Abstract
Microbial respiration-based microbiosensors used for quantification of available dissolved organic carbon (ADOC) instantaneously respired by microorganisms are described. The sensing membranes contained aerobic seawater microorganisms immobilized in a polyurethane hydrogel. Molecular investigations revealed that the bacterial strain used was most closely related to Staphylococcus warneri. This strain was characterized by low substrate selectivity, which was reflected in the response to various mono- and disaccharides, short-chain fatty acids, and amino acids, as determined using Biolog microplates. Specific emphasis was placed on critically assessing biosensor functioning that was affected by preconditioning of the selected bacterial strain, chemical and geometric properties of the sensing membrane (e.g., composition, permeability, and thickness), and the distribution, biomass, and physiological state of immobilized cells, as well as the exposure conditions (e.g., temperature and nutrient supply). The sensors revealed that there was a linear response up to a glucose concentration of 500 μM depending on the type, characteristics, and recent history of the sensors. The detection limit of the sensors was equivalent to about 6 to 10 μM glucose. The 90% response time ranged from 1 to 5 min. Generally, the response of the biosensors became weaker with time. The shelf lives of individual sensors were up to 2 weeks. Measurements based on optical ADOC microbiosensors revealed that in photoautotrophically dominated sandy coastal sediments, the pool sizes and turnover of ADOC were regulated by the photosynthetic activity of benthic microalgae and microbial aerobic respiration. A large increase in ADOC production was observed shortly after the microphytobenthic primary production reached the maximum value at midday, whereas ADOC was consumed by microbial respiration during the night.
A relatively new and challenging technique in microbial ecology is the use of whole-cell microbial biosensors (12, 14, 48) for measurement of various organic and inorganic compounds in aquatic environments. Such biosensors consist of a transducer (e.g., an electrochemical electrode or optode) (for reviews, see references 2, 4, 9) in conjunction with prokaryotic or eukaryotic microorganisms that either are immobilized in a polymeric matrix or are suspended in a reaction chamber in front of the sensor tip. Metabolic functions, such as respiration or the luminescence of cells, are used to monitor the concentration of analytes that are either substrates or inhibitors of these processes. Depending on their selectivity, biosensors enable workers to measure either a specific microbial response to a single compound (specific biosensors) or a common response to groups of environmental compounds (nonspecific biosensors). The rapid and sensitive responses of biosensors have been utilized to determine the presence of a broad spectrum of substances, especially for environmental control (e.g., biosensors for environmental contaminants [47]) and toxic substances [16, 43]) and for use in the food industry (44) and biotechnology. There is an inexhaustible spectrum of microorganisms that have different metabolic capacities (for an overview, see reference 39) which are potentially available for detection of various organic and inorganic chemical substances.
During the last decade there has been special interest in the development of microbial microbiosensors for describing the small-scale distribution of ecologically relevant analytes in benthic microhabitats (6, 23, 38). Bacterium- or yeast-based microbiosensors have been developed for microscale measurement of microbially available dissolved organic carbon (ADOC) (32) in oxic environments, microbially available volatile fatty acids (e.g., acetate, propionate, and lactate) in anoxic environments (29), and nitrate and nitrite (24, 25, 28, 33) and methane (7, 8) in the pore water of sediments. All these microbial biosensors were characterized by measuring tips as small as 25 to 100 μm. Due to their small size and the efficient diffusional transport of substrates at a micrometer scale, microbiosensors respond much faster to changes in analyte concentrations than macrobiosensors respond. They enable us to understand the functioning of benthic microbial communities that interact during production and consumption of ecologically relevant substances, as mentioned above.
In this study we focused on biochemical and physiological properties of respiration-based ADOC microbiosensors that measure all of the dissolved organic carbon compounds that are instantaneously respired by the immobilized cells. Increased knowledge of sensor characteristics allowed us to critically assess sensor functioning and to develop a novel fiber optic-based ADOC microbiosensor. Here we describe the first attempt to use a fiber optic-based ADOC microsensor for continuous online measurement in a microphytobenthic colonized sediment. Combined microsensor-based measurements of ADOC, oxygen, and light intensity obtained during light and dark periods enabled us to obtain insight into short-term variations of ADOC concentrations in a benthic habitat.
MATERIALS AND METHODS
Isolation, selection, and storage of bacteria suitable for microbiosensors.
Aerobic chemoorganotrophic bacteria were isolated from brackish water (depth, 1 m; east coast of the Island of Hiddensee; 54°35.00′N, 13°7.20′E; southern Baltic Sea, Germany). Water samples were plated on modified ZoBell (ZB) medium (49) containing 0.5% (wt/vol) peptone, 0.1% (wt/vol) yeast extract, and 0.85% (wt/vol) NaCl (pH 7.0) and incubated in the dark at 21°C for 3 days. The pure bacterial cultures obtained were grown aerobically in ZB medium with shaking at 21°C. Bacterial strains suitable for microbiosensors were selected on the basis of their substrate utilization potential (selectivity) and oxygen consumption when they were exposed to glucose as a carbon source. To test these physiological parameters, 40 ml of freshly grown bacteria in the early stationary phase (10 h) was harvested by centrifugation at 4°C at 2,500 × g for 10 min using a refrigerated centrifuge (1.0 R Heraeus Megafuge). The pellet was washed twice with 30 ml of mineral salt medium (MSM), which contained 0.5% (wt/vol) NaCl, 0.1% (wt/vol) KCl, 0.1% (wt/vol) NH4Cl, 0.05% (wt/vol) K2HPO4 · 2H2O, 0.02% (wt/vol) MgSO4 · 7H2O, and 0.01% (wt/vol) CaCl2 · 2H2O (pH 7.0), and then it was resuspended in 40 ml of the same medium. The substrate utilization potentials of the bacterial isolates were determined by transferring 150-μl aliquots of cell suspensions (optical density at 578 nm, 1.5) into 96-well Biolog microplates (type GP2) (19). The microplates were incubated in the dark at 37°C for 2 days. Strains with a broad substrate spectrum and a positive response to glucose were selected. For measurement of oxygen consumption, 60-ml glass serum vials with butyl rubber stoppers (Supelco, Bellefonte, PA) were filled with MSM containing 1% (wt/vol) glucose, and then 1.5-ml portions of cell suspensions (optical density at 578 nm, 1.5) of 10 bacterial isolates were added. Bacterial oxygen consumption was measured continuously at 21°C for 6 h with optical oxygen microsensors (type B; PreSens, Regensburg, Germany) mounted in the silicone stoppers of the vials.
Bacterial stock cultures were maintained in cryotubes containing glass beads covered with a cryopreservative solution (Mast Diagnostica, Reinfeld, Germany) as described by Feltham et al. (10) and Jones et al. (18). These tubes were inoculated with cells from a freshly grown culture (1 day) and then stored at −20°C. Bacterial cells in cultures inoculated from these tubes were harvested at the beginning of the stationary phase (10 h) and immobilized in oxygen microsensors.
Phylogenetic analysis of the bacterial strain isolated.
Genomic DNA of the bacterial strain selected was extracted as described by Fesefeldt and Gliesche (11). PCR amplification of the 16S rRNA gene was performed as described by Rainey et al. (35), and sequencing reactions were performed by SEQLAB (Sequence Laboratories, Göttingen, Germany). To determine the closest relatives of the bacterial strain selected, the phylogenetic position (based on positions corresponding to positions 9 to 1510 of the Escherichia coli 16S rRNA gene) (3) was determined using the ARB database (42).
