Abstract
Most polluted sites contain mixed waste. This is especially true of the U.S. Department of Energy (DOE) waste sites which hold a complex mixture of heavy metals, radionuclides, and organic solvents. In such environments enzymes that can remediate multiple pollutants are advantageous. We report here evolution of an enzyme, ChrR6 (formerly referred to as Y6), which shows a markedly enhanced capacity for remediating two of the most serious and prevalent DOE contaminants, chromate and uranyl. ChrR6 is a soluble enzyme and reduces chromate and uranyl intracellularly. Thus, the reduced product is at least partially sequestered and nucleated, minimizing the chances of reoxidation. Only one amino acid change, Tyr128Asn, was responsible for the observed improvement. We show here that ChrR6 makes Pseudomonas putida and Escherichia coli more efficient agents for bioremediation if the cellular permeability barrier to the metals is decreased.
Environmental pollution with toxic agents is a widespread and serious problem that defies simple solutions. The ability of bacteria to mineralize many of these contaminants or to transform them into a valence state that is insoluble and can therefore be localized offers a promising solution. As pointed out elsewhere (2, 3, 8, 11, 17, 27), several measures can increase bacterial effectiveness for bioremediation. These measures include decreasing the toxicity of the pollutants to the remediating bacteria; improving the kinetics of the enzymes of the bacteria for the desired reactions; and, since most polluted environments contain mixed waste, generating individual bacterial enzymes with enhanced capacities for remediating multiple pollutants.
Our work has focused on improving the bacterial capacity for chromate [Cr(VI)] remediation. A by-product of numerous industrial and military projects, such as the manufacture of nuclear weapons, chromate is a ubiquitous environmental pollutant (11, 13). It is soluble and bioavailable and upon cellular uptake leads to toxic effects that include mutagenesis and carcinogenesis (23, 26, 40). Bacteria can reduce chromate to the Cr(III) valence state, which is often less soluble and can be confined to initial contamination sites. Cr(III) is also much less toxic.
The membrane-bound electron transport chain of certain bacteria can reduce Cr(VI) and may enable some of these bacteria to use it as a terminal electron acceptor for energy generation (9). In addition, many soluble enzymes in nearly all bacteria can vicariously reduce Cr(VI). Some examples are lipoyl dehydrogenase and cytochrome c and glutathione reductases, whose physiological roles are to catalyze energetic or biosynthetic reactions. These enzymes reduce chromate by one-electron transfer, generating Cr(V) (18, 33). Cr(V) is a highly reactive radical that redox cycles in the presence of appropriate electron acceptors, such as molecular oxygen. In this process, Cr(V) transfers its electron to dioxygen, regenerating Cr(VI) and producing reactive oxygen species (ROS). With the continued activity of the one-electron reducers, chromate shuttles back and forth between its Cr(VI) and Cr(V) valence states, producing large quantities of ROS and depleting the cell's reducing power. We have presented both in vitro and in vivo evidence that ROS generation has a major role in chromate toxicity to bacteria and in impairing the chromate-remediating efficiency of bacteria (2, 3).
Many soluble oxidoreductases whose physiological role is evidently antioxidant defense (17) exhibit a different mode of chromate reduction. These enzymes are obligatory two-electron reducers of chromate and quinones and, as dimers, can convert Cr(VI) to Cr(III) in one step, minimizing the generation of Cr(V). We have characterized three such bacterial enzymes, ChrR of Pseudomonas putida and Escherichia coli (ChrR of E. coli was formerly referred to as YieF) and NfsA of E. coli (GenBank accession numbers AF375642.1, NC_000913.2 [new number, DQ989184], and P17117, respectively) (1, 2, 27, 28). Overproduction of these enzymes in bacteria decreases chromate toxicity, apparently by minimizing chromate reduction by one-electron reducers and ROS generation. Therefore, a component of our proposed strategy for improving the bacterial chromate remediation capacity is to enhance the kinetics of an obligatory two-electron chromate reducer for chromate reduction (1, 2, 3, 17).
In the work reported in this paper we were concerned with improving the kinetics of E. coli ChrR, one of the obligate two-electron reducers that we have characterized. This enzyme was chosen because (i) it is soluble and thus is easier to manipulate than membrane-bound enzymes; (ii) as it reduces chromate intracellularly, the reduced product is at least partially sequestered and nucleated, minimizing the chances of reoxidation, which is a potential problem with bacterial cell-surface-mediated reduction catalyzed by electron transport chain components; and (iii) it has a broad substrate range and is able to reduce quinones, potassium ferricyanide, 2,6-dichloroindophenol, V(V), Mo(VI), methylene blue, and cytochrome c, as well as the prodrugs mitomycin C and 5-aziridinyl-2,4-dinitrobenzamide (CB 1954) and the drug 17-allylamino-17-demethoxygeldanamycin (1, 4; Barak and Matin, unpublished), which encouraged us to hypothesize that it may also be able to remediate additional contaminants present at waste sites, such as the U.S. Department of Energy (DOE) waste sites. These sites constitute a serious environmental problem and contain, in addition to chromate, radioactive waste, such as uranyl [U(VI)] (12, 15, 24, 36). U(VI), like Cr(VI), is soluble and subject to leaching and therefore is a threat to vital resources like drinking water supplies; its reduced valence state, U(IV), is insoluble. We report here isolation of mutants of the E. coli ChrR enzyme with enhanced kinetics for both Cr(VI) and U(VI) reduction. The effect of in vivo production of an improved enzyme on the chromate- and uranyl-reducing activity of bacteria is also described.
MATERIALS AND METHODS
Strains, plasmids, genes, primers, and growth conditions.
Table 1 shows the strains, plasmids, and primers used in this study. The various E. coli and Salmonella strains overexpressing the wild-type E. coli chrR gene and strains overexpressing the chrR6 gene (which produce the E. coli ChrR enzyme and its evolved version, ChrR6 [see below], respectively), as well as the control strain (E. coli containing the empty pET28a+ vector), were grown aerobically in a 37°C incubator at 225 rpm to the mid-exponential phase, were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and were incubated overnight. Pseudomonas putida KT2440 and the isogenic CRK4 mutant (Table 1) expressing the E. coli chrR or chrR6 gene were grown at 18°C to avoid inclusion body formation following IPTG induction, as previously described (2, 32); CRK4 lacks the P. putida chrR gene and has a decreased capacity to reduce chromate (1).
