms were determined every hour after addition of supernatant using an inverted microscope.
Inhibition of cyst reactivation by CHA0 exoproducts.
Approximately 100 cysts of each of the three protist isolates suspended in the presence of 107 CFU ml−1 of strain CHA19 in microtiter plates were treated with a dilution series of supernatants prepared from different P. fluorescens strains as described above. The highest supernatant concentration (50%) was that of a 1:1 (vol/vol) mixture of a cyst-CHA19 suspension and culture supernatant. After 24 h of incubation, the presence of active protist cells was determined for each treatment, and the highest supernatant concentration that permitted cyst reactivation was recorded for each strain.
Antibiotic extraction and effects of antibiotic extracts on protist isolates.
Extracts in which antibiotics (DAPG, PLT, and PRN) were enriched were prepared from P. fluorescens cultures as previously described (21). Strains CHA631, CHA1012, and CHA1018 were grown for 40 h in 20 ml of medium containing 7.5 g liter−1 malt extract, 10 g liter−1 protease peptone, 750 mg liter−1 MgSO4, and 750 mg liter−1 K2HPO4 in 125-ml flasks at 30°C with agitation (200 rpm). Five milliliters from each culture was acidified with 250 μl of 2 N HCl and extracted with 5 ml of ethyl acetate. After centrifugation (10 min, 5,000 rpm) 4 ml of the organic phase was evaporated in a rotavapor at room temperature. The dry residue was resuspended in 200 μl methanol and stored at −20°C. In order to investigate the relative effect of each antibiotic, a suspension of active protists (100 μl) was mixed with 2-μl portions of the antibiotic extracts diluted 1:1, 1:10, or 1:100 with methanol. Each treatment was done in triplicate. Controls received 2 μl of methanol. Protists were examined using an inverted microscope after 15 min, 1 h, and 12 h to screen for acute intoxication.
Microcosm experiment.
Cucumber (Cucumis sativa cv. Poinset 76) seeds were surface sterilized in a diluted bleach solution (1.1% active Cl) for 10 min, washed six times with sterile H2O for 10 min, and germinated on 1.5% water agar at 28°C in the dark. Microcosms were prepared by filling 60-ml glass tubes with 2.5 g vermiculite and 10 ml AS and were autoclaved before use. Seeds were transferred to microcosms and inoculated using a 2 × 2 factorial design (10 replicates per treatment) with 108 cells of P. fluorescens strain ARQ1 or ARQ2 and, when required, with 104 cysts of C. steinii, Vahlkampfia sp., and Neobodo designis. Equivalent numbers of cysts (3 × 103 cysts) of the three species were added. Aliquots of cyst suspensions were plated on NA to check for the presence of remaining bacteria, which indicated that less than 100 CFU was introduced into each microcosm. Seedlings were grown in a growth chamber by using 16 h of light at 24°C followed by 8 h of darkness at 20°C. Microcosms were randomized three times during the experiment. At 6 days after inoculation, seedlings were separated from the matrix and gently washed in a sterile isotonic saline solution (0.9% NaCl) to release the vermiculite particles, and roots were detached from shoots to determine the fresh weight. Weighed roots were strongly agitated (ca. 300 rpm for 30 min) in 10 ml of a saline solution to recover bacteria from the rhizoplane, and suspensions were plated on NA with 25 μg ml−1 kanamycin to estimate root colonization (CFU g−1).
