Abstract
Yeast cell surface display is a powerful tool for expression and immobilization of biocatalytically active proteins on a unicellular eukaryote. Here bacterial carboxylesterase EstA from Burkholderia gladioli was covalently anchored into the cell wall of Saccharomyces cerevisiae by in-frame fusion to the endogenous yeast proteins Kre1p, Cwp2p, and Flo1p. When p-nitrophenyl acetate was used as a substrate, the esterase specific activities of yeast expressing the protein fusions were 103 mU mg−1 protein for Kre1/EstA/Cwp2p and 72 mU mg−1 protein for Kre1/EstA/Flo1p. In vivo cell wall targeting was confirmed by esterase solubilization after laminarinase treatment and immunofluorescence microscopy. EstA expression resulted in cell wall-associated esterase activities of 2.72 U mg−1 protein for Kre1/EstA/Cwp2p and 1.27 U mg−1 protein for Kre1/EstA/Flo1p. Furthermore, esterase display on the yeast cell surface enabled the cells to effectively grow on the esterase-dependent carbon source glycerol triacetate (Triacetin). In the case of Kre1/EstA/Flo1p, in vivo maturation within the yeast secretory pathway and final incorporation into the wall were further enhanced when there was constitutive activation of the unfolded protein response pathway. Our results demonstrate that esterase cell surface display in yeast, which, as shown here, is remarkably more effective than EstA surface display in Escherichia coli, can be further optimized by activating the protein folding machinery in the eukaryotic secretion pathway.
Since the initial description of protein targeting to the yeast cell surface by fusion of the protein of interest to a natural cell wall protein, yeast cell surface modification has become an attractive tool for immobilization of biologically active enzymes, antibody fragments, or even pathogen-derived antigenic proteins (15-17). Due to the generally regarded as safe status of baker's yeast and because this organism tolerates a variety of cell wall modifications, it has become increasingly attractive as a host for expression of biotechnologically, pharmaceutically, and medicinally relevant proteins. The applications of yeast cell surface display include production of whole-cell biocatalysts and adsorbants which might play a role in the food industry, in the detoxification of wastewater, and in the display of peptide or antibody libraries, as well as in the development of serological diagnostics and vaccines (for a recent review see reference 8).
In biotechnology, enzyme immobilization is an important and often crucial step in the development of tailor-made biocatalysts (1). Thus, construction of novel, recyclable, and regenerative whole-cell biocatalysts by arming the yeast cell surface with proteins having enzymatic activity is of particular interest. Moreover, it has been shown that cell wall immobilization can also significantly enhance the stability and regeneration of a particular enzyme (17). In addition, cell surface display is also attractive for directed evolution as it facilitates both access of a substrate to an enzyme and selection of cells harboring the genetic information for a corresponding enzyme variant displayed on the surface, thus making tedious enzyme preparation and purification largely dispensable (18).
Among the hydrolytic enzymes, the esterases represent a group that has broad natural variety with respect to substrate specificity and reaction type, making them suitable for wider use in biotechnology (2, 9, 10, 14). Recently, a cell surface expression system with both high-level expression and equal and effective distribution of cell wall protein fusions in a yeast cell population was described (3). Here we aimed to refine this system for efficient cell surface display of a bacterial esterase (EstA) by cloning the entire estA gene of Burkholderia gladioli (13) downstream of the sequence encoding the signal peptide sequence of yeast Kre1p and upstream of sequences encoding the glycosylphosphatidylinositol-anchored yeast cell surface proteins Cwp2p and/or Flo1p to ensure in vivo targeting of EstA to the yeast cell wall. We also investigated whether esterase processing within the secretory pathway and subsequent display of the esterase on the yeast cell surface can be further enhanced by simultaneously activating the unfolded protein response (UPR) pathway.
MATERIALS AND METHODS
Yeast strains and culture media.
For in vivo expression of cell wall-anchored esterase A, Saccharomyces cerevisiae strains SEY6210 (MATα leu2-3 his3-Δ200 lys2-801 trp1-Δ901 suc2-Δ9) and BY4742 (MATα leu2-Δ0 his3-Δ1 lys2-Δ0 ura3-Δ0) and the isogenic Δhac1 null mutant BY4742-Δhac1 (Open Biosystems) were transformed with the corresponding vectors and cultivated in synthetic complete (SC) medium (pH 6.5) lacking uracil and/or leucine for up to 30 h in a fermentor at 30°C. Transformation was carried out by the lithium acetate method (7). To allow esterase-dependent cell growth, the SC medium (lacking glucose) contained 50 mM glycerol triacetate (Triacetin; Roth) as the sole carbon source.
Escherichia coli strains, plasmids, and DNA manipulations.