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant characteristics or sequence | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21 | DE3 allowing overexpression of desired protein under IPTG-inducible T7 promoter | Novagen |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 19 |
| NR698 | MC4100 imp4213 | 30 |
| Pseudomonas strains | ||
| KT2440 | Wild-type genome-sequenced strain | 25 |
| chrR mutant (CRK4) | Isogenic with wild-type KT2440 but lacking the chromate reductase gene chrR | 27 |
| CRK4/E. coli chrR | E. coli ChrR (GenBank accession no. NC_000913.2 [new no., DQ989184]) overexpression strain; CRK4 containing plasmid pMMB::E. coli chrRa | This study |
| CRK4 chrR6 | ChrR6 overexpression strain; CRK4 containing plasmid pMMB::chrR6 (GenBank accession no. DQ987901) | This study |
| Salmonella strains | ||
| S. enterica serovar Typhimurium SL 7838 | Attenuated strain containing aroA and sopE gene deletions | S. H. Thorne et al., unpublished |
| SL 7838/E. coli chrR | E. coli ChrR overexpression strain; SL 7838 containing plasmid pET28a+::E. coli chrR | 4 |
| SL 7838/chrR6 | ChrR6 overexpression strain; SL 7838 containing plasmid pET28a+::E. coli chrR6 | 4 |
| Plasmids | ||
| pET28a+ | Overexpression vector | Novagen |
| pET28a+::E. coli chrR | Allows E. coli ChrR overexpression | 1 |
| pET28a+::P. putida chrR | Allows P. putida KT2440 ChrR (GenBank accession no. AF375642.1) overexpression in E. colia | 1 |
| pET28a+::nfsA | Allows E. coli NfsA (GenBank accession no. P17117) overexpressiona | 7 |
| pMMB67EH | Broad-host-range tac expression vector | 16 |
| pMMB67EH::E. coli chrRa | pMMB67EH with BamHI/HindIII His-tagged E. coli chrR insert | This study |
| pMMB67EH::chrR6a | pMMB67EH with BamHI/HindIII His-tagged chrR6 insert | This study |
| Primers | ||
| FE. coli chrR | 5′-CGCGGGGGCATATGTCTGAAAAATTGCAGGT-3′b | 1 |
| RE. coli chrR | 5′-TTTGGGATCCTTAGATCTTAACTCGCTGAA-3′c | 1 |
| FA120N | 5′-GTATTGATTCAGACCAGCTCAATGGGCGTGATTGG-3′ | This study |
| RA120N | 5′-CCAATCACGCCCATTGAGCTGGTCTGAATCAATAC-3′ | This study |
| FN128Y | 5′-TTGGCGGCGCGCGCTGTCAGTATCACCTGCGCCAGA-3′ | This study |
| RN128Y | 5′-TCTGGCGCAGGTGATACTGACAGCGCGCGCCGCCAAT-3′ | This study |
| FN160T | 5′-GTTGATCCGCAAACCGGAGAAGTGATTGA-3′ | This study |
| RN160T | 5′-ATCAATCACTTCTCCGGTTTGCGGATCAAC-3′ | This study |
| RL175G | 5′-TTAACTCGCTGAATAAACTCACCAAATGCGGTCAATTGCCCGGTCAGGTG-3′ | This study |
The accession number is the protein accession number in the PubMed database.
Underlining indicates an NdeI restriction site.
Underlining indicates a BamHI restriction site.
DNA techniques.
Small-scale isolation of plasmid DNA from E. coli was carried out by using the miniprep procedure (QIAGEN Inc., California). Plasmids were transformed into E. coli BL21(DE3) cells (Invitrogen Inc.) and used for protein production. DNA sequencing was conducted by SEQUETECH Corporation (California) using appropriate primers (Table 1).
Enzyme evolution.
Error-prone PCR was used for in vitro evolution. The chrR gene was used as the template. (Unless indicated otherwise, the designations chrR and ChrR refer to the E. coli gene and protein, respectively.) Random mutations were introduced into this gene by error-prone PCR performed as described by Chen and Arnold (10) and Barak et al. (4), using a GeneMorph II random mutagenesis kit (Stratagene Corporation, California). The forward and reverse chrR primers (Table 1) were used to amplify full-length hybrid products.
Screening for shuffled genes encoding high-activity chromate-reducing enzymes.
The shuffled genes were ligated into the pET28a+ plasmid and transformed into E. coli BL21(DE3) to allow overexpression. Recombinants were selected on plates containing kanamycin (50 μg ml−1). High-throughput screening of 6,000 recombinants was performed by inoculating colonies into individual wells of 96-well microtiter plates containing 200 μl LB medium and kanamycin. After growth to the stationary phase (overnight incubation; final A660, 1.0 to 1.5), 20-μl aliquots from each well were used to inoculate a second series of plates, using M9 minimal medium (Sigma Co.). Each well received the same initial inoculum density. The first set of plates was stored at −80°C after addition of glycerol. Cells in the second inoculation series were allowed to grow to the mid-exponential phase and then exposed to 0.5 mM IPTG to induce expression of the recombinant gene. After overnight incubation, cells were lysed by addition of 30 μl BugBuster (Novagen Inc.), incubated for 20 min at room temperature, and centrifuged for 20 min at 3,000 × g. One hundred microliters of the supernatant was mixed with 100 μl of a solution containing 500 μM potassium chromate or uranyl acetate, 2 mM NADH, 100 mM Tris-HCl (pH, 7), and double-distilled H2O (4).
Protein purification.
The most efficient enzymes for Cr(VI) reduction activity were purified on nickel columns, as previously described (27), using inocula obtained from the frozen plates. These enzymes were His tagged and therefore bound effectively to the nickel column. Protein concentrations were determined with a Bio-Rad Dc protein assay kit, using bovine serum albumin as a standard.
Site-directed mutagenesis.
Appropriate primers (Table 1) were used for site-directed mutagenesis. These primers were designed to create single-codon mutations using the method of Kuipers et al. (22). The modified PCR products were cloned into pET28a+ and transformed into E. coli BL21(DE3). The desired mutations were verified by sequencing.