Morphological and physiological studies of the bacterial isolate.
Morphological characteristics, including the cell size, shape, and arrangement, of the bacterial isolate were determined microscopically (Axiophot; Zeiss). Determination of relevant physiological parameters involved testing the tolerance of the isolate to salinity and temperature by growing cells on agar plates. For this, freshly grown bacterial cells were plated on agar plates containing modified ZB medium containing 0.5 to 1.0% (wt/vol) NaCl and incubated aerobically at different temperatures (21, 48, and 60°C) for 3 days. Bacterial growth was monitored by recording the optical densities at 578 nm of the bacterial cultures in ZB medium containing 0.85% (wt/vol) NaCl incubated at 21°C at 30-min intervals up to 7 h. The percentages of cells with active respiration based on the total number of bacteria during different growth phases were determined using 200-μl subsamples and double staining with the fluorescent electron transport system-specific reagent 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) (final concentration, 5 mM) and 4,6-diamidino-2-phenylindole (DAPI) (final concentration, 5 μg ml−1) (37, 41). The total number of cells and the number of cells with active respiration were determined by epifluorescence microscopy using UV and green excitation light (wavelengths, 365 nm and 510 to 560 nm, respectively). The potential responses of the isolate to different carbon sources were analyzed using Biolog GP2 microplates (19) as previously described. API ZYM test strips (bioMérieux, Lyon, France) were used to screen the bacterial isolate for a range of hydrolytic enzymatic activities. Freshly grown cells in the early stationary phase were transferred to the solution delivered by the manufacturer of the API ZYM strips. The API ZYM strips were incubated in a humid chamber at 37°C for 4 h.
Construction of sensing membranes.
Polyurethane (PU) (NeoRez R-970; NeoResins, Waalwijk, The Netherlands) membranes containing microorganisms were constructed by mixing viable cells with a mixture of PU and polyvinylalcohol (PVA) (Elvanol 71-30; DuPont, Germany) (36). Bacterial cells were grown in 40 ml of modified ZB medium supplemented with 1% (wt/vol) glucose at 21°C. Cells in the early stationary growth phase (10 h) were harvested by centrifugation at 2,500 × g for 10 min (1.0 R Heraeus Megafuge), washed twice with 30 ml of MSM, and resuspended in approximately 0.4 ml of MSM. Then 100 to 200 μl of the bacterial suspension was thoroughly mixed with 180 μl of 12% (vol/vol) PU and 20 μl of 5% (wt/vol) PVA. Sensing membranes used for amperometric ADOC microbiosensors were prepared by adding 20 to 50 μl of the mixture to tapered tips of silane-coated glass micropipettes (inside diameter, 0.5 to 0.8 mm). The resulting cell-loaded PU membranes were dried for 1 h at room temperature and then stored at 4°C until they were used. The thickness of the soaked membranes used for amperometric microbiosensors ranged from 850 to 1,500 μm.
Visualization of immobilized cells in PU membranes.
Glass slides (5 mm by 5 mm) were covered with a thin layer of cell immobilisate prepared as described above. The cell immobilisate was dried for 1 to 2 h at room temperature, frozen for 5 to 7 h at −20°C, and then soaked in carbon-free MSM for 0.5 h at room temperature. Cell immobilisates were processed by sequential fixation with 2% (vol/vol) glutaraldehyde and 1.3% (wt/vol) osmium tetroxide and several washes with carbon-free MSM. To reduce cell shrinkage, an additional sequential fixation with 2% (wt/vol) tannic acid and 2% (wt/vol) uranyl acetate was performed (13), followed by sequential dehydration with ethanol at concentrations increasing from 10 to 60% (vol/vol). Samples were gold coated (Polaron SC 7640 sputter coater) and viewed with a Carl Zeiss scanning electron microscope (model DSM 940 A).
Microbiosensor construction.
To construct amperometric microbiosensors, a micropipette tip containing a dried sensing PU membrane was cut off and fastened with a microclamp in a stative holder (Fig. 1A). The sensing membrane was soaked in air-saturated carbon-free MSM for at least 2 h at 21°C. A Clark-type oxygen microelectrode constructed in our laboratory was connected to a picoamperemeter (JKT Technology, Kiel, Germany) and calibrated in oxygen-free and air-saturated carbon-free MSM. Using a micromanipulator and a stereomicroscope, the measuring tip of the oxygen microelectrode was inserted stepwise into the membrane. Oxygen concentrations in the matrix were determined each 10 μm. The optimal position of the sensor tip (Fig. 1B) was found to be close to the membrane-medium interface and was indicated by a sensor signal that was approximately 70 to 90% of the signal obtained at the membrane-air interface. After the optimal sensor position was determined, the glass housing of the sensing membrane was glued to the tip of the oxygen microelectrode. Then the amperometric ADOC microbiosensor was provided with a small amount of glucose (10 μM glucose solution) for 2 to 3 h before calibration was performed.
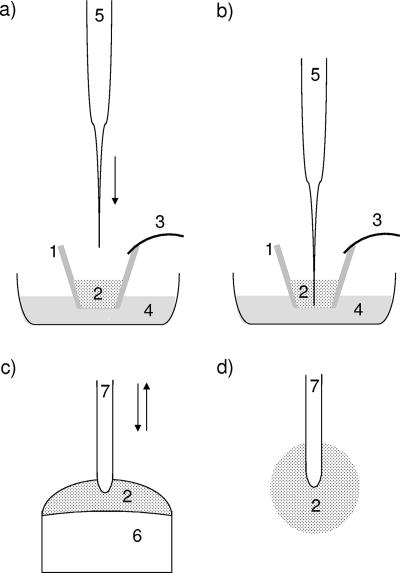
FIG. 1.
Schematic diagram of the construction of amperometric (a and b) and fiber optic (c and d) ADOC microbiosensors. 1, glass wall of a cut-off micropipette; 2, cell-loaded polyurethane hydrogel; 3, microclamp; 4, carbon-free mineral salt medium; 5, amperometric oxygen microelectrode; 6, tip of a plastic syringe; 7, oxygen microoptode. The arrows indicate how the oxygen microsensors are inserted into the cell-loaded hydrogel. For an amperometric ADOC sensor, the tip of an oxygen microelectrode was inserted stepwise into the gel, which was kept in carbon-free mineral salt medium. For a fiber optic ADOC sensor, the tip of an oxygen microoptode was repeatedly inserted into and withdrawn from the cell-loaded hydrogel until the optode was coated with a spherical sensing layer. (b and d) Final positions of the oxygen microsensors in the hydrogel. The drawings are not to scale.
For construction of a fiber optic-based microbiosensor, commercially available oxygen optodes with tip diameters of 30 to 140 μm (type B2; PreSens, Regensburg, Germany) and an oxygen meter (Microx TX2; PreSens) were used. Using a micromanipulator, the tip of the microoptode was initially inserted into a droplet of pure bacterial cell mass until a thin cell coating was formed. The cell layer was cured for 10 min. Then the tip of the optode was inserted 5 to 10 times into a droplet of cell-loaded PU hydrogel in the opening (inside diameter, 2 mm) of a single-use syringe (Fig. 1C and D). The procedure was complete when a cell-loaded hydrogel microsphere (diameter, approximately <300 μm) was formed. The gel microsphere was cured for at least 30 min at room temperature and then stored in carbon-free mineral salt medium. After 1 h, the optical ADOC microbiosensor was ready to use.