Standard molecular manipulations were performed as described by Sambrook et al. (11). For cloning, E. coli strain DH5α [F− recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(argF-lacZYA)U169 (Φ80dlacZΔM15) λ−] was grown at 37°C in LB medium supplemented with 100 μg ampicillin ml−1 when necessary. PCR amplification was performed using high-fidelity Taq DNA polymerase (Roche) according to the instructions of the manufacturer, and all products were routinely sequenced by fluorescent cycle sequencing with an automated DNA sequencer (LI-COR4200; MWG Biotech). EstA was PCR amplified from plasmid pMSH81 using primers 5′-Est (5′-GAATTCATGGTCCAGCTCCATATGG GCGGCGGTGACGAC-3′) and 3′-Est (5′-GTCGACCTTGGTGACGCCGGCCGCCGCG ATCTGCTG-3′) introducing either an EcoRI cleavage site (5′) or a SalI cleavage site (3′) (underlined). The intragenic cleavage site for SalI was removed by PCR-mediated overlap extension (3, 6) using 5′-Est (SalX) (5′-CGCTCGCCGCGCTGCAGGTAGACACCAGCAAGGTCAAGGTG-3′) and 3′-Est (SalX) (5′-CACCTTGACCTTGCTGGTGTCTACCTGCAGCGCGGCGAGCG-3′) as the primers. After amplification, the corresponding DNA fragment was subcloned into pCR2.1-TOPO used according to the instructions of the manufacturer (Invitrogen, Groningen, Germany), routinely sequenced, and finally cloned as an XhoI/BglII fragment into pFB2 and pFB3 (3) to obtain pFB2-EstA and pFB3-EstA, respectively. For cotransformation and coexpression of both EstA cell wall fusions (Kre1/EstA/Cwp2p and Kre1/EstA/Flo1p) in a single cell, the Kre1/EstA/Cwp2p cassette in plasmid pFB2-EstA was excised with XhoI and BglII and subsequently cloned into the 2μ multicopy yeast expression vector pYX242 (R&D Systems). The resulting plasmid, pFB4-EstA, contained LEU2 as a selectable marker and therefore could be cotransformed in yeast together with either pFB2 or pFB3, both containing URA3 as a selectable marker gene. The HAC1 expression plasmid pMS109 resembled a single-copy centromere vector that was kindly provided by M. Valkonen (20). Unless indicated otherwise, all vectors were transformed into S. cerevisiae SEY6210, and EstA expression and cell surface targeting were subsequently analyzed.
Indirect immunofluorescence and yeast cell wall preparation.
Exponentially growing yeast cells were harvested, washed with phosphate-buffered saline (PBS) (pH 7.4), and incubated with a polyclonal anti-EstA antibody (raised in a rabbit against the purified esterase A protein) diluted 1/100 in PBS for 1 h at room temperature. Subsequently, the cells were washed three times with PBS and incubated with fluorescein isothiocyanate-labeled anti-rabbit immunoglobulin G (Dianova) for 1 h at room temperature in the dark. After washing, the cells were resuspended in PBS containing 3.7% formaldehyde for fixation and examined with a fluorescence microscope (Olympus BX51) at an excitation wavelength of 488 nm. Isolation of yeast cell walls and glucanase extraction were performed as previously described (3, 15). The protein contents of whole cells and isolated cell walls were determined using a bicinchoninic acid protein assay kit (Sigma) with bovine serum albumin as the standard.
Enzyme activity assay.
To determine the esterase activity of whole cells, yeast transformants were grown in SC medium lacking uracil until the mid-exponential phase. Then a cell culture sample was pelleted and resuspended in PBS (pH 8.0) at a final optical density at 600 nm of 10; 32 μl of this suspension was added to 968 μl of 0.1 M Tris-HCl buffer (pH 7.0) containing p-nitrophenyl acetate (final concentration, 4.5 mM) as the substrate. The release of p-nitrophenol was measured at 32°C by determining the increase in absorbance at 405 nm with a spectrophotometer (UVmini 1240; Shimadzu). One unit of esterase activity was defined as the amount of enzyme releasing 1 μmol min−1 p-nitrophenol under the assay conditions used; under these conditions, the molar absorbance coefficient was determined to be 5.144 ml (μmol cm)−1.
RESULTS
Cell surface expression of EstA protein fusions.