Cr(VI) assays.
Cr(VI) quantification, transformation of Cr(VI) by whole cells and cell extracts, the cell extract preparation, and chromate reductase assays were conducted as described previously (1, 27). Kinetic measurements of enzyme activity were obtained (in quadruplicate) at pH 7 and 37°C unless indicated otherwise.
U(VI) determination.
U(VI) was quantified as described by Teixeira et al. (38), as follows. Samples were collected after incubation for a specified time. A 120-μl sample was mixed with 130 μl of a reagent mixture containing a complexing solution consisting of 2-(2-thiazolyazo-p-cresol), Triton X-100 (0.15 M), N-cetyl-N,N,N-trimethyammonium bromide, and triethanolamine buffer (pH 6.5) in proportions of 5:1:1:1:5. This method depends on the binding of 2-(2-thiazolyazo-p-cresol) to U(VI), which is aided by Triton X-100 and N-cetyl-N,N,N-trimethyammonium bromide. After 15 min of color development, the A588 of samples were determined using a microplate reader (model EL311sx; BIO-TEK Inc.).
U(IV) determination.
Uranium(IV) production was determined (G. J. Vazquez and A. J. Francis, unpublished) as follows. One hundred microliters of a freshly prepared reaction solution was added to a 100-μl sample. The reaction solution was prepared by mixing 3.5 ml of FeCl3 (1 mM, pH, 2), 0.75 ml of 1,10 phenanthroline (10 mM), and 0.75 ml of acetate buffer (1 M, pH, 4). One mole of uranyl(IV) reduces 2 mol of Fe3+ to Fe2+; the latter complexes with the 1,10-phenanthroline, producing a red or orange color with absorbance at 510 nm. The U(IV) concentration was determined using a standard curve prepared with different concentrations of Fe(NH4)2(SO4)2.
ROS generation assay.
The ROS generation assay was performed as previously described (1, 2). The reaction mixtures contained 100 mM Tris-HCl (pH 7), 125 μM NADH, 250 μM K2CrO4, and 25 μg ml−1 E. coli ChrR or 8 μg ml−1 ChrR6. The two enzymes gave similar rates of NADH reduction at these concentrations. H2O2 formation was quantified using an Amplex-Red kit (Molecular Probes).
XANES analysis.
X-ray absorption near-edge spectroscopy (XANES) was performed for chromate (0.5 to 1 mM) reduced by E. coli ChrR or ChrR6 at the Cr K edge (5,989 eV). The samples were suspended in Tris-HCl buffer (pH 6.8) containing 3 mM NADH. Samples were placed in a heat-sealed polyethylene bag and mounted on an Al sample holder having a cutout that was 2 mm high by 20 mm long by 1.5 mm thick, and the analysis was performed on beamline X10C at the National Synchrotron Light Source in the fluorescence mode using a 13-element Ge detector. The standards were Cr(VI) (potassium chromate; K2CrO4) and Cr(III) [chromium hydroxide; Cr(OH)3]. Chromium hydroxide was prepared by dissolving Cr(NO3)3 · 9H2O in deionized water and slowly increasing the pH to 11 with sodium hydroxide. The resulting precipitate was allowed to settle overnight, washed twice with deionized water, and allowed to air dry.
Spectra (five scans per sample) were collected from 200 eV below to 300 eV above the absorption edge. Data in the XANES region were collected with 0.5-eV energy steps at 2.0 s per interval. Chromium metal foil was placed in the reference channel and was examined simultaneously with each sample to monitor shifts in the beamline energy. The XANES spectra were background subtracted and normalized to the edge jump using the suite of programs described by Ravel and Newville (29). The first derivative of the absorption edge energy was used to determine the oxidation state.
Cell permeabilization.
The method of Belli and Fryklund (5) was used for cell permeabilization. Briefly, cells were grown overnight, harvested by centrifugation (1,700 × g, 10 min), and resuspended in 1.0 ml of 75 mM Tris-HCl (pH 7)-10 mM MgSO4. Chloroform was added to a concentration of 1.5%, and the cell suspension was vortexed and incubated at 37°C for 30 min.
Computer program.
Sequences were aligned with Clustal W (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html).
RESULTS
Obligate two-electron reducers can convert U(VI) to U(IV).
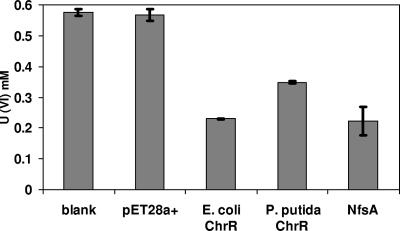
As mentioned above, enzymes with the capacity to remediate multiple contaminants are highly desirable, and we focused on E. coli ChrR as a candidate because of its broad substrate range. We found, however, that crude extracts of E. coli cells transformed with the pET28a+ vector containing the E. coli chrR, P. putida chrR, or E. coli nfsA gene could all reduce U(VI) (Fig. 1); 90 to 95% of the U(VI) disappearing from the reaction mixture was detectable as U(IV) (not shown). Appropriate controls established that enzymes encoded by these genes were responsible for the observed conversion. Thus, no activity was detected in reaction mixtures containing LB medium alone or in extracts of the strain containing the empty plasmid. The latter finding also shows that the normal levels of ChrR and NfsA present in E. coli wild-type strain BL21 are not sufficient to catalyze this reaction at an appreciable rate. When overproduced, E. coli ChrR and NfsA were more active in this conversion than P. putida ChrR.
FIG. 1.
Uranyl disappearance catalyzed by crude extracts of recombinant E. coli strains expressing different two-electron reducers. Reactions catalyzed in LB medium alone (blank) and by extracts of the strain transformed with the empty pET28+ vector were included as controls. The residual level of uranyl was determined after 6 h of incubation. The initial uranyl acetate concentration was 500 μM.
Evolved E. coli ChrR enzyme mutant exhibits improved Cr(VI)- and U(VI)-reducing activities.