To maintain function, the tips of ADOC microbiosensors were inserted into a 10 μM glucose solution for 2 h at 21°C twice a week to provide the immobilized cells with organic carbon.
Calibration.
ADOC microbiosensors were calibrated in MSM supplemented with concentrations of glucose ranging from 0.01 to 2 mM. Standard solutions were air saturated and used at 21°C and a salinity of 8.5‰. After each measurement in glucose solution, sensors were kept in MSM until the signal returned to the initial value obtained before carbon addition. The insertion time was restricted to a maximum of 6 min to avoid carbon loading of the sensing membrane.
The 90% response time (t90)was calculated from the time-dependent linear decrease in the sensor signal immediately after the addition of glucose. Calibration curves were derived from the difference between the signal measured in the MSM and the signal measured in glucose containing standard solutions after 2 min (fiber optic-based sensors) and 4 min (amperometric sensors). Calibration curves were normalized to carbon equivalents. Sensor sensitivity was determined from the slopes of the linear parts of the calibration curves.
Reproducibility and shelf life.
To test the reproducibility of the measurements, we measured concentrations of glucose (0.1, 0.2, 0.5, and 1 mM) in duplicate. The stability of the sensor signal was examined during calibration measurement performed for different times (from hours to days).
Environmental application.
Samples of photoautotrophically dominated sediments from shallow-water coastal inlets (Libben; 54°35′N, 13°09′E; water depth, 3.4 m; Nordrügensche Bodden; southern Baltic Sea, Germany) were obtained with a multicorer (1) in September 2004. The water temperature and salinity determined with probes (LF 196 microprocessor conductivity meter with integrated temperature sensor; Wissenschaftliche Technische Werkstätten GmbH, Weilheim, Germany) were 15.9°C and 8.5‰, respectively. The light intensity determined with a light quantum sensor (LICOR LI-190SZ) directly above the seafloor at the time of sampling (2 p.m.) was 56 μmol photons m−2 s−1. The upper 2 mm of the sediment column (diameter, 10 cm; length, 20 cm) was sliced and placed in a plastic dish (diameter, 10 cm). Sediments were overlaid with a 3-mm layer of air-saturated carbon-free MSM. Three microoptodes for measurement of ADOC, oxygen, and light intensity (26) with tip diameters of approximately 400 μm, 140 μm, and 150 μm were used. Microsensors were positioned near each other in a 0.5-cm2 area at the sediment surface. The ADOC and oxygen sensors were connected to oxygen meters (type TX2; PreSens), and the light sensor was connected to an additional picoamperemeter (JKT Technology, Kiel, Germany). Continuous online measurements of ADOC, oxygen, and light intensity were obtained during light and dark periods at 21°C for 3 days. Data were recorded every 2 min. At the end of the experiment, sediments were thoroughly mixed to determine the water content (21), the total organic carbon and nitrogen contents with an elemental analyzer (Heraeus vario-el) (20), and the chlorophyll a content as a measure of microphytobenthic biomass (15).
RESULTS AND DISCUSSION
Our goal was to describe the characteristics of an ADOC microbiosensor that was first used by Neudörfer and Meyer-Reil (32) to obtain small-scale measurements in oxic sediments. This study was the first step in describing the properties and behavior of the bacteria immobilized and the matrix in order to improve our understanding of biosensor functioning. In addition, we used the fiber optic technology for construction of an ADOC microbiosensor for the first time. Our initial efforts involved monitoring ADOC concentrations with fiber optic-based biosensors in aquatic environments. In a case study simultaneous microsensor measurements of the ADOC and oxygen concentrations and the light intensity at the sediment surface provided detailed insight into the short-term dynamics of ADOC during light and dark periods.
Isolation of bacterial strains suitable for ADOC microbiosensors.
All 10 isolates investigated originated from brackish waters (salinity, 8.5‰). For construction of ADOC sensors we selected only isolates that exhibited aerobic growth in MSM supplemented with 1% (wt/wt) glucose as the sole carbon source. Of the three isolates characterized by rapid growth and a high oxygen consumption rate, the one (strain GB1) with the broadest substrate spectrum (lowest substrate selectivity) was chosen. This strain had an oxygen consumption rate of 6.4 to 8.7 μmol O2 h−1 in MSM at 21°C.
Phylogenetic and physiological characteristics of the bacterial isolate.
Phylogenetic analyses revealed that the chemoorganotrophic bacterial strain selected was closely related to the aerobic gram-positive organism Staphylococcus warneri (level of similarity, 99.07%). This conclusion was supported by morphological and physiological data (Table 1).
TABLE 1.
Morphological and physiological characteristics of S. warneri GB1
| Characteristic | Strain GB1a |
|---|---|
| Cell shape | Coccus |
| Cell size | 1 μm |
| Cell arrangement | Individual cells, diplococci, tetrads, short chains |
| Colony diam on 0.85% (wt/vol) NaCl agar | <1 mm (after 1 week of incubation at 21°C) |
| Colony color | Beige |
| Gram staining | Positive |
| Motility | − |
| Growth on agar containing: | |
| 0.5% (wt/vol) NaCl | + |
| 0.65% (wt/vol) NaCl | + |
| 0.85% (wt/vol) NaCl | + |
| 1.0% (wt/vol) NaCl | + |
| Growth at: | |
| 21°C | + |
| 45°C | + |
| 60°C | − |
| Aerobic growth | + |
| Acid production from: | |
| Glucose | + |
| Sucrose | (+) |
| Galactose | (+) |
| Maltose | (+) |
| Enzymatic activities | |
| Catalase | + |
| Arginine dihydrolase | + |
| Alkaline phosphataseb | + |
| Acid phosphataseb | + |
| Esteraseb | + |
| Esterase-lipaseb | + |
| Naphthol-AS-Bl-phosphohydrolaseb | + |
−, negative; +, positive; (+), weak positive.
Analyses were performed with API ZYM test strips.
Preconditioning of cells.
An important criterion for the functioning of an ADOC biosensor is that the microbial cells that are immobilized are preadapted to easily degradable organic carbon compounds (e.g., glucose). It is assumed that preincubation with substrates results in induction of microbial transport proteins and stimulation of degrading enzymes (39). To ensure that the bacteria that were going to be immobilized had high metabolic activity, cells were grown in glucose-supplemented ZB medium. Freshly grown cells of S. warneri GB1 had an oxygen consumption rate of 46 μmol O2 h−1 in ZB medium at 21°C 2 h after inoculation. The cells used for immobilization ideally should be repeatedly cultured on glucose-supplemented ZB medium and harvested during the late exponential or early stationary growth phase (optical density at 578 nm, >0.4). Harvesting cells from these growth phases is thought to be advantageous as (i) the cells exhibit relatively high metabolic activity and (ii) there is high enzyme stability.