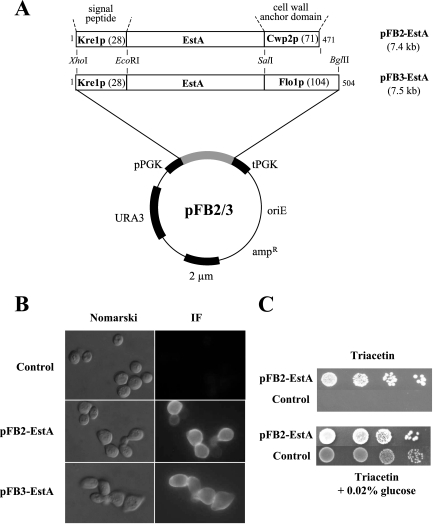
A technique to anchor the 9-amino-acid hemagglutinin epitope HA to the yeast cell surface by fusion to endogenous cell wall proteins (Cwp2p, Flo1p) was recently described (3), and we used this system for surface display of a bacterial esterase (EstA) in yeast. The estA gene of B. gladioli was PCR amplified, introducing an EcoRI site at the 5′ end and a SalI site at the 3′ end. Simultaneously, the internal SalI site was removed by splicing by overlap extension (SOE)-PCR (6), and the resulting fragment was ligated into pFB2 and pFB3 (3), yielding expression vectors pFB2-EstA and pFB3-EstA (Fig. 1A). In each plasmid, estA was under transcriptional control of the constitutive PGK1 promoter, encoding 43-kDa and 47-kDa protein fusions consisting of the Kre1p signal peptide (for import into the endoplasmic reticulum [ER] lumen), EstA, and the C-terminal part of either Cwp2p (pFB2-EstA) or Flo1p (pFB3-EstA). In vivo cell wall targeting, analyzed by indirect immunofluorescence, indicated that both EstA fusions were localized to the outer yeast cell surface, whereas control cells harboring the empty vector produced almost no fluorescent signal (Fig. 1B). As previously shown for cell surface display of HA (3), no obvious difference in labeling intensity or distribution was detectable for the two anchoring domains of Cwp2p or Flo1p.
FIG. 1.
(A) Schematic diagram of the cell wall fusion proteins used in this study. The number of amino acids for each fusion is indicated, and the length of the anchoring part (in amino acids) is shown in parentheses in each case. The fusion genes are under transcriptional control of the phosphoglycerate kinase promoter (pPGK) and terminator (tPGK), allowing constitutive in vivo expression. (B) Labeling of EstA-expressing cells of S. cerevisiae SEY6210 by indirect immunofluorescence using polyclonal anti-EstA and fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G. Nomarski and matching immunofluorescence (IF) micrographs are shown. (C) Esterase-dependent growth of S. cerevisiae expressing surface-anchored EstA from plasmid pFB2-EstA. Different dilutions of yeast transformants were spotted onto agar containing either 50 mM glycerol triacetate (Triacetin) or Triacetin with 0.02% glucose as the sole carbon source. Growth was recorded after 3 days of incubation at 30°C. In negative control cells, EstA was expressed in the cytosol.
To take advantage of EstA expressed on the yeast cell surface, the yeast transformants were tested to determine whether they grew on 50 mM glycerol triacetate (Triacetin) as a sole carbon source. Cell surface-immobilized EstA should enable yeast cells to hydrolyze Triacetin into glycerol and acetate, which in turn can be further metabolized as a carbon source. As shown in Fig. 1C, yeast cells expressing Kre1/EstA/Cwp2p were capable of growing on Triacetin, whereas negative control cells transformed with the empty vector (or cells expressing EstA in the cytosol [data not shown]) were not. Exactly the same phenotype was observed for yeast expressing Kre1/EstA/Flo1p on the cell surface (data not shown). Although addition of 0.02% glucose to the medium restored growth on Triacetin-containing agar for all cells, expression of EstA on the yeast cell surface provided a significant growth advantage compared with negative control cells (Fig. 1C). These data demonstrate that EstA expressed on the yeast cell surface accepts substrates other than p-nitrophenyl acetate, indicating that the active center of the enzyme must be accessible to even larger substrates.
Activity of cell surface-expressed EstA.