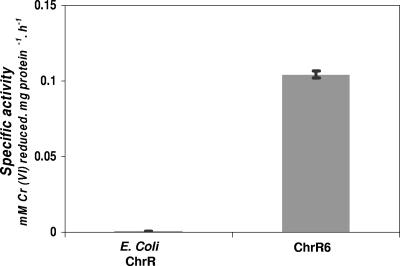
Using directed evolution employing error-prone PCR, we generated mutations in the E. coli chrR gene, and the products were screened to determine the chromate-reducing activities of the proteins that the genes encoded. Of the proteins showing improved activity, the mutant enzyme termed ChrR6 (formerly Y6 [4]) was the most active; crude extracts of the strain overproducing this enzyme showed ca. 200-fold-greater chromate-reducing activity than the extracts of the strain overproducing the wild-type E. coli ChrR enzyme showed (Fig. 2). We also screened the mutant gene library directly for uranyl-reducing activity. Although the products of many mutant genes showed improved uranyl reductase activity, ChrR6 proved to be the most effective in this respect also.
FIG. 2.
Chromate reductase specific activities of crude extracts of recombinant E. coli cells expressing E. coli ChrR and ChrR6.
Further characterization of ChrR6 involved work with the pure protein. Compared to E. coli ChrR, ChrR6 exhibited markedly improved kinetics for chromate reduction. These kinetics included a 30-fold-higher Vmax, a ninefold-lower Km, and hence close to a 300-fold-higher kcat/Km (Table 2), consistent with the crude extract results described above. For uranyl reduction, ChrR6 exhibited higher Vmax, kcat, and kcat/Km values (Table 3). The Km of ChrR6 for uranyl was, however, higher than that of E. coli ChrR. The high rate of reduction of Cr(VI) or U(VI) was not noticeably affected in the presence of the two metal species simultaneously, indicating that ChrR6 was indeed capable of remediating both species in a mixed setting (not shown). It is noteworthy that in crude extracts, ChrR6 showed a much greater increase in activity than E. coli ChrR showed than when pure proteins of these enzymes were compared. The reason for this is not known but may be related to the ability of obligate two-electron reducing oxidoreductases to interact with membranes (34), suggesting that ChrR6 and E. coli ChrR may interact with membranes differently.
TABLE 2.
Kinetics of Cr(VI) reduction for wild-type E. coli ChrR and the evolved ChrR6 enzyme
| Enzyme | Vmax [nmol Cr(VI) mg protein−1 min−1] | Km (μM) | kcat (s−1)a | kcat/Km |
|---|---|---|---|---|
| E. coli ChrR | 295 ± 27 | 376 ± 14 | 30 ± 2 | 4.5 × 104 ± 3 × 103 |
| ChrR6 | 8,812 ± 611 | 41 ± 5 | 521 ± 18 | 1.3 × 107 ± 3 × 105 |
Based on a dimeric enzyme molecular mass of 50 kDa (1).
TABLE 3.
Kinetics of U(VI) reduction for wild-type E. coli ChrR and the evolved ChrR6 enzyme
| Enzyme | Vmax [nmol U(VI) mg protein−1 min−1] | Km (μM) | kcat (s−1)a | kcat/Km |
|---|---|---|---|---|
| E. coli ChrR | 213 ± 17 | 108 ± 49 | 29 ± 11 | 1.6 × 104 ± 1.7 × 103 |
| ChrR6 | 2,511 ± 421 | 779 ± 40 | 331 ± 39 | 5 × 105 ± 2 × 104 |
Based on a dimeric enzyme molecular mass of 50 kDa (1).
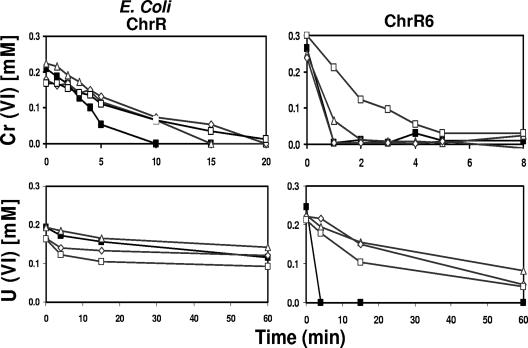
ChrR6 exhibited a different pH activity profile than E. coli ChrR (Fig. 3). For example, at pH 5 the Cr(VI) and U(VI) reduction rates of ChrR6 were improved compared to those of E. coli ChrR.
FIG. 3.
Kinetics of Cr(VI) and U(VI) reduction at different pH values. Symbols: ▪, pH 5; ▵, pH 7; ⋄, pH 8; □, pH 9.5. The pH 5 preparation was obtained using acetic acid; the preparations at other pHs were obtained using Tris-HCl. The reactions were performed using 1-ml mixtures containing 250 μM potassium chromate or uranyl acetate, 2 mM NADH, 100 μg ml−1 enzyme, and the appropriate buffer. The experiment was conducted in triplicate; the differences between the mean values for the runs were less than 10%, as determined by analysis of covariance.
End products of ChrR6 Cr(VI) and U(VI) reduction.
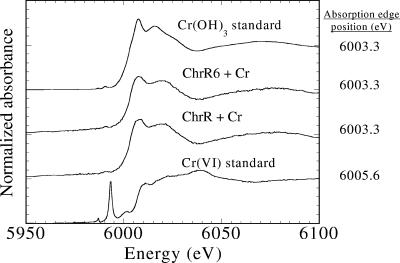
To ascertain that ChrR6 retained the capacity of wild-type E. coli ChrR to quantitatively convert Cr(VI) to Cr(III) (27), XANES analyses were performed. The XANES spectrum is sensitive to oxidation state changes in the target atom, and the absorption edge energy increases with a higher oxidation state. XANES spectra for the standards and samples are shown in Fig. 4. The first derivatives of the absorption edge energy for Cr(VI) and Cr(III) standards were at 6,005.6 and 6,003.3 eV, respectively. In addition, the Cr(VI) standard had a pre-edge peak at 5,993.3 eV. Upon complete reduction by the E. coli ChrR or ChrR6 enzyme, the derivative of the absorption edge energy was at 6,003.3 eV. This shift to an energy lower than the energy of the chromate standard was identical to the shift observed for the Cr(III) standard and confirmed that Cr(VI) was reduced to Cr(III). The absence of a pre-edge peak at 5,993.0 eV for the samples compared to the Cr(VI) spectrum also indicated that Cr(VI) was not present in the samples.