RESULTS
Extracellular products of P. fluorescens CHA0 inhibit growth of protists.
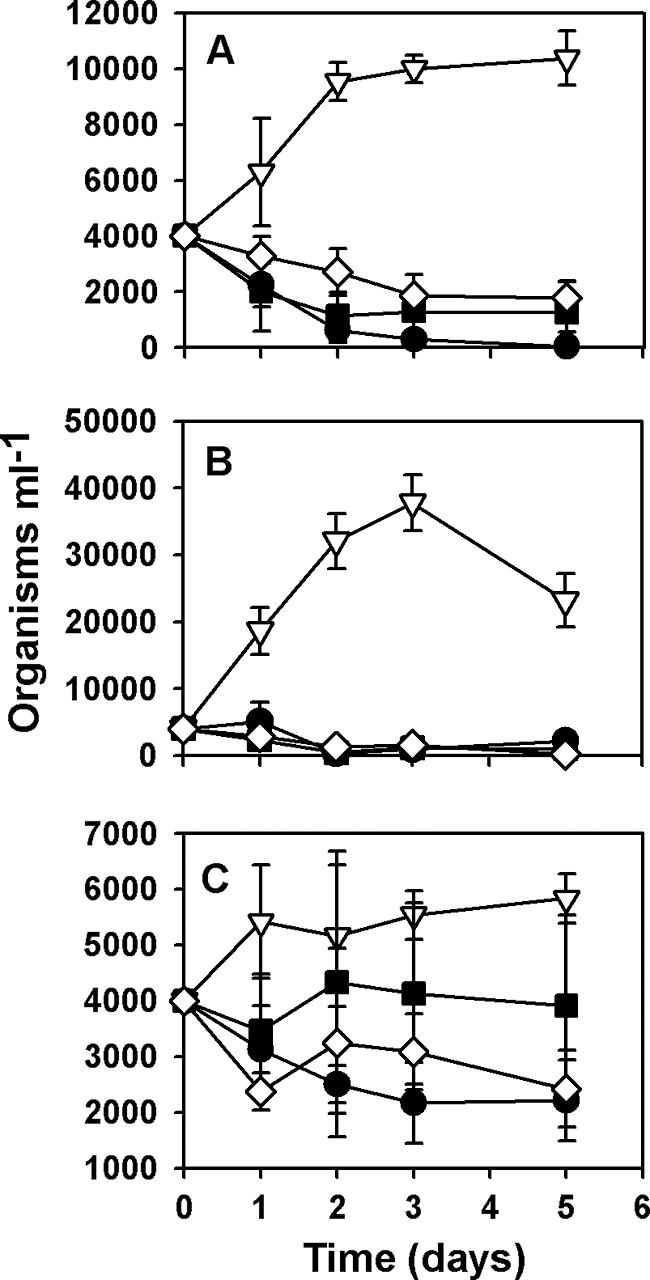
P. fluorescens strain CHA0 and several isogenic mutants with mutations affecting the production of one or several extracellular products were offered as a prey to the amoeba Vahlkampfia sp. strain And12, the flagellate N. designis And31, and the ciliate C. steinii Sp1. None of these organisms was able to grow on CHA0 cells in the experimental conditions used for cocultivation (Fig. 1), and the organisms rapidly encysted or died (although a few active C. steinii cells could be observed in all experiments). However, when the protists were cultivated with CHA19 cells, which lacks the membrane sensor protein GacS required for concerted activation of antibiotic, HCN, and extracellular protease synthesis (43), marked differences in their growth responses to this mutant strain were observed (Fig. 1). The size of the population of the flagellate N. designis increased up to 10-fold after 3 days, and active populations were maintained until the end of the experiment when the organisms were feeding on CHA19. For Vahlkampfia sp. there was a moderate increase in the size of the population (ca. 250%), and the organisms finally encysted with CHA19 as food. The ciliate C. steinii grew modestly with CHA19, and the final increase in the cell number was 50% (Fig. 1). These results suggest that CHA0 extracellular compounds have a role in the inhibition of protist development. Strain CHA1144 is a regulatory mutant lacking the small RNA triad required for GacS-dependent activation of mRNAs for exoproduct formation (20). The effect of CHA1144 on protist growth was similar to that of CHA19 (Table 2); however, CHA1144 was not as innocuous as CHA19 as the growth of each protist isolate was systematically better with CHA19 than with CHA1144 as prey (Table 2).
FIG. 1.
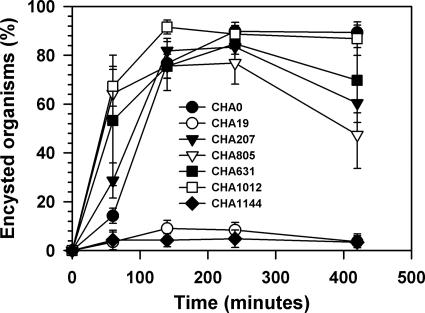
Growth of the protists N. designis And31 (A), Vahlkampfia sp. strain And12 (B), and C. steinii Sp1 (C) on P. fluorescens CHA0 and different exoproduct-deficient mutants. •, CHA0; ▿, CHA19; ⋄, CHA631; ▪, CHA805. The values are means ± standard errors.
TABLE 2.
Responses of the protists Vahlkampfia sp. strain And12, N. designis And31, and C. steiniiSp1 to P. fluorescens CHA0 isogenic mutants whose production of extracellular compounds is affecteda
| Strain | Known extracellular productsb | % Increase in cell no.c
|
||
|---|---|---|---|---|
| Vahlkampfia sp. strain And12 | N. designis And31 | C. steinii Sp1 | ||
| None | 12 | 225 | 1 | |
| CHA0 | DAPG, PLT, PRN, HCN, AprA | −99 | −77 | −45 |
| CHA19 | None of the CHA0 products | 159*** | 480*** | 46*** |
| CHA1144 | None of the CHA0 products | 130*** | 161*** | −18* |
| CHA207 | DAPG, PLT, PRN, AprA | −99NS | −92* | −8* |
| CHA631 | PLT++, PRN, HCN, AprA | −69* | −76* | −2* |
| CHA805 | DAPG, PLT, PRN, HCN | −55*** | −95*** | −40NS |
| CHA1012 | DAPG++, PRN, HCN, AprA | −89NS | −87*** | −9NS |
The numbers of protists (active cells and cysts) were determined after 5 days of cocultivation with P. fluorescens strains.
PLT++ and DAPG++, higher levels of PLT and DAPG, respectively, than the levels in CHA0.
Increase relative to the initial number of organisms. *, P < 0.05; ***, P < 0.01; NS, not statistically significantly different from CHA0.