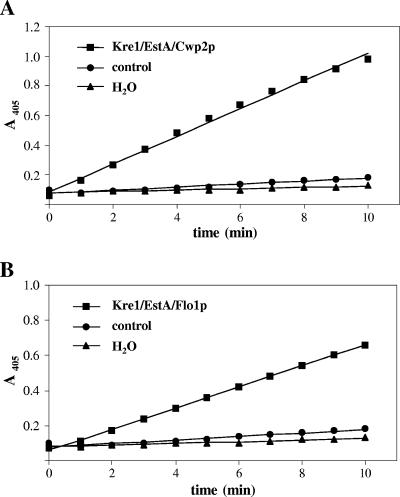
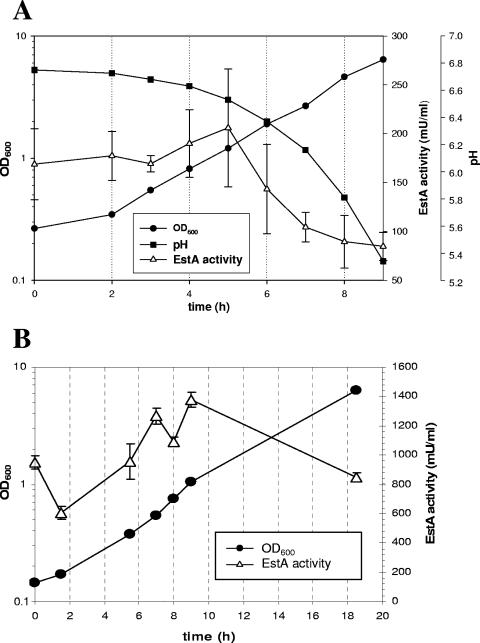
Esterase activity in EstA-expressing cells was determined by measuring the release of p-nitrophenol from nitrophenyl acetate as described previously (18). To ensure that the presence of cells did not interfere with the photometric assay of p-nitrophenol release at 405 nm, a control reaction was performed with cells transformed with the empty vector. The resulting activity was related to the protein content of the cells, which yielded a specific esterase activity of about 1 mU mg−1 protein. As shown in Fig. 2, the release of p-nitrophenol by control cells was not much greater than the release by a second control, illustrating that there was spontaneous hydrolysis of nitrophenyl acetate to p-nitrophenol and acetate. Having confirmed this, we next determined the release of p-nitrophenol by EstA-expressing yeast cells in a batch culture over a 30-h period. The esterase activity was highest in the early and mid-exponential growth phases (Fig. 3A [only data for Kre1/EstA/Cwp2p are shown]) independent of the protein that was used for cell wall anchoring, indicating that biologically active EstA is anchored to the wall. During exponential growth, however, a rapid decrease in enzyme activity was observed (Fig. 3A), which was not due to release of EstA from the cell wall as esterase activity was not detectable in the cell-free culture supernatant (data not shown). Rather, the loss of activity resulted from rapid inactivation of the enzyme due to a substantial decrease in the medium pH during cultivation (Fig. 3A). Consistently, esterase activity could be maintained at an almost constant level when the pH of the growth medium was controlled and continuously adjusted to pH 6.5, the optimum pH for EstA activity (13), by addition of 25% (wt/vol) NH4OH (Fig. 3B). To determine enzyme activity under these conditions (at constant pH 6.5), transformants were grown to the mid-log phase, and p-nitrophenol release was measured, which yielded specific activities of 103 mU mg−1 protein for Kre1/EstA/Cwp2p and 73 mU mg−1 protein for Kre1/EstA/Flo1p. Thus, at the cellular level, yeast cells expressing Kre1/EstA/Cwp2p and yeast cells expressing Kre1/EstA/Flo1p had specific esterase activities of 1.12 nU and 0.80 nU, respectively, while the activity in negative control cells was approximately 0.01 nU. Taken together, these data demonstrate that active B. gladioli EstA can be targeted equally effectively to the cell wall by using either Cwp2p or Flo1p as the anchoring domain.
FIG. 2.
Esterase activity in EstA-expressing yeast measured by release of p-nitrophenol from p-nitrophenyl acetate. The controls were yeast cells transformed with the corresponding empty vector or spontaneous hydrolysis of p-nitrophenol in water.
FIG. 3.
(A) Influence of medium pH on the activity of Kre1/EstA/Cwp2p-expressing yeast during growth in batch culture. Yeast cells were grown in a batch culture at pH 6.5 and harvested. Then 32 μl of a cell suspension having an optical density at 600 nm (OD600) of 10 was added to 968 μl of a reaction mixture in a photometer cuvette, and the formation of p-nitrophenol was measured for 2 min at 405 nm. (B) Activity of EstA-expressing cells of S. cerevisiae strain BY4742 during growth in a pH-regulated fermentor. Activity was measured as described above for panel A. Since the relative EstA activity did not differ significantly if Cwp2p or Flo1p was used as the C-terminal cell wall-anchoring domain, only results for Kre1p/EstA/Flo1p-expressing yeast transformants are shown.
EstA is covalently incorporated into the cell wall.