FIG. 4.
Comparison of absorption edge positions at the Cr K edge for Cr(III) and Cr(VI) standards and chromate (indicated by Cr in the figure) samples reduced by the E. coli ChrR and ChrR6 enzymes.
U(IV) generation by pure ChrR6 was determined chemically, as described above for measurements with crude extracts (Fig. 1). Again, more than 90% of the uranyl disappearing from the reaction mixture was recovered as U(IV) (not shown).
ROS generation during chromate reduction by ChrR6.
The XANES analysis, while showing that there was complete conversion of Cr(VI) to Cr(III) by ChrR6, did not rule out the possibility that there was transient generation of Cr(V). We previously established that Cr(V) is not generated during Cr(VI) reduction by E. coli ChrR. Some of the evidence for this conclusion is that only 25% of the NADH electrons consumed during E. coli ChrR-catalyzed chromate reduction were utilized in ROS generation. Thus, the four-electron-reduced E. coli ChrR dimer donated three electrons to chromate and one electron to dioxygen in a one-step reaction, avoiding Cr(V) generation and redox cycling. If Cr(V) had been generated and cycling had occurred, much more ROS would have been formed.
To test if ChrR6 was able to convert Cr(VI) to Cr(III) and U(VI) to U(IV) in one step, we compared the portions of the NADH electrons utilized in ROS generation by E. coli ChrR and ChrR6 during chromate or uranyl reduction. As expected in light of our previous results (1), approximately 25% and 32% of the NADH electrons were consumed in generation of ROS during the E. coli ChrR-catalyzed reactions for Cr(VI) and U(VI), respectively. The corresponding values for ChrR6 were 12.5% and 16%, respectively. The data strongly suggest that like E. coli ChrR, ChrR6 is able to convert Cr(VI) to Cr(III) and U(VI) to U(IV) in one step and does not generate oxidative intermediates.
Amino acid sequences of the improved enzymes.
Four substitutions occurred in the amino acid sequence of ChrR6 (Val120Ala, Tyr128Asn, Thr160Asn, and Glu175Leu) compared to the E. coli ChrR sequence. When each altered amino acid was individually changed to the original residue (see Materials and Methods), only the Asn128Tyr change diminished ChrR6 activity (Table 4). Furthermore, the single Tyr128Asn substitution in the E. coli ChrR protein led to even greater increases in the Cr(VI) and U(VI) reduction rates compared to the rates for ChrR6 (147,619 ± 46,576 and 6,007 ± 226 nmol mg protein−1 min−1, respectively). To understand the chemical basis of these findings, attempts are currently under way to crystallize the E. coli ChrR and ChrR6 proteins and to apply computational models that predict protein sequence-structure relationships (Y. Barak, Y. Nov, D. Ackerley, and A. Matin, unpublished).
TABLE 4.
Site-directed mutagenesis variants of ChrR6 and their specific Cr(VI) reduction rates
| Enzyme | Sp act [μM Cr(VI) mg protein−1 h−1] |
|---|---|
| E. coli ChrR | 295 ± 27 |
| ChrR6 | 8,812 ± 611 |
| Ala120Val | 9,200 ± 545 |
| Asn128Tyr | 200 ± 53 |
| Asn160Thr | 7,836 ± 321 |
| Leu175Glu | 10,205 ± 487 |
Effect of ChrR6 expression on chromate reduction by whole cells.
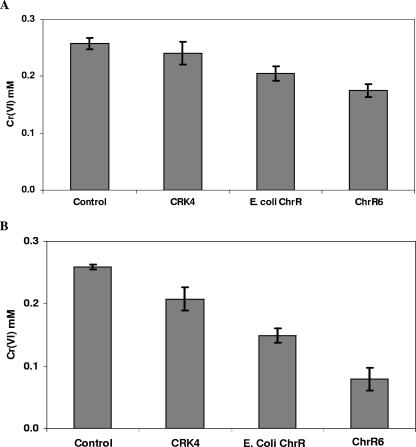
As our long-term objective is to generate bacterial strains capable of superior remediation in situ, we tested the efficacy of ChrR6 in P. putida, which is an indigenous organism at polluted sites and is thus a suitable candidate for progress toward this end. The E. coli ChrR- and ChrR6-encoding genes (E. coli chrR and chrR6, respectively) were cloned in the broad-range vector pMMB67EH, and the recombinant plasmids were introduced into P. putida KT2440 mutant CRK4. This strain lacks the chrR gene that encodes the P. putida ChrR protein (27) and has decreased chromate-reducing ability compared to the wild-type strain (1). Neither the E. coli ChrR-expressing recombinant strain nor the E. coli ChrR6-expressing recombinant strain exhibited any significant improvement in chromate reduction capacity (Fig. 5A). However, cell extracts of the E. coli chrR-transformed strain did exhibit improved chromate reductase activity, and further improvement was seen in extracts of cells expressing the ChrR6 enzyme (Fig. 5B). The results indicate that the E. coli chrR gene and its evolved chrR6 version were expressed in P. putida and suggest that the permeability barrier to chromate masked the enhanced cellular chromate reductase activity of the transformed strains. To further test the involvement of the transport barrier, the capacities of the transformed cells to reduce chromate were determined following permeabilization of the cells by chloroform treatment. The recombinant strains expressing E. coli ChrR or ChrR6 showed greater reductase activity than CRK4, and the ChrR6-expressing strain showed the highest activity (Fig. 6A).
FIG. 5.
(A) Cr(VI) disappearance caused by whole cells of P. putida CRK4 strains transformed with the empty plasmid or a plasmid containing the E. coli ChrR or ChrR6 gene. The control data are the data for chromate disappearance in LB medium alone. (B) Chromate disappearance caused by crude extracts of the strains described above. See Materials and Methods for further details.
FIG. 6.