In order to investigate the relative contribution of each of the known extracellular toxins produced by CHA0 to the inhibition of protist development, we analyzed the growth responses of Vahlkampfia sp., N. designis, and C. steinii fed different mutants derived from strain CHA0 (Fig. 1 and Table 2). Vahlkampfia sp. encysted or died when it was fed all mutants, but at different rates (Fig. 1A and Table 2). The lack of exoprotease AprA or DAPG synthesis seemed to reduce bacterial toxicity for Vahlkampfia sp. (Fig. 1A and Table 2). Addition of the PLT mutant and DAPG-overproducing strain CHA1012 resulted in a higher death rate than addition of CHA0 resulted in (data not shown), although the final number of organisms was not different from the final number obtained with the CHA0 treatment (Table 2). After 5 days of cocultivation, active amoebae were observed only with CHA805 (AprA−) or CHA631 (DAPG−) as prey. N. designis died rapidly and at similar rates in the presence of all mutants tested except the exoproduct-deficient strains CHA1144 and CHA19 (Fig. 1B and Table 2). By contrast, the ciliate C. steinii seemed to be better able to resist the different bacterial toxins as the decrease in the number of active organisms was markedly lower with all the mutants studied (Fig. 1C and Table 2). Nevertheless, DAPG and HCN together appeared to contribute to CHA0 toxicity for this ciliate (Table 2).
Short-term responses of active protist cells to P. fluorescens exoproducts.
Active Vahlkampfia cells were challenged with supernatants from cultures of the different P. fluorescens mutants. The amoebae were rapidly induced to encyst in the presence of supernatants from cultures of wild-type or mutant P. fluorescens strains that did not synthesize either AprA, HCN, DAPG, or PLT (Fig. 2). After 2 h of treatment, more than 75% of the amoebae had encysted. However, Vahlkampfia cysts were able to partially reactivate after 6 h of treatment in the presence of supernatants that did not contain AprA or HCN (Fig. 2). Interestingly, the supernatants from wild-type strain CHA0 and from the DAPG-overproducing strain CHA1012 did not allow cysts to reactivate (Fig. 2). Supernatants from nontoxic strains CHA1144 and CHA19 did not affect amoeba viability during the experiment (Fig. 2), which is consistent with our previous observation that CHA19 and CHA1144 are good sources of food for Vahlkampfia sp. (Fig. 1). Surprisingly, the flagellate And31 was hardly affected by bacterial supernatants, and we did not detect any significant effect on protist behavior compared to the control treatment (data not shown). Thus, it was not possible to discriminate between the contributions of the secondary metabolites. Similarly, the ciliate C. steinii was not affected at all by the different supernatants, suggesting that the inhibition of flagellate and ciliate growth (Fig. 1) may be due to ingested bacteria and not to dissolved metabolites.
FIG. 2.
Influence of exoproducts present in bacterial supernatants on the encystment behavior of Vahlkampfia sp. strain And12.
Predation of GFP-tagged P. fluorescens cells.
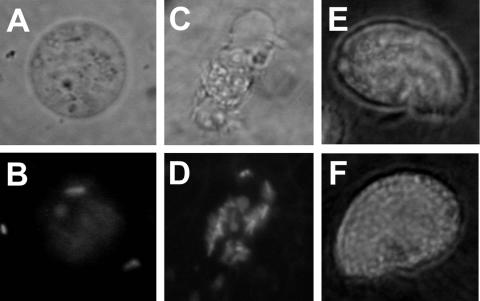
In order to correlate the growth responses of protists to P. fluorescens cells with cellular responses, we monitored the fate of GFP-tagged bacteria offered to each of the organisms as prey. After 30 min of contact with strain ARQ1 (i.e., GFP-tagged CHA0), the amoeba Vahlkampfia became round, was immobile, and contained a few ingested bacterial cells, which resulted in faint fluorescence (Fig. 3A and B). By contrast, amoebae fed ARQ2 (i.e., GFP-tagged CHA19) had a normal amoeboid shape, normal cellular activity, and strong fluorescence concentrated in vacuoles, and they contained several intact fluorescent bacterial cells (Fig. 3C and D). These results indicate that Vahlkampfia may loose its viability after direct contact with CHA0 toxins or after ingestion of a few bacterial cells. In agreement with the moderate response of the ciliate C. steinii to CHA0 cells (Fig. 1), there was not a significant difference in GFP labeling of ciliate cells after brief exposure to strains ARQ1 and ARQ2; in both cases, the protists were found to contain many fluorescent cells after 30 min of contact (Fig. 3). For the flagellate N. designis fluorescence or fluorescent bacterial cells were not detected in cells when the organism was fed either ARQ1 or ARQ2. It is possible that the level of fluorescence was too low to be detected, as this organism ingests only a few bacteria (18) and might have a high turnover rate. The results obtained with ARQ3 (i.e., GFP-tagged CHA1144) were similar to those obtained with ARQ2 (data not shown), and there was not any evident difference in the behavior of protists with either disarmed strain.
FIG. 3.