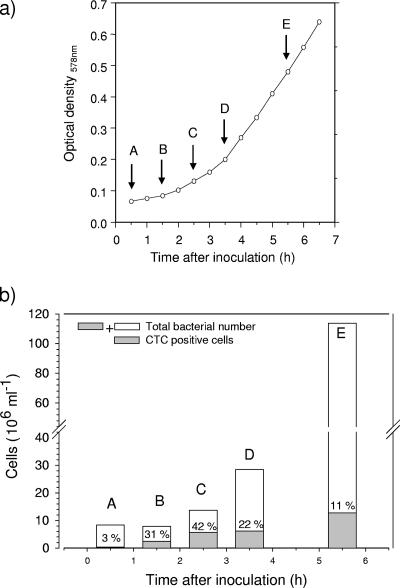
Electron transport system-based analyses of preconditioned cell cultures showed that the percentage of cells with active respiration varied during growth (Fig. 2). The percentage of CTC-positive cells was highest (42%) in the early exponential growth phase (2.5 h after inoculation) and decreased to 22% in the late exponential growth phase (3.5 h after inoculation). As the cells of S. warneri GB1 were relatively small (≤1 μm), formazane granules, formed by the reduction of CTC, might have been too small to be detected, and therefore the number of CTC-positive cells may have been underestimated. Despite this limitation, our results show that the respiration-based analysis which we used was suitable for revealing changes in bacterial respiratory activity in different growth phases.
FIG. 2.
(a) Growth curve for S. warneri GB1 in ZB medium at 21°C. Times A and B, lag phase; times C to E, exponential phase. (b) Total numbers of bacteria (DAPI counts) and numbers of bacteria with active respiration (CTC-positive cells) during different growth phases (see above) (b). The numbers above the gray bars indicate the percentages of CTC-positive cells based on the total numbers of bacterial cells.
Chemical and geometric properties of sensing membranes.
Besides the biological component, the chemical composition and the geometric features (e.g., “bioactive” surface area, shape, thickness, and pore volume) of the sensing membranes are important factors that influence the response and sensitivity of biosensors. The first ADOC microbiosensor described by Neudörfer and Meyer-Reil (32) contained aerobic chemoorganotrophic bacteria or yeast cells that were immobilized in a matrix consisting of PVA. To improve the diffusion and permeability characteristics of the sensing membrane of the ADOC sensor, microorganisms were entrapped in PU-based hydrogels in later studies (21, 22), as they were in this study. Compared to conventional PVA immobilization methods, PU-based membranes provide greater mechanical stability and elasticity (31) and greater porosity and are thought to be chemically inert and not degradable by microorganisms.
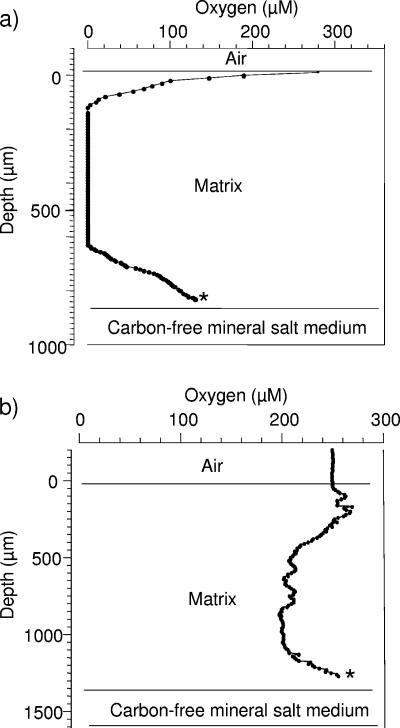
Comparing PVA membranes and PU-based membranes with comparable cell loading characteristics (the wet weight of cell material accounted for 35% of the total wet weight of the membrane), we found that the concentrations and distributions of oxygen differed inside the sensing membranes (Fig. 3). In gels consisting of PVA exclusively, steep gradients of O2 were established at the interfaces of the sensing membrane. At the upper interface (membrane-air interface) the O2 concentration rapidly decreased from 280 μM at the membrane surface to 0 μM at a depth of 120 μm. In the inner part of the membrane oxygen was absent. At 200 μm from the membrane's lower interface (membrane-aqueous medium interface) the O2 concentration started to increase again and reached a maximum of 130 μM. In contrast, in cell-loaded PU-based hydrogels, the O2 gradients at the membrane's interfaces were much less pronounced. In the upper 200 μm of the sensing membrane the O2 concentration ranged from 250 to 274 μM. Below this zone, the O2 concentration decreased slightly with increasing depth until a minimum concentration of 198 μM was reached. O2 concentrations between 198 and 212 μM in the inner part of the membrane (depth, 500 to 1,100 μm) indicated the high O2 permeability of PU-based membranes.
FIG. 3.
Oxygen permeability in cell-loaded PVA-based membranes (a) and PU-based membranes (b). The cell loads in the different matrices were comparable and accounted for 35% of the total membrane wet weight. For biosensor construction, the O2 microelectrode was fixed close to the membrane-medium interface, where the O2 concentrations reached approximately 70 to 90% of the value at the matrix-air interface. The final positions of the O2 microelectrodes are indicated by asterisks.
Whereas the chemical composition primarily determines the diffusion characteristics of the sensing membrane, the thickness and surface area influences the response time of the sensor. The matrices should be as thin as possible, resulting in a short diffusion distance and providing a fast response time. In addition, thin matrices exhibit less “memory effect” resulting from carbon remnants than thick matrices exhibit. In our study, fiber optic-based biosensors were coated with spherical sensing membranes that were approximately 100 to 200 μm thick, whereas amperometric biosensors had a planar sensing membrane whose thickness ranged from 850 to 1,500 μm (Fig. 1). The 90% response times of the fiber optic-based microsensors (t90, 1.0 to 1.5 min) were generally two- to threefold less than those of the amperometric microsensors (t90, 3 to 5 min), reflecting the shorter diffusion paths between the surfaces of the matrices and the reactive surfaces of the sensors.
Microbial biomass and cell distribution in sensing membranes.
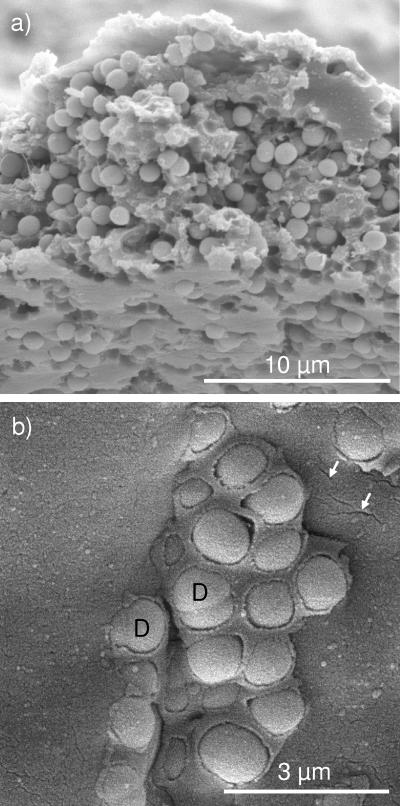
Besides the chemical and geometric properties of the membrane, the density and distribution of the cells entrapped in the sensing membrane are essential factors which influence the response of a sensor. Scanning electron micrographs (Fig. 4) revealed that immobilized cells of S. warneri GB1 were loosely packed and homogeneously distributed in freshly constructed membranes. Only a very few dividing cells were entrapped in the membranes. Small spaces around the cells and microchannels in the hydrogel were thought to enhance the porosity of the matrix and to guarantee that gases and solutes could be easily delivered to the cells and that wastes could be efficiently removed.