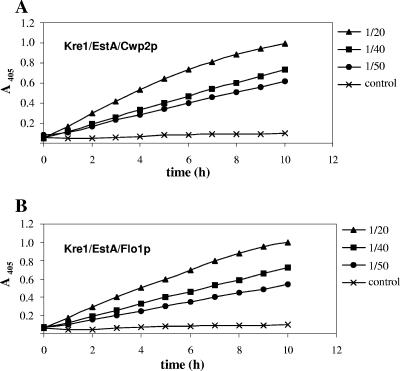
To verify that EstA is immobilized on the yeast cell surface, cell walls were isolated from yeast transformants and specific esterase activities were determined. As shown in Fig. 4, isolated cell walls were also capable of effectively hydrolyzing p-nitrophenyl acetate. The EstA specific activity was determined with samples diluted 1/40 and 1/50 since with the 1/20 dilution the cell wall-associated enzyme concentration was so high that the course of p-nitrophenol release became nonlinear (Fig. 4). While cell walls from Kre1/EstA/Cwp2p-expressing yeast exhibited esterase activity of 2.10 U mg−1 protein, the EstA activity in cell walls from Kre1/EstA/Flo1p-expressing cells was 1.17 U mg−1 protein. In contrast, the activity in isolated cell walls from negative control cells was only approximately 0.05 U mg−1 protein. The observed 20-fold (Kre1/EstA/Cwp2p) and 16-fold (Kre1/EstA/Flo1p) increases in specific esterase activity compared to whole cells (103 and 73 mU mg−1 protein) provided the first evidence that each protein fusion was integrated into the wall and exhibited esterase activity. To confirm that there was EstA cell surface anchoring through covalent linkage to the cell wall β-1,3-d-glucan network (21), isolated cell walls were extracted with the β-1,3-glucanase laminarinase to remove any covalently bound EstA from the yeast cell surface. As expected, laminarinase-extracted cell walls lacked any residual esterase activity (data not shown), confirming that the active form of EstA was originally embedded covalently in the cell wall. Consistently, the esterase activity was present exclusively in the culture supernatant after glucanase treatment, again demonstrating that EstA can be completely solubilized from the cell surface by laminarinase treatment.
FIG. 4.
Release of p-nitrophenol from p-nitrophenyl acetate by isolated cell walls of S. cerevisiae. Yeast cells were grown until the mid-exponential phase and harvested. Different dilutions of isolated cell wall fractions were added to 968-μl reaction mixtures in a photometer cuvette, and the formation of p-nitrophenol was determined at different times for 10 min at 405 nm. (A) Kre1/EstA/Cwp2p. (B) Kre1/EstA/Flo1p.
Activation of cellular UPR enhances EstA cell surface display.
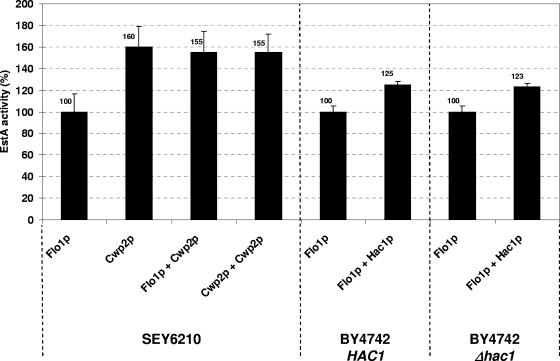
Sato and colleagues have recently shown that the C-terminal portion of Flo1p is crucial for anchoring in the cell wall, and consequently, the C-terminal 102 amino acids of Flo1p proved to be less effective than longer parts of the protein (12). Since in the present study a comparable portion of Flo1p (104 amino acids) was used, we asked if it was possible to improve its anchoring efficacy (without further elongating its C terminus) by constitutive activation of the yeast UPR pathway through single-copy expression of the UPR regulator Hac1p (4). Activated Hac1p has been shown previously to positively affect foreign protein secretion in S. cerevisiae by increasing the amount of ER chaperones required for proper protein folding in the secretory pathway (20). To analyze whether UPR activation also enhances cell surface expression in yeast, we constitutively coexpressed Hac1p on the single-copy yeast centromere vector pMS109 (20) together with either Kre1/EstA/Cwp2p or Kre1/EstA/Flo1p. As shown in Fig. 5, UPR activation through Hac1p single-copy expression increased the overall activity of Kre1/EstA/Flo1p on the yeast cell surface by about 25%. Interestingly, the specific esterase activity in a Kre1/EstA/Flo1p-expressing Δhac1 null mutant was not negatively affected and was the same as the EstA activity in the isogenic HAC1 wild-type strain (Fig. 5). Analogous to the situation in wild-type yeast, Hac1p single-copy expression in the Δhac1 mutant caused a comparable increase in EstA activity (about 23%) (Fig. 5), indicating that under normal growth conditions in a fermentor Hac1p and consequently the UPR are not induced. Attempts to further enhance EstA cell surface expression by overexpressing Hac1p from a 2μ multicopy vector resulted in a severe decrease in the growth rate of the corresponding yeast transformant (data not shown), most likely reflecting the additional burden on the cells when Hac1p and UPR were overexpressed. Despite this and in contrast to EstA cell surface anchoring via Flo1p (Kre1/EstA/Flo1p), the esterase activity in Kre1/EstA/Cwp2p-expressing wild-type yeast cells slightly but reproducibly increased when Hac1p was overexpressed from a 2μ multicopy vector, while a statistically significant increase in EstA activity was not observed after single-copy expression of Hac1p from pMS109 (data not shown). Nevertheless, the positive effect of UPR induction by single-copy expression of a constitutive active form of the UPR regulator Hac1p indicates that EstA cell surface expression can be enhanced by chaperone upregulation in the yeast secretory pathway, particularly when Flo1p provides the cell wall anchoring domain.