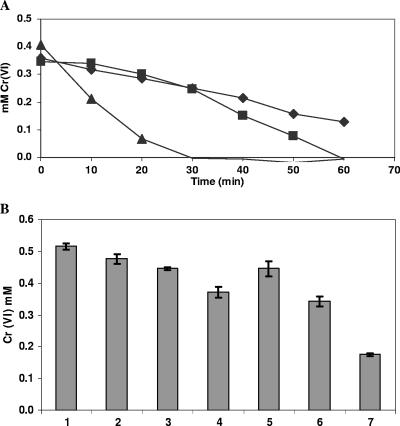
(A) Cr(VI) disappearance caused by whole cells of chloroform-permeabilized P. putida CRK4 strains transformed with the empty plasmid or a plasmid containing the E. coli ChrR or ChrR6 gene. The initial concentration of potassium chromate was 500 μM. The experiment was conducted in triplicate; the differences between the mean values for the runs were less than 10%, as determined by analysis of covariance. Symbols: ⧫, CRK4; ▪, CRK4 expressing E. coli ChrR; ▴, CRK4 expressing ChrR6. (B) Cr(VI) disappearance caused by whole cells of E. coli MC4100 and NR698 transformed with the empty plasmid or a plasmid containing the E. coli ChrR or ChrR6 gene. The numbers on the abscissa indicate the following: 1, control (LB medium alone); 2, MC4100/pET; 3, MC4100 with E. coli ChrR; 4, MC4100 with ChrR6; 5, NR698/pET; 6, NR698 with E. coli ChrR; 7, NR698 with ChrR6.
To further establish that the permeability barrier to chromate may be a limiting factor in chromate reduction in bacteria, we took advantage of the availability of E. coli mutant NR698, whose outer membrane permeability is impaired (30). The plasmids mentioned above were transformed in this mutant and the isogenic wild-type strain, MC4100, and the chromate reductase activities of the two strains were compared. Again, as was observed with the intact P. putida strain, there was little difference between the reduction capacity of the MC4100 parent strain and the reduction capacities of the E. coli ChrR- and ChrR6-expressing transformants. However, with the permeability barrier breached in the case of strain NR698, a clear improvement in chromate reduction was evident in the ChrR6-expressing strain (Fig. 6B). Furthermore, chloroform-treated MC4100 and NR698 strains (not expressing or overexpressing E. coli ChrR or ChrR6) exhibited even greater reduction rates (not shown). However, not all bacteria were impermeant. Thus, when strain SL 7838 of Salmonella enterica serovar Typhimurium was transformed with a plasmid bearing the chrR6 gene, the chromate reduction activity exhibited by whole cells was high compared to that of the E. coli chrR-transformed strain even without cell permeabilization.
Effect of ChrR6 expression on uranyl reduction by whole cells.
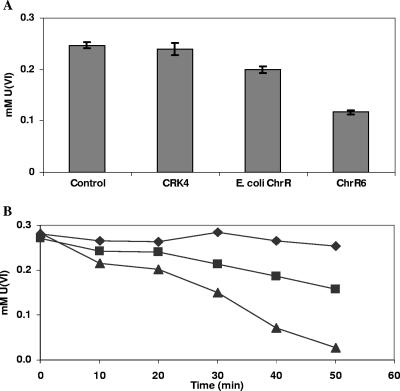
Nonpermeabilized P. putida CRK4 cells showed marginally greater uranyl reduction when they were transformed with the chrR6 gene than when they were transformed with the E. coli chrR gene (Fig. 7A). Again, however, the difference was more pronounced with permeabilized cells (Fig. 7B), showing that the full potential of ChrR6 for uranyl reduction is also masked by the cell permeability barrier.
FIG. 7.
(A) U(VI) disappearance caused by whole cells of P. putida CRK4 strains transformed with the empty plasmid or a plasmid containing the E. coli chrR or chrR6 gene. The control data are for uranyl disappearance in LB medium alone. (B) Uranyl disappearance caused by chloroform-treated cells of the strains described above. The experiment was conducted in triplicate. The differences between the means for the runs were less than 5%, as determined by analysis of covariance. Symbols: ⧫, CRK4; ▪, CRK4 expressing E. coli ChrR; ▴, CRK4 expressing ChrR6.
DISCUSSION
We focused on soluble bacterial two-electron oxidoreductases for improving bacterial chromate bioremediation capacity for several reasons. One reason is that these enzymes provide a safe pathway for chromate reduction which, since it is intracellular, minimizes the chances of reoxidation and resolubilization of the reduced product; the second reason is that since the enzymes are soluble, they are relatively easy to manipulate; and the third reason is that the broad substrate ranges of the enzymes, particularly that of E. coli ChrR, encouraged the notion that their reducing activities could be harnessed to remediate multiple pollutants. We show here that the improved version of E. coli ChrR, the ChrR6 enzyme, does indeed have the potential to confer on bacteria the capacity for superior intracellular reduction of two serious pollutants, chromate and uranyl. An additional important reason for working with these enzymes is that they are NAD(P)H oxidoreductases and are active under both aerobic and anaerobic conditions as long as the cofactors are available. Thus, expression of ChrR6 in anaerobic dissimilatory metal reducers, such as species of Shewanella, Desulfovibrio, and several other nitrate reducers, can enhance their metal bioremediation efficiency as well.
ChrR6 showed superior kinetics for chromate reduction in all respects tested, including a 30-fold-improved Vmax, a severalfold-lower Km, and kcat/Km values that were increased by orders of magnitude. It retained the capacity to quantitatively convert Cr(VI) to Cr(III), and as it generated very little ROS during this conversion, it seems reasonable to conclude that it did so in one step, avoiding the generation of Cr(V), which is a major source of toxicity to bacteria during chromate reduction. The structural basis for the increase in enzyme activity with the Tyr128-to-Asn substitution remains to be explored. However, since cysteines have been implicated to be important in protein catalytic sites (31), it may be significant that this substitution is close to Cys126.
Expression in P. putida resulted in a markedly enhanced chromate reduction rate when assays were performed with cell extracts, indicating that ChrR6 is potentially useful for enhancing the chromate remediation activity of a bacterium native to contaminated sites that could be used for in situ bioremediation. This potential, however, was not evident with intact cells, so cells not expressing the E. coli ChrR enzyme and cells expressing this enzyme or ChrR6 showed about the same level of chromate reduction. The hypothesis that the difference between the activity of intact cells and the activity of cell extracts was indeed due to a permeability barrier was confirmed by the demonstration that cells permeabilized by chloroform treatment showed the advantage of ChrR6 expression, as revealed by cell extract measurements.