FIG. 4.
Scanning electron micrographs of freshly constructed PU membranes with S. warneri cells (original magnification, ×2,000 [a]). The membrane was prepared as described in the text for scanning electron microscopy and cut to visualize the embedded cells, and 10,000-fold magnification of the surface of the membrane revealed small fissures (arrows) in the gel (b). D, dividing cells. The micrographs were provided by R. Hanschke.
We found that the optimal cell load was in the range from 35 to 55% of the total membrane wet weight. A lower cell load resulted in a weak response, whereas a higher cell load resulted in a nonresponding sensor (data not shown) due to the limited permeation of gases and substrates. From these observations, we concluded that the amount of cells immobilized in the sensing membrane plays an important role in the response of a microbiosensor. A similar trend has been reported for macroscale biological oxygen demand sensors. Previously, some authors (5, 40) showed that with immobilized Bacillus subtilis cells (on dialysis membranes) and Pseudomonas sp. cells (on cellulose nitrate membranes) there was an initial increase in the response of the sensors with increasing cell loads and that the sensor signal subsequently decreased with higher cell loads.
Exposure of sensing membranes to changing environmental conditions (such as changes in the nutrient supply in calibration solutions and natural water samples, temperature, and salinity) influences the biomass and distribution of immobilized cells. Substrates added to the sensing membranes are potential nutrient sources that could induce growth of the immobilized bacteria (45) and might limit diffusion due to biomass accumulation. With prolonged exposure time decaying cells release nutrients that might be an additional nutrient source for viable bacteria in the membrane. Wijffels (46) reported changes in bacterial biomass and distribution in carrageenan gel beads (radius, 1 mm). Microscopic images revealed that initially small microcolonies of entrapped Nitrosomonas europaea were homogeneously distributed over the gel beads. With increasing duration of cultivation, pronounced bacterial biomass gradients were formed. After 49 days of cultivation, about 90% of the total biomass was localized in large colonies near the bead surface down to a depth of 100 μm.
Response of the sensors to labile organic carbon.
The response of an ADOC sensor to labile organic carbon is specific for the bacterial strain used and depends on the physiological potential (substrate spectrum and sensitivity) of the immobilized bacteria. In this study we defined ADOC as all of the labile organic compounds that were respired by the S. warneri strain used within a few minutes. This means that some substrates that are rapidly used by other bacterial strains might not have been included in the ADOC. Additionally, it is necessary to consider the fact that natural ADOC consists of a complex mixture of organic substances that are different sizes and have different diffusion coefficients, resulting in a highly variable distribution and concentration of organic substances in the hydrogel, thus influencing the response of the bacteria.
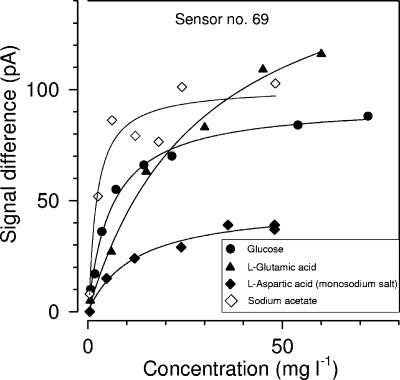
The responses of ADOC microbiosensors to selected dissolved organic compounds are shown in Fig. 5. There was a strong response to glucose, l-glutamic acid, and sodium acetate. The response to l-aspartic acid was weakened, and no reaction to glycine or serine was detected. The responses of the ADOC sensors coincided with the results of the Biolog microplate analyses (Table 2), showing that Biolog microplates are suitable tools for screening the spectrum of substrates that are immediately utilized by the cells used in the ADOC sensors.
FIG. 5.
Response of an amperometric ADOC sensor to different instantaneously utilizable carbon substrates.
TABLE 2.
Response of S. warneri GB1 to various carbon sourcesa
| Carbon source | Reactionb |
|---|---|
| Mono-, di-, and trisaccharides | |
| d-Glucose | + |
| d-Fructose | + |
| Maltose | + |
| d-Mannose | + |
| d-Psicose | + |
| 3-Methyl-glucose | + |
| β-Methyl-d-glucoside | + |
| N-Acetyl-d-glucosamine | + |
| Palatinose | + |
| Gentiobiose | (+) |
| d-Trehalose | (+) |
| Turanose | (+) |
| Sucrosec | (+) |
| Maltotriose | (+) |
| Polysaccharide | |
| Dextrin | (+) |
| Alcohols | |
| d-Arabitol | + |
| d-Mannitol | (+) |
| Glycerol | + |
| Carboxylic acids | |
| Formic acidc | (+) |
| N-Acetyl-l-glutamic acid | + |
| α-Ketobutyric acidc | (+) |
| α-Ketoglutaric acid | (+) |
| α-Ketovaleric acid | + |
| l-Lactic acid | (+) |
| Ester | |
| Methyl pyruvate | + |
| Polysorbate | |
| Tween 80c | (+) |
| Amino acids | |
| d-Alanine | (+) |
| l-Alanine | + |
| l-Serine | + |
| l-Glutamic acid | + |
| l-Threoninec | (+) |
| Dipeptide | |
| l-Alanyl-glycine | + |
| Phosphates | |
| Glucose-6-phosphate | (+) |
| dl-α-Glycerol-phosphate | + |
Unless indicated otherwise, substrates were tested by using type GP2 microplates (for gram-positive bacteria).
+, positive reaction; (+), weak reaction.
Substrate tested by using type GN2 microplates (for gram-negative bacteria).
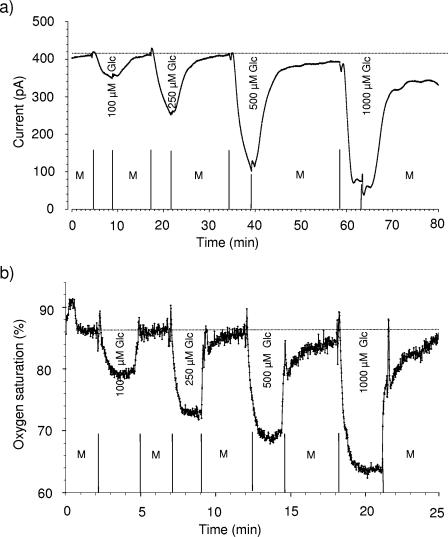
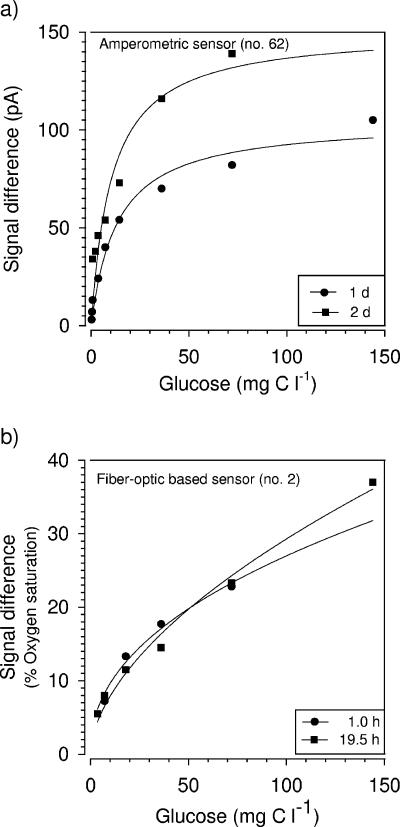
To calibrate ADOC sensors, a mixture of different carbon sources that is similar to the natural composition of ADOC is desirable. However, as analysis of ADOC is still a challenge in microbial ecology and we focused on a preliminary approach to ADOC quantification, the sensor's response was standardized using only a single carbon source (glucose). The ADOC microbiosensors generally exhibited a linear response at low glucose concentrations (for fiber optic-based sensors, 0 to 200 μM, equivalent to 0 to 14.4 mg C liter−1; for amperometric sensors, 0 to 300 μM, equivalent to 0 to 36.0 mg C liter−1) and a nonlinear response at higher concentrations (Fig. 6 and 7). The nonlinear response at high glucose concentrations might be explained by substrate saturation of the immobilized cells. Prolonged recovery phases of the sensors revealed that more time was required by the cells to consume the excess organic carbon (“memory effect”). Sensors responding to relatively high glucose concentrations reached the steady state only after a long time (15 min to 2 h), whereas sensors responding to low glucose concentrations returned to a steady state in carbon-free MSM within a few minutes.