Ingestion of GFP-tagged P. fluorescens cells by protists. Protists were offered ARQ1 or ARQ2 cells and examined with a microscope after 30 min. (A) Severely affected round Vahlkampfia sp. strain And12 cell fed ARQ1 cells (white light; magnification, ×400). (B) Same cell as the cell in panel A viewed with UV light. A few fluorescent ARQ1 cells are present (magnification, ×400). (C) Active amoeboid Vahlkampfia sp. strain And12 cell fed ARQ2 cells (white light; magnification, ×400). (D) Same cell as the cell in panel C viewed with UV light. Many fluorescent ingested ARQ2 cells and digestive vacuoles are present (magnification, ×400). (E) Active C. steinii Sp1 cell full of ARQ1 cells or their cellular contents (UV light; magnification, ×400). (F) Active C. steinii Sp1 cell full of ARQ2 cells or their cellular contents (UV light; magnification, ×400).
DAPG and PLT together inhibit cyst reactivation.
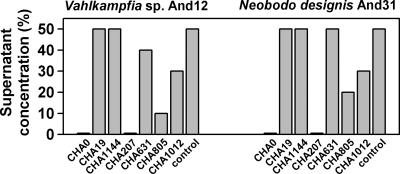
Encystation is a typical protist response that has been suggested to be a cell density-dependent phenomenon (14), whereas exit from this dormant state (i.e., cyst reactivation) is thought to be induced by the presence of food. We studied reactivation of Vahlkampfia sp. and N. designis cysts cultivated with CHA19, which was demonstrated to be a good source of food (Fig. 1), in the presence of different P. fluorescens extracellular products provided in culture supernatants. The two protists exhibited comparable responses to supernatants from different P. fluorescens isogenic mutants, although the flagellate was slightly less sensitive than the amoeba to the extracellular compounds (Fig. 4). In line with results described above (Fig. 1 and 2), CHA0 supernatant did not allow cyst reactivation, whereas supernatants from CHA19 or CHA1144 permitted reactivation of cysts of both protist isolates (Fig. 4). Of the known CHA0 exoproducts regulated by Gac, DAPG was the most potent inhibitor of cyst reactivation (Fig. 4). PLT was less important than DAPG, and the exoprotease AprA had only a minor role in the inhibition of cyst reactivation (Fig. 4). Finally, the absence of HCN production did not modify the response of protist cysts.
FIG. 4.
Inhibition of reactivation of Vahlkampfia sp. strain And12 and N. designis And31 cysts by culture supernatants of CHA0 and related mutants. The bars indicate the highest concentrations of culture supernatants present in the assays that allowed cyst reactivation. A value of 50% corresponds to a 1:1 (vol/vol) mixture of protist cysts and culture supernatant.
Toxicity of antibiotics for active protists.
Extracts in which antibiotics produced by CHA0 (DAPG, PLT, and PRN) were enriched were prepared from CHA1012 and CHA631 cultures. Strain CHA1018 was used as a negative control for DAPG and PLT effects. As DAPG and PLT mutually exert negative transcriptional cross-regulation (8, 36), extracts of strain CHA1012 contain more DAPG than extracts of CHA0 contain, whereas the opposite is true for extracts of CHA631 (36). When Vahlkampfia sp. and C. steinii active cells were exposed to a DAPG-enriched extract of CHA1012, they exhibited effects of acute toxicity, which resulted in rapid cell lysis within 1 h after exposure (Fig. 5). Loss of motility was observed within minutes. These two protists had different responses to DAPG; 10-fold less extract was required for Vahlkampfia to produce the response that was observed for ciliates. Extracts lacking DAPG, such as extracts of CHA631 (enriched in PLT) and CHA1018 (no DAPG or PLT), induced encystation of Vahlkampfia cells, but lysis was not observed. This response suggests that PRN may have a role as an inducer of encystation, although the possibility that another, unidentified extracted compound was responsible for this effect cannot be discarded. On the other hand, addition of the PLT-enriched CHA631 extract to C. steinii cells also resulted in rapid death of a minor proportion of the organisms (ca. 10%), whereas the rest of the organisms had a distorted morphology, similar to the morphology observed when the organisms were exposed to a hypertonic solution. Observation after 12 h of exposure to a PLT-containing extract indicated that the organisms that survived had normal morphology and viability. The CHA1018 extract did not have any effect on C. steinii. The flagellate N. designis exhibited a high level of sensitivity even to the CHA1018 extract (containing no DAPG or PLT) but not to methanol, so it was not possible to discriminate separate effects of DAPG, PLT, and PRN.
FIG. 5.
Effect of a DAPG-enriched culture extract on C. steinii Sp1 cells. Magnification, ×400.
Microcosm experiment.