Chloroform treatment impairs the permeability barrier of both the outer membrane and the cytoplasmic membrane, and to determine whether the primary barrier resided in the outer membrane, the cytoplasmic membrane, or both, we used E. coli strain NR698, which has a lesion in the imp4213 gene and therefore has impaired outer membrane permeability. This strain did indeed unmask cellular ChrR6 activity, indicating that the outer membrane permeability barrier has a role. Furthermore, since chloroform-treated cells of isogenic wild-type strain MC4100 exhibited this unmasking to a greater extent, it is clear that the lack of efficiency of chromate transport across the cytoplasmic membrane also has a role. We are currently attempting to enhance metal transport through the P. putida cell envelope by overproducing the OprF porin, as well as the sulfate transporter (41; B. Salles and A. Matin, unpublished). As shown by E. coli mutant NR698 used in these studies, it is possible to enhance envelope permeability to metals by genetic manipulation.
The broad substrate range of the two-electron reducers, such as E. coli ChrR, is potentially useful since an improved version could simultaneously have superior capacities to remediate more than one contaminant at waste sites. As most waste sites, especially those of the DOE, contain mixed waste (14, 20, 21, 35, 37), an enzyme with superior capacities to remediate multiple pollutants has obvious advantages. So far, E. coli ChrR and other two-electron-reducing wild-type enzymes tested in this study have been of interest in bioremediation because of their chromate reductase activity, but as shown here, they are also possess able to convert soluble U(VI) to insoluble U(IV), which is desirable. Both these activities were greatly enhanced in ChrR6. Since uranyl also is a serious pollutant, the ability of ChrR6 to convert two serious DOE contaminants to their innocuous valence states is a valuable trait.
It is noteworthy that a single amino acid change in E. coli ChrR, Tyr128Asn, resulted in a marked improvement in the kinetics of the enzyme for both chromate reduction and uranyl reduction. Indeed, as we have reported elsewhere (4), the same change dramatically increased the effectiveness of this enzyme for reduction of prodrugs, such as mitomycin C and 5-aziridinyl-2,4-dinitrobenzamide (CB 1954), making the evolved enzyme useful also in the context of improving reductive prodrug cancer chemotherapy. The crystal structure of E. coli ChrR is not available yet, and it is not known whether the amino acid at position 128 is part of the active site of the enzyme. Enzyme activity, however, can also be influenced by amino acids not residing in the active site (6, 39). Whatever the mechanism, it is evident that the change affects the activity of the enzyme with substrates whose chemical structures are very different. In this respect ChrR6 seems to intensify an inherent characteristic of E. coli ChrR, i.e., being active with structurally different substrates. This raises the possibility that ChrR6 may also have improved kinetics for reducing other metals or radionuclides, such as plutonium and technetium. This possibility is under investigation.
Acknowledgments
We are grateful to Aviva Levina from University of Sydney for assistance with Cr(V) compounds; Thomas J. Silhavy of Princeton University for providing the MC4100 and NR698 strains; and Bruno Salles, Mike Benoit, and Mimi Keyhan for their useful advice and stimulating discussions.
This work was supported by grant DE-FG02-03ER63627 from the Natural and Accelerated Bioremediation Program of the U.S. Department of Energy. Y.B. and D.F.A. were supported in part by Lady Davis and FRST New Zealand (STAX0101) fellowships, respectively.
REFERENCES
- 1.Ackerley, D. F., C. F. Gonzalez, C. H. Park, R. Blake, M. Keyhan, and A. Matin. 2004. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl. Environ. Microbiol. 70:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerley, D. F., C. F. Gonzalez, C. H. Park, R. Blake, M. Keyhan, and A. Matin. 2004. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ. Microbiol. 6:851-860. [DOI] [PubMed] [Google Scholar]
- 3.Ackerley, D. F., Y. Barak, S. V. Lynch, J. Curtin, and A. Matin. 2006. Effect of chromate stress on Escherichia coli K-12. J. Bacteriol. 188:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barak, Y., S. H. Thorne, D. F. Ackerley, S. V. Lynch, C. H. Contag, and A. Matin. 2006. Enzyme improvement by directed evolution for reductive cancer chemotherapy. Mol. Cancer Ther. 5:97-103. [DOI] [PubMed] [Google Scholar]
- 5.Belli, W. A., and J. Fryklund. 1995. Partial characterization and effect of omeprazole on ATPase activity in Helicobacter pylori by using permeabilized cells. Antimicrob. Agents Chemother. 39:1717-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts, M. J., and R. B. Russel. 2003. Amino acid properties and consequences of substitutions, p. 175-183. In M. R. Barnes and I. C. Gray (ed.), Bioinformatics for geneticists. Wiley, New York, NY.
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. J. Riley, J. D. Collado-Vides, C. K. Glasner, G. F. Rode, J. Mayhew, N. W. Gregor, H. A. Davis, M. A. Kirkpatrick, D. J. Goeden, M. B. Rose, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 8.Cervantes, C. J., S. Campos-Garcia, F. Devars, H. Gutierrez-Corona, J. C. Loza-Tavera, T. Guzman, and R. Moreno-Sanchez. 2001. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25:335-347. [DOI] [PubMed] [Google Scholar]
- 9.Chardin, B., M. T. Giudici-Orticoni, G. De Luca, B. Guigliarelli, and M. Bruschi. 2003. Hydrogenases in sulfate-reducing bacteria function as chromium reductase. Appl. Microbiol. Biotechnol. 63:315-321. [DOI] [PubMed] [Google Scholar]
- 10.Chen, K., and F. H. Arnold. 1993. Turning the activity of an enzyme for unusual environments: sequential random mutagenesis of sublitisin E for catalysis in dimethylformamide. Proc. Natl. Acad. Sci. USA 90:5618-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa, M. 2003. Potential hazards of hexavalent chromate in our drinking water. Toxicol. Appl. Pharmacol. 188:1-5. [DOI] [PubMed] [Google Scholar]
- 12.Environmental Protection Agency. 2004. Radionuclide biological remediation resource guide. Environmental Protection Agency, Washington, D.C.