FIG. 6.
Responses of amperometric (a) and fiber optic-based (b) ADOC sensors to glucose at 21°C. The sensors were alternately inserted in carbon-free mineral salt medium (M) and solutions containing glucose at concentrations ranging from 100 to 1,000 μM. After addition of glucose, the time-dependent current signal of the sensor decreased as oxygen was consumed by the immobilized aerobic bacteria responding to glucose. The dotted line indicates the sensor signal before glucose addition.
FIG. 7.
Calibration curves for amperometric (a) and fiber optic-based (b) ADOC sensors responding to glucose at 21°C. The response of the amperometric sensor was recorded 1 and 2 days after sensor construction, whereas the response of the fiber optic-based sensor was detectable 1 and 19.5 h after it was manufactured. The calibration curves were derived from the differences in the sensor signals measured in the air-saturated carbon-free medium and the glucose solution.
Generally, fiber optic-based biosensors and amperometric microsensors had t90 of 1.0 to 1.5 min and 3 to 5 min, respectively (Fig. 6). The detection limit of ADOC microbiosensors was equivalent to approximately 6 to 10 μM glucose, depending on the type and individual characteristics of the sensors. Duplicate measurements for 0.1, 0.2, 0.5, and 1 mM glucose solutions deviated less than 10% (data not shown). In addition, newly constructed fiber optic ADOC microbiosensors responded immediately (within 1 h) to glucose, whereas amperometric sensors responded to glucose after approximately 5 h of preconditioning, which was necessary for membrane soaking and activation of the immobilized cells in a carbon-containing medium.
Unsoaked cell-loaded membranes used for amperometric ADOC biosensors could be stored separately for 6 to 8 weeks at 4°C. As soon as the membranes were soaked in carbon-containing medium for 1 to 2 h at 21°C, the immobilized bacteria were reactivated and responded immediately to labile organic carbon. The preparation of high numbers of membranes could facilitate ADOC sensor construction as time-consuming processes such as cultivation and preconditioning of the cells are not necessary. A promising idea for further sensor development is to have ADOC sensors with replaceable membranes, which would allow (i) replacement of only the biological component while the same transducer (O2 microelectrode) was used and (ii) varying the substrate response of the immobilized bacteria by using different bacterial strains or natural microbial populations.
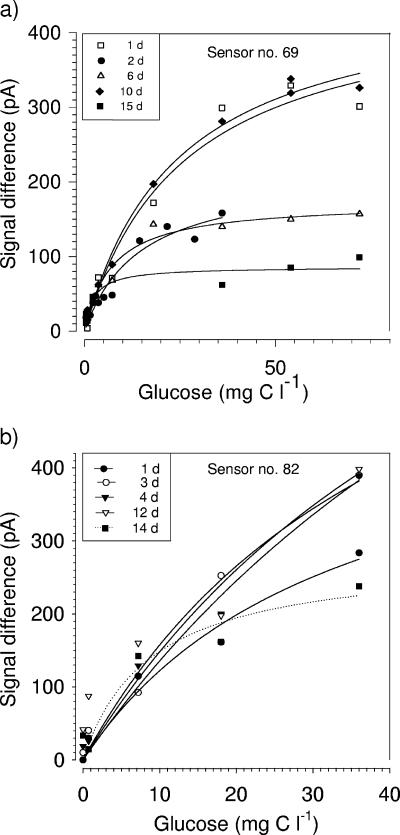
Shelf life and sensitivity of sensors.
The shelf life of ADOC microbiosensors was up to 2 weeks when sensors used for ADOC measurements were kept in carbon-free MSM at 21°C during nonmeasuring periods. The sensitivity of the sensors decreased with prolonged shelf life, as shown for two examples in Fig. 8. The highest sensitivities of amperometric sensors 69 and 82 were observed on days 1 and 10 and on days 3, 4, and 12, respectively, after construction. The reason for the temporary decreases in the sensitivity of sensor 69 on days 2 and 6 is unclear as the individual “prehistory” of each sensor was not studied in detail. A temporary excess of organic carbon, as well as unfavorable microenvironmental conditions (e.g., microbial production of inhibiting substances), might have been responsible for reduced aerobic respiration of the immobilized cells. After 2 weeks, the signals of sensors 69 and 82 decreased by approximately 10 to 80% compared with the initial signal depending on the glucose concentration. Alterations in the distribution of microbial biomass, the cell viability, and the permeability properties of the sensing membrane might have caused the decrease in sensitivity with time. The sensitivity also drastically decreased when the sensor was continuously supplied with high glucose concentrations (>1 mM; 72 mg C liter−1) for several days (data not shown). The initial sensitivity of the sensor was retained when glucose was added after 3 days of nutrient depletion. These observations revealed that the respiration activity of the immobilized cells was influenced by the nutrient availability and the duration of nutrient exposure. Similar observations were reported by Leenen et al. (27), who studied the dynamics of growth and death of immobilized nitrifying cells (N. europaea) in carrageenan gel beads during 40 days of cultivation. These authors found that the number of viable cells was reduced at high substrate concentrations, which led to decreasing aerobic respiration rates, whereas the number of viable cells near the surface of the gel beads increased again after a period of nutrient depletion. Other investigators have also reported reduced sensitivity of immobilized cells after only a few days. Hyde et al. (17) described reduced efficiency for Pseudomonas aeruginosa entrapped in polytetrafluoroethylene, which was reflected in a 26% reduction in substrate turnover on day 2. After 5 days substrate turnover was not detectable.
FIG. 8.
Long-term stability of amperometric ADOC sensors. The responses of sensors 69 (a) and 82 (b) to glucose were recorded for 2 weeks.
Environmental application of ADOC microbiosensors.
Although numerous novel microbial biosensors have been developed for measurement of various organic and inorganic compounds during the last decade, the environmental application of biosensors is still limited. In particular, determination of the amount of organic carbon in aquatic habitats that is instantaneously utilizable by microbes is a key variable that can be monitored with microbial ADOC biosensors. The information obtained provides valuable information concerning spatiotemporal variations in ADOC and thus contributes to our understanding of the carbon flux through microbial communities.