FIG. 5.
In vivo esterase activity of cell surface-expressed EstA is enhanced by constitutive activation of the cellular UPR. Wild-type yeast strains SEY6210 and BY4742 and an isogenic Δhac1 null mutant were cotransformed with EstA expression constructs, and overall esterase activities were determined before and after constitutive coexpression of Hac1p from the single-copy centromere vector pMS109. Average values were determined, and for comparison, the esterase activity in each wild-type transformant was defined as 100%.
EstA coanchoring by Cwp2p and Flo1p does not have a synergistic effect on cell wall-associated esterase activity.
In an attempt to further increase in vivo esterase activity, two EstA fusions were simultaneously coexpressed in a single cell by using 2μ vectors containing two different genetic markers (URA3 in plasmid pFB3-EstA and LEU2 in plasmid pFB4-EstA) that allowed selection of Ura+/Leu+ yeast transformants. To ensure that there were equal plasmid copy numbers in the single and double transformants, we included controls resembling yeasts that had been transformed with pFB3-EstA and simultaneously cotransformed with the empty vector pFB4 or had been transformed with pFB4-EstA and simultaneously cotransformed with the empty vector pFB3. Furthermore, the presence of equal plasmid copy numbers in the EstA-coexpressing transformants and in the EstA-expressing controls was confirmed by PCR (data not shown). Since in the double transformants the two fusions were anchored by different carrier proteins, we speculated that esterase activity might increase under such conditions. However, this was not the case, and EstA coanchoring by Flo1p and Cwp2p yielded an overall activity that roughly corresponded to the activity in yeast cells expressing Kre1/EstA/Cwp2p (Fig. 5). This indicates that the mechanism (or a critical step in the mechanism) responsible for incorporating EstA into the wall might be saturated or simply overloaded by coexpression of EstA from two different anchoring domains at a time. In support of this, constitutive coexpression of Kre1/EstA/Cwp2p from two different plasmids did not increase in vivo EstA activity (Fig. 5).
DISCUSSION
Cell surface expression has become a well-established method to immobilize biotechnologically relevant proteins on the surface of a unicellular microorganism. Besides phages, several gram-negative and gram-positive bacteria are capable of anchoring heterologous proteins to the cell surface (5). Because of safety demands in bioindustrial processes, yeast has been used for cell surface expression due to its generally regarded as safe status and the ease with which it can be genetically manipulated. Based on its widespread use in the production of beer, bread, and wine for centuries, S. cerevisiae is a suitable host and is particularly useful in the development of tailor-made cell surface modifications. Consequently, a variety of cell surface expression systems and applications of these systems have been designed in the past decade (8, 15, 17). For instance, effective incorporation of the viral HA epitope into the cell wall was achieved by using the natural yeast proteins Cwp2p, Flo1p, and Kre1p for intracellular fusion protein targeting (3). Here, we used this system to covalently attach a hydrolytic enzyme of biotechnological interest, esterase A of B. gladioli (13), to the yeast cell wall. Both anchoring proteins used, Cwp2p and Flo1p, were suitable for targeting EstA effectively to the yeast cell surface, resulting in esterase specific activities of 103 mU mg−1 protein in Kre1/EstA/Cwp2p-expressing yeast whole cells and 2.1 U mg−1 protein in isolated cell walls. In comparison, however, the esterase activity in Kre1/EstA/Flo1p-expressing yeast cells was significantly lower, and the levels of activity were only about 30% of the activity observed for Kre1/EstA/Cwp2p-expressing cells in whole cells and 45% in isolated cell walls. This finding is in agreement with the results of a previous study which showed that an α-galactosidase/Cwp2p fusion resulted in maximum hydrolytic activity compared to the activities observed with several other carrier proteins (21). The number of protein fusions embedded in the wall was probably responsible for this observation. However, exact quantification of cell wall-associated enzyme activity is somewhat hampered by the substantial glycosylation of yeast cell wall (manno)proteins, making it difficult to precisely determine the protein content of isolated cell walls; thus, specific activities determined for enzymes displayed on the yeast cell surface might be somewhat overestimated. Nevertheless, in the present study we found that EstA protein fusions could be released from the cell surface only by enzymatic digestion of the cell wall glucan layer, confirming that in either case EstA became tightly linked to the cell wall.