- 13.Fendorf, S., B. W. Wielinga, and C. M. Hansel. 2000. Chromium transformations in natural environments: the role of biological and abiological processes in chromium(VI) reduction. Int. Geolog. Rev. 42:691-701. [Google Scholar]
- 14.Francis, A. J., and C. J. Dodge. 1998. Remediation of soils and wastes contaminated with uranium and toxic metals. Environ. Sci. Technol. 32:3993-3998. [Google Scholar]
- 15.Francis, A. J., C. J. Dodge, and G. E. Meinken. 2002. Biotransformation of pertechnetate by clostridia. Radiochim. Acta 90:791-797. [Google Scholar]
- 16.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez, C. F., D. F. Ackerley, S. V. Lynch, and A. Matin. 2005. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J. Biol. Chem. 280:22590-22595. [DOI] [PubMed] [Google Scholar]
- 18.Goodgame, D. M., and A. M. Joy. 1986. Relatively long-lived chromium(V) species are produced by the action of glutathione on carcinogenic chromium(VI). J. Inorg. Biochem. 26:219-224. [DOI] [PubMed] [Google Scholar]
- 19.Harris, C. R., and T. J. Silhavy. 1999. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 181:3438-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalin, M., W. N. Wheeler, and G. Meinrath. 2005. The removal of uranium from mining waste water using algal/microbial biomass. J. Environ. Radioact. 78:151-177. [DOI] [PubMed] [Google Scholar]
- 21.Keyhan, M., D. F. Ackerley, and A. Matin. 2003. Targets of improvement in bacterial chromate bioremediation, p. 143-144. In M. Pellei and A. Porta (ed.), Remediation of contaminated sediments. Proceedings of the Second International Conference on Remediation of Contaminated Sediments. Battelle Press, Columbus, Ohio.
- 22.Kuipers, O. P., H. J. Boot, and W. M. De Vos. 1991. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 19:4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levina, A., P. A. Lay, and N. E. Dixon. 2001. Disproportionation of a model chromium(V) complex causes extensive chromium(III)-DNA binding in vitro. Chem. Res. Toxicol. 14:946-950. [DOI] [PubMed] [Google Scholar]
- 24.Lovley, D., P. Widman, J. Woodward, and E. Phillips. 1993. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl. Environ. Microbiol. 59:3572-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Düsterhöft, B. Tümmler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 26.O'Brien, T. J., B. R. Brooks, and S. R. Patierno. 2005. Nucleotide excision repair functions in the removal of chromium-induced DNA damage in mammalian cells. Mol. Cell. Biochem. 279:85-95. [DOI] [PubMed] [Google Scholar]
- 27.Park, C.-H., M. Keyhan, B. Wielinga, S. Fendorf, and A. Matin. 2000. Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl. Environ. Microbiol. 66:1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, C. H., C. Gonzalez, D. F. Ackerley, M. Keyhan, and A. Matin. 2002. Molecular engineering of soluble bacterial proteins with chromate reductase activity, p. 103-111. In R. E. Hinchee, A. Porta, and M. Pellei (ed.), Remediation and reuse of contaminated sediments. Proceedings of the First International Conference on Remediation of Chlorinated Sediments, vol. S1-3. Battelle Press, Columbus, Ohio. [Google Scholar]
- 29.Ravel, B., and M. Newville. 2005. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 12:537-541. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz, N., B. Falcone, D. Kahne, and T. J. Silhavy. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307-317. [DOI] [PubMed] [Google Scholar]
- 31.Sandy, J., A. Mushtaq, S. J. Holton, P. Schartau, M. E. M. Noble, and E. Sim. 2005. Investigation of the catalytic triad of arylamine N-acetyltransferases: essential residues required for acetyl transfer to arylamines. Biochem. J. 15:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schein, C. H. 1989. Production of soluble recombinant proteins in bacteria. Bio/Technology 7:1141-1148. [Google Scholar]
- 33.Shi, X. L., and N. S. Dalal. 1988. On the mechanism of the chromate reduction by glutathione: ESR evidence for the glutathionyl radical and an isolable Cr(V) intermediate. Biochem. Biophys. Res. Commun. 156:137-142. [DOI] [PubMed] [Google Scholar]
- 34.Sparla, F., G. Tendeschi, P. Pupillo, and P. Trost. 1999. Cloning and heterologous. expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase. FEBS Lett. 463:382-386. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki, T., N. Miyata, H. Horitsu, K. Kawai, K. Takamizawa, and Y. Tai. 1992. NAD(P)H-dependent chromium (VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate is formed during the reduction of Cr(VI) to Cr(III). J. Bacteriol. 174:5340-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, Y., and J. F. Banfield. 1999. The geomicrobiology of uranium. Uranium—mineralogy, geochemistry, and the environment. Mineralog. Soc. Am. Rev. Mineralog. 38:393-432. [Google Scholar]
- 37.Suzuki, Y., D. Shelly, K. Kelly, M. Kemner, and J. F. Banfield. 2005. Direct microbial reduction and subsequent preservation of uranium in natural near-surface sediment. Appl. Environ. Microbiol. 71:1790-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teixeira, L. S. G., A. C. S. Costa, S. L. C. Ferreira, M. L. M. Freitas, and S. Carvalho. 1999. Spectrophotometric determination of uranium using 2-(2-thiazolylazo)-p-cresol (TAC) in the presence of surfactants. J. Braz. Chem. Soc. 10:519-522. [Google Scholar]
- 39.Wu, K., R. Eng, R. J. Knox, and S. Chen. 2001. Demonstration of the activation of prodrug CB 1954 using human DT-diaphorase mutant Q104Y-transfected MDA-MB-231 cells and mouse xenograft model. Arch. Biochem. Biophys. 385:203-208. [DOI] [PubMed] [Google Scholar]
- 40.Ye, J., S. Wang, S. S. Leonard, Y. Sun, L. Butterworth, J. Antonini, M. Ding, Y. Rojanasakul, V. Vallyathan, V. Castranova, and X. Shi. 1999. Role of reactive oxygen species and p53 in chromium(VI)-induced apoptosis. J. Biol. Chem. 274:34974-34980. [DOI] [PubMed] [Google Scholar]
- 41.Yoshimura, F., and H. Nikaido. 1982. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J. Bacteriol. 152:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]