Until now, data based on the use of ADOC microbiosensors for environmental monitoring have been very scarce. Using amperometric ADOC microbiosensors, the concentrations and pool sizes of labile organic carbon have been determined in coastal sediments of the Nordrügensche Bodden (southern Baltic Sea) (21, 22). These studies indicated that spatial variations in ADOC concentrations were related to sediment characteristics and microbial activity.
The general limitations of amperometric sensors (e.g., oxygen consumption, signal drift, fragility, time-consuming construction) prompted the development of optically based microbial macrobiosensors that have been used recently for measurement of biological oxygen demand (5, 34). This study was the first attempt to use a newly developed optical ADOC microbiosensor that has numerous advantages compared to electrochemical sensors (e.g., easier construction and handling, no polarization, no signal drift, no electrical disturbance). The goal of this study was to use fiber optic-based microsensors to monitor short-term variations in ADOC in a laboratory experiment.
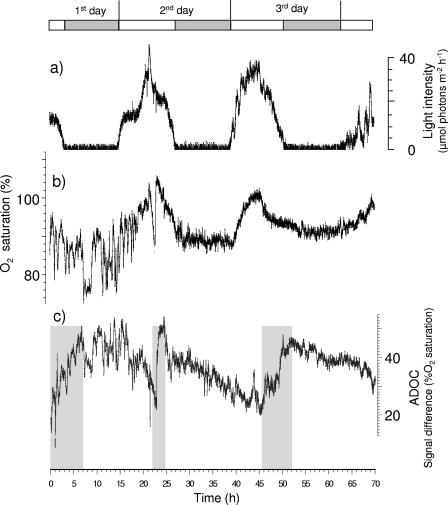
Microsensor-based measurements were obtained for sandy photoautotrophically dominated surface sediments (depth, 0 to 2 mm) of the Nordrügensche Bodden (southern Baltic Sea). These sediments had a water content of 24% and a total organic carbon content of 0.2%. A chlorophyll a concentration of 15.0 μg cm−3 indicated that there was a relatively large active microphytobenthic biomass. Variations in ADOC, oxygen, and light intensity were measured by microoptodes during cycles consisting of 12 h of light and 12 h of darkness. The light intensities measured at the sediment-water interface during the light periods ranged from 1 to 39 μmol photons m−2 s−1 and were similar to in situ light intensities (Fig. 9A). Increases in light intensity were paralleled by increases in oxygen concentration, indicating that there was photosynthetic oxygen production by benthic microalgae (Fig. 9B). In the dark, decreases in the oxygen concentration were caused by microbial aerobic respiration.
FIG. 9.
Short-term variations in light intensity (a), oxygen (b), and ADOC (c) measured with fiber optic-based microoptodes at the sediment surface of photoautotrophically dominated sediments. The data show that the ADOC concentration (expressed as the difference in oxygen saturation signal between the oxygen sensor and the ADOC sensor) increased shortly after microphytobenthic oxygen production reached its maximum value at 1:00 p.m. ADOC continuously accumulated for several hours (shaded areas). Then the O2 demand of respiring bacteria caused a decrease in O2 saturation coinciding with a decrease in the net concentration of ADOC. Whereas ADOC was immediately consumed during the dark period on days 2 and 3, relatively high ADOC concentrations remained during the dark period on day 1, probably because of additional sources of dissolved organic substrates (e.g., release of ADOC by dead and decaying animals) that counteracted the immediate consumption of photoautotrophically produced ADOC. The open and shaded bars above the curves indicate the lengths of the light and dark periods, respectively.
The ADOC concentrations above the sediment surface ranged from 7.8 to 32.5 mg liter−1. These values were similar to ADOC concentrations measured for the pore water of estuarine surface sediments (depth, 0 to 1 cm) of the southern Baltic Sea (slightly muddy sand), where the maximum ADOC concentration was 37.7 mg liter−1 (corresponding to 20.8 μg cm−3) (22). It was especially interesting that the ADOC concentration increased shortly after the maximum oxygen production was observed. This increase lasted for approximately 6, 3, and 7 h on days 1 to 3 before the ADOC was consumed by microbial aerobic respiration in the following 16, 20, and 17 h, respectively. The rapid consumption of microbially available organic carbon observed on day 1 was due to high ADOC turnover, whereas the decreasing ADOC consumption rates observed on days 2 and 3 were due to a decrease in ADOC turnover. It is assumed that incubation effects might be responsible for changes in ADOC turnover; the signal fluctuations of the oxygen sensor and ADOC sensor were much greater on the first day and decreased on the second and third days, when the benthic system became more stabilized. Our results revealed turnover times for newly produced ADOC of about 7 to 20 h for the sediment investigated. This is in the range for the turnover times of easily degradable organic in coastal sediments analyzed by Meyer-Reil and Faubel (30) using radiolabeled substrates.
The preliminary results of our time series experiment enabled us to obtain insight into the temporal scales of major microbial processes that influence the pool size and the turnover of labile organic carbon in benthic (micro)habitats. For ongoing spatiotemporal investigations of ADOC it is very important to utilize the numerous advantages of fiber optic-based biosensors and to improve their biochemical and physiological characteristics. Information concerning the influence of various environmental parameters on the biomass distribution, activity, and physiological state of the immobilized cells in biosensor membranes is essential for understanding their functioning. Based on the results of this study, improved ADOC microsensors with defined characteristics will be used for ongoing investigations that focus on short-term observations of carbon that is instantaneously available to microbes and its turnover in various pelagic and benthic (micro)environments.
Acknowledgments
We are grateful to F. Schauer and his undergraduate students (Department of Microbiology, Ernst-Moritz-Arndt-Universität Greifswald) for performing morphological and physiological analyses of S. warneri GB1. We also thank R. Hanschke for her valuable help with scanning electron microscopy and S. Schalla for performing the PCR analyses.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Black, H. J., M. Dainat, M. Köster, and L.-A. Meyer-Reil. 2002. A multiple corer for taking virtually undisturbed samples from shallow water sediments. Estuar. Coast. Shelf Sci. 54:45-50. [Google Scholar]
- 2.Blum, L. J., and P. R. Coulet. 1991. Biosensor principles and applications. Marcel Dekker, New York, NY.