Because of our in vitro observations that protist isolates feed on exoproduct-deficient P. fluorescens strains but have difficulty coping with wild-type exoproduct-producing strain CHA0 (Fig. 1) and because of previous reports of the low fitness and reduced colonization of roots in natural soil observed for CHA0 mutants not producing extracellular toxins (20, 26, 29), we examined the abilities of P. fluorescens strains ARQ1 (gfp-tagged CHA0 derivative) and ARQ2 (gfp-tagged exoproduct-deficient derivative of CHA19) to colonize the rhizoplane of cucumber seedlings in an axenic microcosm experiment under grazing pressure created by introduction of a mixture of the three protist isolates used in this study (Table 3). Strain ARQ1 performed as well in the presence of protists (8.7 × 108 CFU g−1) as in the absence of protists (7.9 × 108 CFU g−1). On the other hand, ARQ2 was slightly handicapped for colonization of seedling roots compared to wild-type strain ARQ1 (5.2 × 108 CFU g−1). However, introduction of the protist population into ARQ2 microcosms resulted in a further significant decrease (ca. 56%) in the colonization rate (2.3 × 108 CFU g root−1) (Table 3). A pairwise multiple-comparison test used after a two-way analysis of variance indicated that the effect of the gacS mutation on cucumber colonization depended on the presence of the protist mixture as there was a statistically significant interaction between these two factors (P = 0.0004).
TABLE 3.
Colonization of cucumber roots by P. fluorescens in the presence of protist predation pressure
| P. fluorescens strain | Log CFU g−1a
|
|
|---|---|---|
| Without protists | With protists | |
| ARQ1 (wild type) | 7.90 ± 0.18 a | 7.94 ± 0.13 a |
| ARQ2 (gacS; no exoproducts) | 7.72 ± 0.17 b | 7.36 ± 0.08 c |
The logarithm of raw data was used to perform a two-way analysis of variance (n = 8 or 9). Different letters after values indicate that treatment averages ± standard errors were significantly different, as determined by a pairwise multiple-comparison Student-Newman-Keuls test (P < 0.05). The experiment was repeated once with essentially the same results.
DISCUSSION
Toxicity of CHA0 for soil protists.
Selective grazing pressure has led to the development of various adaptation mechanisms in bacteria, such as increased motility and changes in cell wall and membrane properties (28). Toxin production has recently been recognized as an additional mechanism leading to enhanced persistence of bacteria in soil (27, 28). Clearly, a strain like P. fluorescens CHA0 that synthesizes a set of toxic extracellular compounds does not represent a good source of food for protists belonging to different taxa, such as the flagellate N. designis And31, the amoeba Vahlkampfia sp. strain And12, and the ciliate C. steinii Sp1 (Fig. 1). CHA0 has also been demonstrated to have a toxic effect on the protist Tetrahymena pyriformis (34). A single mutation in the membrane-bound sensor gacS that abolished exoproduct formation (43) transformed strain CHA19 into an edible organism, enabling protists to grow on the disarmed cells (Fig. 1). Strain CHA19 does not produce the antibiotics DAPG, PLT, and PRN, HCN, or the exoprotease AprA. It is also possible that another unidentified extracellular metabolite is also absent in CHA19 cultures. This is suggested by the low, but still residual, toxicity of CHA1144 compared to the toxicity of CHA19 (Table 2). In any case, these strains are not able to synthesize several compounds. In order to obtain insight into the relative contribution of each of these products to protist toxicity, we used isogenic mutants derived from CHA0 (Table 1), which allowed more accurate identification of differences in grazing efficiency due to the lack of one of the exometabolites, minimizing the possible artifacts that may arise from comparisons of different but related strains distinguished by their abilities to produce antibiotics (7). Although studying the roles of metabolites by subtraction results in relatively high background noise due to possible remaining toxic compounds, it was possible to observe differential and stronger effects of CHA631 (which lacks DAPG) and CHA805 (which lacks the exoprotease AprA) on amoebae, whereas the other single mutants induced similar protist responses (Table 2 and Fig. 2). The marked inhibition of cyst reactivation (Fig. 4) and the effects on active protist cells of culture extracts enriched in the antibiotics DAPG and PLT (Fig. 5) confirmed the relatively strong toxicity of DAPG. This effect of DAPG is not surprising, as this compound is known to be required for suppression of root disease by P. fluorescens (21) and it has been reported to affect a wide range of organisms, such as the plant pathogen Pythium (12), protozoa (34), and nematodes (7). The wide range of organisms that seem to be susceptible to DAPG suggests that this low-molecular-weight phenolic derivative may interfere with basic cellular processes, such as electron transport or membrane permeability. The role of the CHA0 extracellular protease has been characterized even less. It has been reported that the lack of the exoprotease results in reduced biocontrol activity of CHA0 against nematodes (38). The exoprotease and DAPG also seem to complement each other, increasing the biocontrol activity against Pythium (13). It is likely that key membrane proteins, such as those involved in transport or signal transduction, are targeted by the AprA protease and result in abnormal cellular responses. Based on our results, the effect of HCN on the studied protists seems to be minor. Bacteria unable to synthesize HCN were less aggressive only with the ciliate C. steinii in cocultivation experiments (Table 2), and culture supernatants of HCN− cells allowed partial reactivation of Vahlkampfia sp. cysts only during short-term incubation (Fig. 2). Due to the volatile nature of HCN and perhaps to the low rate of production under the experimental conditions used here, HCN toxicity may have been obscured by the effects of the other exometabolites.
Acute toxicity after cell ingestion or intoxication with extracellular products.