Interestingly, due to a substantial decrease in the pH during batch cultivation, the esterase activity of EstA-expressing yeast cells decreased rapidly after the cells entered the mid-exponential growth phase. Consequently, cultivation in a regulated fermentor under constant pH conditions restored EstA activity and ensured almost constant enzyme activity during the entire yeast cultivation process. In particular, for Flo1p-anchored EstA, the overall efficacy of cell wall targeting could be further increased by single-copy coexpression of a constitutive active variant of the transcription factor Hac1p, the major and central regulator of the cellular UPR in yeast (4). This is consistent with the results of a previous study demonstrating that providing an extra copy of Hac1p resulted in a 70% increase in α-amylase secretion, while endoglucanase secretion was not affected (20). In the case of Cwp2-anchored EstA, a slight but statistically significant increase in esterase cell surface display was observed only when Hac1p was overexpressed from a 2μ multicopy vector. The observed difference in cell wall-anchoring capacity between Cwp2p and Flo1p might be caused by the fact that full-length Cwp2p (i.e., the C-terminal 71 amino acids without the signal peptide) is already somehow optimized for or adapted to the cellular anchoring machinery and thus its efficacy or capacity for foreign protein anchoring to the wall cannot be increased further by UPR activation. In contrast, using only the C-terminal 104 amino acids of Flo1p as the carrier domain resembled using a rather drastic truncation of Flo1p since the natural length of Flo1p is 1,536 amino acids (19). Thus, any shortage of Flo1p might lead to a decrease in its anchoring efficacy caused by less effective accessibility of the anchored protein on the cell surface. In direct support of this hypothesis, a comparable 102-amino-acid domain has been proven to be less effective in anchoring foreign proteins to the yeast cell surface than Flo1p variants containing an extended C-terminal domain are (12). Furthermore, addition of the natural intramolecular spacer region of wild-type Flo1p to the truncated 104-amino-acid variant used in this study significantly optimized the accessibility of an HA peptide on the yeast cell surface (3). Likewise, it was shown previously that the length of the carrier domain in Flo1p has a direct effect on Flo1p-mediated cell wall expression (21). However, we show here that the anchoring efficacy of such an “artificial” domain can be further increased by enhancing cellular production of ER-resident chaperones (such as Pdi1p and Kar2p) due to Hac1p-mediated induction of the yeast unfolded protein response pathway. Interestingly and unexpectedly, we were unable to further increase the level of esterase activity by coexpressing the two fusions simultaneously in a single cell. Since the plasmid copy number in the EstA-coexpressing double transformants was the same as that in yeast transformants expressing only a single EstA construct and also containing the corresponding second plasmid as a control, this observation suggests that either the cellular anchoring machinery is overloaded or some critical step during secretion and/or cell wall anchoring (such as glycosylphosphatidylinositol anchor biosynthesis) is saturated. However, since we did not observe any significant EstA secretion into the culture supernatant, it can be assumed that the EstA fusion is stably embedded in the cell wall.
For many applications the cell surface-expressed protein must be accessible for large substrates. However, effective expression on the cell surface does not necessarily imply that there is a high level of enzyme activity (17). In the present study, the accessibility of immobilized EstA was analyzed indirectly by spotting esterase-expressing cells onto agar plates containing glycerol triacetate as the sole carbon source. If cell surface-immobilized EstA was active, it should have been able to hydrolyze glycerol triacetate into acetate and glycerol, both of which can be used for cell growth. In either case, regardless of the immobilizing protein used, the yeast was able to grow on Triacetin, whereas empty vector controls or cells expressing EstA in the yeast cell cytosol did not grow on Triacetin. Apparently, this substrate is easily accessible to immobilized esterase, and the resulting breakdown products are readily available for cell growth (although glycerol triacetate is considerably larger than p-nitrophenyl acetate, which was used as a substrate in the esterase activity assays). This interesting observation suggests that yeast cells expressing EstA on the surface could be used to cleave esterase-susceptible substrates of biotechnical interest. Thus, the first breakdown product is subsequently used or at least taken up by the cells, while the second product can be easily purified or at least enriched outside the cell.
By using the bacterial autotransporter system, Schultheiss et al. (18) succeeded in displaying esterase A on the cell surface of E. coli, yielding specific EstA activities of 1.7 mU mg−1 protein for whole cells and 23 mU mg−1 protein for the isolated outer membrane fraction. In comparison, the specific activities determined in the present study for yeast expressing Kre1/EstA/Cwp2p were roughly 60-fold higher for whole cells and several orders of magnitude higher for isolated cell walls (compared to the bacterial outer membrane fraction). This remarkable difference in efficacy demonstrates the tremendous capacity of S. cerevisiae for use in cell surface expression of biotechnologically relevant enzymes. Thus, our data indicate that esterase-displaying yeast could be used for a variety of biotechnological purposes; currently, a multistep reaction process is being developed to use esterases having different substrate specificities coexpressed on the surface of a single yeast as a cell-based biosensor.