Acute toxicity for protists during ingestion has been reported for violacein-producing bacteria (27). Our experiments revealed great differences in the sensitivities to P. fluorescens exoproducts of the protist species tested in this study. The amoeba Vahlkampfia sp. strain And12 was very sensitive to dissolved exometabolites (Fig. 2), whereas the flagellate N. designis And31 was almost not affected by P. fluorescens supernatants. By contrast, we observed a very rapid death of flagellates when they were fed the same bacteria (Fig. 1). This suggests that the mechanisms of toxicity may differ depending on the predator. The acute toxicity of ingested cells for the flagellate N. designis may be due to the production of toxins inside the predator, as proposed by other workers (27). The ciliate C. steinii also exhibited strong resistance to the exometabolites present in culture supernatants and was able to survive even with the toxin-producing strain CHA0 as a food source, although at lower densities. Interestingly, this ciliate isolate has been obtained from a heavily contaminated soil (23). C. steinii belongs to the order Colpodea, in which typical colonizer strains that readily adapt to adverse environments are found (25).
Indeed, during cocultivation experiments toxicity may arise as a combined effect of bacterial ingestion and the progressive release of bacterial toxins in the culture medium. By feeding the protists gfp-tagged bacteria, we observed that the ciliate C. steinii did not show a preference for the toxin producer (ARQ1) or an innocuous strain (ARQ2 or ARQ3), whereas the amoeba did not feed on ARQ1 (gfp-tagged wild-type CHA0) or died rapidly after ingestion of a few cells (Fig. 3). This confirms that C. steinii is resistant to CHA0 exometabolites and illustrates that CHA0 products inhibit predation by Vahlkampfia sp. The mechanisms for this inhibition still must be investigated. The difference observed in resistance to CHA0 extracellular products may arise from the fact that some protists absorb nutrients osmotrophically, which may explain their greater susceptibility to dissolved antibiotics. These organisms can be cultivated axenically, like the heterolobosean Naegleria (37), which is phylogenetically related to Vahlkampfia sp. strain And12 (23). Other protists are strictly phagotrophic, and they are likely to absorb the bacterial toxins only after bacterial ingestion.
Effects of exometabolites on protist encystation and cyst reactivation.
All known CHA0 exoproducts induced rapid encystation of the amoeba Vahlkampfia sp. strain And12 (Fig. 2). Notably, the absence in culture supernatants of AprA (CHA805) or enhanced levels of DAPG (CHA1012) and PLT (CHA631) resulted in faster encystation of Vahlkampfia cells (Fig. 2), as if the prolonged half-life of a putative membrane target for DAPG and/or PLT made amoebae more sensitive to these antibiotics. Once Vahlkampfia cells were encysted, incubation with various supernatants and edible food (CHA19) showed that reactivation of Vahlkampfia cysts and cysts of the flagellate N. designis was inhibited at low concentrations of dissolved exometabolites (Fig. 4). One of these exometabolites, DAPG, was particularly responsible for sustaining the dormant state (Fig. 2 and 4). It is interesting that the AprA-deficient CHA805 culture supernatant allowed amoebal cysts to recover faster than cysts recovered after they were exposed to supernatants from other P. fluorescens strains (Fig. 2). This is consistent with the lower level of susceptibility determined for the same organism feeding on active bacteria (Table 2). If cyst reactivation requires the activity of another membrane protein that may be targeted by AprA, the absence of the exoprotease would stimulate cyst reactivation. Thus, AprA activity may perturb amoebal membrane receptors, thus resulting in an altered ability to sense environmental or chemical signals and modification of the encystation-reactivation cycle.
Extracellular compounds may prevent predation of P. fluorescens CHA0 by protists in the rhizosphere.
P. fluorescens CHA0 isogenic mutants that do not produce exometabolites have reduced colonization ability when they are inoculated into a natural soil (20), but no difference from wild-type CHA0 was observed in sterile microcosms (43). In this study we analyzed one of the possible interactions between P. fluorescens and other soil organisms, i.e., the susceptibility to predation by protists, as protists have been found to determine the survival of inoculated bacteria (32, 39). Under our experimental conditions, the production of extracellular compounds appears to help bacteria avoid predation (Table 3). The ability of exometabolite-producing strain ARQ1 to colonize was not affected by the presence of protists, but the disarmed strain ARQ2 could not produce a comparable cell density on cucumber roots when protists were introduced into the microcosms. As described above, our cocultivation experiments suggested that toxin production has a different impact on protists if the toxins are dissolved in the surrounding media or if they originate from eaten bacteria. However, due to the GacS/GacA regulation of exoproduct synthesis in strain CHA0 (20, 43), toxin production is maximal when bacteria colonize the roots and a certain cell density is reached. Such a mechanism protects the whole bacterial population but leaves individual bacteria vulnerable to predation during the precolonization phase. The results of the microcosm experiment are less marked than the results obtained with natural soil (20). We attribute this to the fact that the inoculated bacteria did not face competition with other rhizosphere bacteria. Thus, even if grazed, ARQ2 is still able to profit from nutrients present in root exudates without being outcompeted by other bacterial species inhabiting natural soil.
Toxin production as a mechanism to avoid grazing pressure.
In pseudomonads, the GacS/GacA regulatory system contributes to adaptation to a wide range of environmental stresses (35), and it has been shown to increase survival in natural soil (9, 29). Our results suggest that exoproduct synthesis is part of CHA0 adaptation and fitness in natural soil. Until now, the effects of exoproducts of biocontrol organisms on protists have been considered chiefly secondary effects (1, 7). However, here we stress that the production of these compounds may also be part of an active defense of the bacteria against predation, as demonstrated with the biofilm-forming organism Pseudomonas aeruginosa (42) and the soil bacterium Chromobacterium violaceum (27). Since it is a biocontrol species, it seems logical to hypothesize that P. fluorescens CHA0 evolved to produce a battery of exoproducts to fight off plant parasites. We analyzed the effects of P. fluorescens CHA0 on three different soil protists having different feeding strategies. For the flagellate and the ciliate, but not for the amoeba, it seems that it is ingestion of CHA0 cells that poisons the grazer, but this does not save the ingested cells. This may be viewed as a defense mechanism to save the population at the expense of individual bacterial cells. We should remember that the suite of CHA0 biocontrol factors is produced mainly as a result of activation of a sensory system that responds to a quorum-sensing-like signal (5, 43), which occurs at high cell densities, such as those attained in the rhizosphere. Thus, the “sacrifice” of some bacteria, which kill the predator while they are being digested, can be considered an expression of kin selection (11) that can also contribute to reduced grazing pressure on related disarmed bacteria sharing the same niche. Antipredator strategies, such as that displayed by strain CHA0 against different soil protists, may contribute to biocontrol activity as it may help to maintain larger populations in the rhizosphere of colonized plant species.
Acknowledgments
We are grateful to Dieter Haas and Christoph Keel (DMF, Université de Lausanne, Switzerland) for providing P. fluorescens strains. C.V. and L.G.W. are members of CONICET (Argentina).
REFERENCES
- 1.Andersen, K. S., and A. Winding. 2004. Non-target effect of bacterial biological control agents on soil protozoa. Biol. Fertil. Soils 40:230-236. [Google Scholar]
- 2.Baehler, E., M. Bottiglieri, M. Pechy-Tarr, M. Maurhofer, and C. Keel. 2005. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99:24-38. [DOI] [PubMed] [Google Scholar]
- 3.Bamforth, S. S. 1988. Interactions between protozoa and other organisms. Agric. Ecosyst. Environ. 24:229-234. [Google Scholar]
- 4.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 5.Blumer, C., S. Heeb, G. Pessi, and D. Haas. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 96:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonkowski, M. 2004. Protozoa and plant growth: the microbial loop in soil revisited. New Phytol. 162:617-631. [DOI] [PubMed] [Google Scholar]
- 7.Brimecombe, M. J., F. A. A. M. De Leij, and J. M. Lynch. 2000. Effect of introduced Pseudomonas fluorescens strains on nematode and protozoan populations in the rhizosphere of wheat and pea. Microb. Ecol. 38:387-397. [DOI] [PubMed] [Google Scholar]
- 8.Brodhagen, M., M. Henkels, and J. E. Loper. 2004. Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 70:1758-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chancey, S. T., D. W. Wood, E. A. Pierson, and L. S. Pierson III. 2002. Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. Appl. Environ. Microbiol. 68:3308-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coûteaux, M.-M., and J. F. Darbyshire. 1998. Functional diversity amongst soil protozoa. Appl. Soil Ecol. 10:229-237. [Google Scholar]
- 11.Denison, R. F., C. Bledsoe, M. Kahn, F. O'Gara, E. L. Simms, and L. S. Thomashow. 2003. Cooperation in the rhizosphere and the “free rider” problem. Ecology 84:838-845. [Google Scholar]
- 12.De Souza, J. T., C. Arnould, C. Deulvot, P. Lemanceau, V. Gianinazzi-Pearson, and J. M. Raaijmakers. 2003. Effect of 2,4-diacetylphloroglucinol on Pythium: cellular responses and variation in sensitivity among propagules and species. Phytopathology 93:966-975. [DOI] [PubMed] [Google Scholar]
- 13.Dunne, C., Y. Moenne-Loccoz, F. J. de Bruijn, and F. O'Gara. 2000. Overproduction of an inducible extracellular serine protease improves biological control of Pythium ultimum by Stenotrophomonas maltophilia strain W81. Microbiology 146:2069-2078. [DOI] [PubMed] [Google Scholar]
- 14.Ekelund, F., H. B. Frederiksen, and R. Rønn. 2002. Population dynamics of active and total ciliate populations in arable soil amended with wheat. Appl. Environ. Microbiol. 68:1096-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girlanda, M., S. Perotto, Y. Moenne-Loccoz, R. Bergero, A. Lazzari, G. Défago, P. Bonfante, and A. Luppi. 2001. Impact of biocontrol Pseudomonas fluorescens CHA0 and a genetically modified derivative on the diversity of culturable fungi in the cucumber rhizosphere. Appl. Environ. Microbiol. 67:1851-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 17.Haas, D., C. Keel, and C. Reimmann. 2002. Signal transduction in plant-beneficial rhizobacteria with biocontrol properties. Antonie Leeuwenhoek 81:385-395. [DOI] [PubMed] [Google Scholar]