Acknowledgments
We are grateful to M. Valkonen for providing the HAC1 expression vector pMS109.
We appreciate financial support provided by grant E-TTT from the Ministerium für Bildung, Kultur und Wissenschaft of the Saarland.
Footnotes
Published ahead of print on 15 September 2006.
Dedicated to Ferdinand Radler on the occasion of his 77th birthday.
REFERENCES
- 1.Bornscheuer, U. T. 2003. Immobilizing enzymes: how to create more suitable biocatalysts. Angew. Chem. Int. Ed. Engl. 42:3336-3337. [DOI] [PubMed] [Google Scholar]
- 2.Bornscheuer, U. T. 2002. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 26:73-81. [DOI] [PubMed] [Google Scholar]
- 3.Breinig, F., and M. J. Schmitt. 2002. Spacer-elongated cell wall fusion proteins improve cell surface expression in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 58:637-644. [DOI] [PubMed] [Google Scholar]
- 4.Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391-404. [DOI] [PubMed] [Google Scholar]
- 5.Georgiou, G., C. Stathopoulos, P. S. Daugherty, A. R. Nayak, B. L. Iverson, and R. Curtiss III. 1997. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15:29-34. [DOI] [PubMed] [Google Scholar]
- 6.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 7.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo, A., and M. Ueda. 2004. Yeast cell-surface display—applications of molecular display. Appl. Microbiol. Biotechnol. 64:28-40. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto, T., H. Fukuda, M. Ueda, A. Tanaka, and A. Kondo. 2002. Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Appl. Environ. Microbiol. 68:4517-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiter, B., A. Glieder, D. Talker, and H. Schwab. 2000. Cloning and characterization of EstC from Burkholderia gladioli, a novel-type esterase related to plant enzymes. Appl. Microbiol. Biotechnol. 54:778-785. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Sato, N., T. Matsumoto, M. Ueda, A. Tanaka, H. Fukuda, and A. Kondo. 2002. Long anchor using Flo1 protein enhances reactivity of cell surface-displayed glucoamylase to polymer substrates. Appl. Microbiol. Biotechnol. 60:469-474. [DOI] [PubMed] [Google Scholar]
- 13.Schlacher, A., T. Stanzer, I. Osprian, M. Mischitz, E. Klingsbichel, K. Faber, and H. Schwab. 1998. Detection of a new enzyme for stereoselective hydrolysis of linalyl acetate using simple plate assays for the characterization of cloned esterases from Burkholderia gladioli. J. Biotechnol. 62:47-54. [DOI] [PubMed] [Google Scholar]
- 14.Schoemaker, H. E., D. Mink, and M. G. Wubbolts. 2003. Dispelling the myths—biocatalysis in industrial synthesis. Science 299:1694-1697. [DOI] [PubMed] [Google Scholar]
- 15.Schreuder, M. P., S. Brekelmans, H. van den Ende, and F. M. Klis. 1993. Targeting of a heterologous protein to the cell wall of Saccharomyces cerevisiae. Yeast 9:399-409. [DOI] [PubMed] [Google Scholar]
- 16.Schreuder, M. P., C. Deen, W. J. Boersma, P. H. Pouwels, and F. M. Klis. 1996. Yeast expressing hepatitis B virus surface antigen determinants on its surface: implications for a possible oral vaccine. Vaccine 14:383-388. [DOI] [PubMed] [Google Scholar]
- 17.Schreuder, M. P., A. T. Mooren, H. Y. Toschka, C. T. Verrips, and F. M. Klis. 1996. Immobilizing proteins on the surface of yeast cells. Trends Biotechnol. 14:115-120. [DOI] [PubMed] [Google Scholar]
- 18.Schultheiss, E., C. Paar, H. Schwab, and J. Jose. 2002. Functional esterase surface display by the autotransporter pathway in Escherichia coli. J. Mol. Catal. B. Enzym. 18:89-97. [Google Scholar]
- 19.Teunissen, A. W., E. Holub, J. van der Hucht, J. A. van den Berg, and H. Y. Steensma. 1993. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast 9:423-427. [DOI] [PubMed] [Google Scholar]
- 20.Valkonen, M., M. Penttila, and M. Saloheimo. 2003. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Vaart, J. M., R. te Biesebeke, J. W. Chapman, H. Y. Toschka, F. M. Klis, and C. T. Verrips. 1997. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl. Environ. Microbiol. 63:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]