- 18.Jezbera, J., K. Hornak, and K. Simek. 2005. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol. Ecol. 52:351-363. [DOI] [PubMed] [Google Scholar]
- 19.Johansen, J. E., S. J. Binnerup, K. B. Lejbølle, F. Mascher, J. Sorensen, and C. Keel. 2002. Impact of biocontrol strain Pseudomonas fluorescens CHA0 on rhizosphere bacteria isolated from barley (Hordeum vulgare L.) with special reference to Cytophaga-like bacteria. J. Appl. Microbiol. 93:1065-1074. [DOI] [PubMed] [Google Scholar]
- 20.Kay, E., C. Dubuis, and D. Haas. 2005. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl. Acad. Sci. USA 102:17136-17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keel, C., U. Schnider, M. Maurhofer, C. Voisard, J. Laville, U. Burger, P. Wirthner, D. Haas, and G. Défago. 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant-Microbe Interact. 5:4-13. [Google Scholar]
- 22.Koch, B., L. Jensen, and O. Nybrøe. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187-195. [DOI] [PubMed] [Google Scholar]
- 23.Lara, E., C. Berney, F. Ekelund, H. Harms, and A. Chatzinotas. 7 August 2006, posting date. Molecular comparison of cultivable protozoa from a pristine and a polycyclic aromatic hydrocarbons polluted site. Soil Biol. Biochem. [Online.] doi: 10.1016/j.soilbio.2006.06.017. [DOI]
- 24.Laville, J., C. Blumer, C. von Schroetter, V. Gaia, and G. Défago. 1998. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 180:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luftenegger, G., W. Foissner, and H. Adam. 1985. R-selection and K-selection in soil ciliates—a field and experimental approach. Oecologia 66:574-579. [DOI] [PubMed] [Google Scholar]
- 26.Mascher, F., Y. Moënne-Loccoz, U. Schnider-Keel, C. Keel, and D. Haas. 2002. Inactivation of the regulatory gene algU or gacA can affect the ability of biocontrol Pseudomonas fluorescens CHA0 to persist as culturable cells in nonsterile soil. Appl. Environ. Microbiol. 68:2085-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matz, C., P. Deines, J. Boenigk, H. Arndt, L. Eberl, S. Kjelleberg, and K. Jürgens. 2004. Impact of violacein-producing bacteria on survival and feeding of bacterivorous nanoflagellates. Appl. Environ. Microbiol. 70:1593-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matz, C., and S. Kjelleberg. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302-307. [DOI] [PubMed] [Google Scholar]
- 29.Natsch, A., C. Keel, H. A. Pfirter, D. Haas, and G. Défago. 1992. Contribution of the global regulator gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 into soil microcosms. Appl. Environ. Microbiol. 60:2553-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page, F. C. 1988. A new key to freshwater and soil Gymnaboeae. Freshwater Biological Association, Ambleside, United Kingdom.
- 31.Reimmann, C., C. Valverde, E. Kay, and D. Haas. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 187:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rønn, R., J. Grunert, and F. Ekelund. 2001. Protozoan response to addition of the bacteria Mycobacterium chlorophenolicum and Pseudomonas chlororaphis to soil microcosms. Biol. Fertil. Soils 33:126-131. [Google Scholar]
- 33.Rønn, R., A. McCaig, B. Griffiths, and J. Prosser. 2002. Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl. Environ. Microbiol. 68:6094-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlimme, W., M. Marchiani, K. Hanselmann, and B. Jenni. 1999. BACTOX, a rapid bioassay that uses protozoa to assess the toxicity of bacteria. Appl. Environ. Microbiol. 65:2754-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnider-Keel, U., K. B. Lejbølle, E. Baehler, D. Haas, and C. Keel. 2001. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 67:5683-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, and C. Gigot-Bonnefoy. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuster, F. L. 1979. Small amoebas and amoeboflagellates, p. 215-285. In M. Levandowsky and S. Hutmer (ed.), Biochemistry and physiology of protozoa, 2nd ed., vol. 1. Academic Press, New York, NY. [Google Scholar]
- 38.Siddiqui, I. A., D. Haas, and S. Heeb. 2005. Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. Appl. Environ. Microbiol. 71:5646-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sørensen, J. S., T. Schyberg, and R. Rønn. 1998. Predation by protozoa on Escherichia coli K12 in soil and transfer of resistance plasmid RP4 to indigenous bacteria in soil. Appl. Soil Ecol. 11:79-90. [Google Scholar]
- 40.Valverde, C., S. Heeb, C. Keel, and D. Haas. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361-1379. [DOI] [PubMed] [Google Scholar]
- 41.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, and M. Schnider. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. N. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Verlagsgesellschaft GmbH, Weinheim, Germany.
- 42.Weitere, M., T. Bergfeld, S. A. Rice, C. Matz, and S. Kjelleberg. 2005. Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ. Microbiol. 7:1593-1601. [DOI] [PubMed] [Google Scholar]
- 43.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]