- 3.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of the 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byfield, M. P., and R. A. Abuknesha. 1994. Biochemical aspects of biosensors. Biosens. Bioelectron. 9:373-400. [DOI] [PubMed] [Google Scholar]
- 5.Chee, G. J., Y. Nomura, K. Ikebukuro, and I. Karube. 2000. Optical fiber biosensor for the determination of low biochemical oxygen demand. Biosens. Bioelectron. 15:371-376. [DOI] [PubMed] [Google Scholar]
- 6.Damgaard, L. R., L. H. Larsen, and N. P. Revsbech. 1995. Microscale biosensors for environmental monitoring. Trends Anal. Chem. 14:300-303. [Google Scholar]
- 7.Damgaard, L. R., and N. P. Revsbech. 1997. A microscale biosensor for methane containing methanotrophic bacteria and an internal oxygen reservoir. Anal. Chem. 69:2262-2267. [DOI] [PubMed] [Google Scholar]
- 8.Damgaard, L. R., N. P. Revsbech, and W. Reichardt. 1998. Use of an oxygen-insensitive microscale biosensor for methane to measure methane concentration profiles in a rice paddy. Appl. Environ. Microbiol. 64:864-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Souza, S. F. 2001. Microbial biosensors (review). Biosens. Bioelectron. 16:337-353. [DOI] [PubMed] [Google Scholar]
- 10.Feltham, R. K. A., A. K. Power, P. A. Pell, and P. H. A. Sneath. 1978. A simple method for storage of bacteria at −76°C. J. Appl. Bacteriol. 44:313-316. [DOI] [PubMed] [Google Scholar]
- 11.Fesefeldt, A., and C. G. Gliesche. 1997. Identification of Hyphomicrobium spp. using PCR-amplified fragments of the mxaF gene as a molecular marker. Syst. Appl. Microbiol. 20:387-396. [Google Scholar]
- 12.Frense, D., A. Müller, D. Beckham, and U. Klingebiel. 1997. Mikrobielle Sensoren für die Umweltanalytik. Biotec 6:36-38. [Google Scholar]
- 13.Hanschke, R., and F. Schauer. 1996. Improved ultrastructural preservation of yeast cells for scanning electron microscopy. J. Microsc. 184:81-87. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, L. H., and S. J. Sørensen. 2001. The use of whole-cell biosensors to detect and quantify compounds or conditions affecting biological systems. Microb. Ecol. 42:483-494. [DOI] [PubMed] [Google Scholar]
- 15.Helsinki Commission. 1988. Guidelines for the Baltic monitoring programme for the third stage. Baltic Sea Environmental Proceedings, vol. 27D. Biological determinants. Helsinki Commission, Helsinki, Finland.
- 16.Hollis, R. P., K. Killham, and L. A. Glover. 2000. Design and application of a biosensor for monitoring toxicity of compounds to eukaryotes. Appl. Environ. Microbiol. 66:1676-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde, F. W., G. R. Hunt, and L. A. Errede. 1991. Immobilization of bacteria and Saccharomyces cerevisiae in poly(tetrafluoroethylene) membranes. Appl. Environ. Microbiol. 57:219-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, D., P. A. Pell, and P. H. A. Sneath. 1991. Maintenance of bacteria on glass beads at −60°C to −76°C, p. 45-50. In B. E. Kirsop and A. Doyle (ed.), Maintenance of microorganisms and cultures cells. Academic Press, London, United Kingdom.
- 19.Konopka, A., L. Oliver, and R. F. Turco. 1998. The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb. Ecol. 35:103-115. [DOI] [PubMed] [Google Scholar]
- 20.Köster, M., S. Dahlke, and L.-A. Meyer-Reil. 1997. Microbiological studies along a gradient of eutrophication in a shallow coastal inlet in the southern Baltic Sea (Nordrügensche Bodden). Mar. Ecol. Prog. Ser. 152:27-39. [Google Scholar]
- 21.Köster, M., S. Dahlke, and L.-A. Meyer-Reil. 2005. Microbial colonization and activity in relation to organic carbon in sediments of hypertrophic coastal waters (Nordrügensche Bodden; Southern Baltic Sea). Aquat. Microb. Ecol. 39:69-83. [Google Scholar]
- 22.Köster, M., and L.-A. Meyer-Reil. 2001. Characterization of carbon and microbial biomass pools in shallow water coastal sediments of the southern Baltic Sea (Nordrügensche Bodden). Mar. Ecol. Prog. Ser. 214:25-41. [Google Scholar]
- 23.Kühl, M., and N. P. Revsbech. 2001. Biogeochemical microsensors for boundary layer studies, p. 180-210. In B. P. Boudreau and B. B. Jørgensen (ed.), The benthic boundary layer. Transport processes and biogeochemistry. Oxford University Press, Oxford, United Kingdom.
- 24.Larsen, L. H., N. P. Revsbech, and S. J. Binnerup. 1996. A microsensor for nitrate based on immobilized denitrifying bacteria. Appl. Environ. Microbiol. 62:1248-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen, L. H., T. Kjær, and N. P. Revsbech. 1997. A microscale NO3-biosensor for environmental applications. Anal. Chem. 69:3527-3531. [DOI] [PubMed] [Google Scholar]
- 26.Lassen, C., H. Plough, and B. B. Jørgensen. 1992. A fibre-optic scalar irradiance microsensor: application for spectral light measurements in sediments. FEMS Microbiol. Ecol. 86:247-254. [Google Scholar]
- 27.Leenen, E. J. T. M., A. A. Boogert, A. A. M. van Lammeren, J. Tramper, and R. H. Wijffels. 1996. Dynamics of artificially immobilized Nitrosomonas europaea: effect of biomass decay. Biotechnol. Bioeng. 55:630-641. [DOI] [PubMed] [Google Scholar]
- 28.Lorenzen, J. L., L. H. Larsen, T. Kjær, and N. P. Revsbech. 1998. Biosensor determination of the microscale distribution of nitrate, nitrate assimilation, nitrification, and denitrification in a diatom-inhabited freshwater sediment. Appl. Environ. Microbiol. 64:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, R. L., L. H. Larsen, and N. P. Revsbech. 2002. Microscale biosensor for measurement of volatile fatty acids in anoxic environments. Appl. Environ. Microbiol. 68:1204-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer-Reil, L.-A., and A. Faubel. 1980. Uptake of organic matter by meiofauna and interrelationships with bacteria. Mar. Ecol. Prog. Ser. 3:251-256. [Google Scholar]
- 31.Muscat, A., U. Prüße, and K.-D. Vorlop. 1996. Stable support materials for the immobilization of viable cells, p. 55-60. In R. H. Wijffels, R. M. Buitelaar, C. Bucke, and J. Tramper (ed.), Immobilized cells: basis and applications. Elsevier Science, Tokyo, Japan.
- 32.Neudörfer, F., and L.-A. Meyer-Reil. 1997. A microbial biosensor for the microscale measurement of bioavailable organic carbon in oxic sediments. Mar. Ecol. Prog. Ser. 147:295-300. [Google Scholar]
- 33.Nielsen, M., L. H. Larsen, M. S. M. Jetten, and N. P. Revsbech. 2004. Bacterium-based NO2 biosensor for environmental applications. Appl. Environ. Microbiol. 70:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preininger, C., I. Klimant, and O. S. Wolfbeis. 1994. Optical fiber sensor for biological oxygen demand. Anal. Chem. 66:1841-1846. [Google Scholar]
- 35.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1092. [DOI] [PubMed] [Google Scholar]
- 36.Ramsch, B. 1993. Erfahrungen mit künstlichen Polymeren als Matrix für Mikroorganismen in der Abwasserbehandlung. Bioengineering 6:23-30. [Google Scholar]
- 37.Relexans, J. C. 1996. Measurement of the respiratory electron transport system (ETS) activity in marine sediments: state-of-the-art and interpretation. I. Methodology and review of literature data. Mar. Ecol. Prog. Ser. 136:277-287. [Google Scholar]
- 38.Revsbech, N. P., T. Kjær, L. R. Damgaard, J. Lorenzen, and L. H. Larsen. 2000. Biosensors for analysis of water, sludge and sediments with emphasis on microscale biosensors., p. 195-222. In J. Buffle and G. Horvai (ed.), In situ monitoring of aquatic systems: chemical analysis and speciation. John Wiley, New York, NY.
- 39.Riedel, K. 1991. Biochemical fundamentals and improvement of the selectivity of microbial sensors—a minireview. Bioelectrochem. Bioenerg. 25:19-31. [Google Scholar]
- 40.Riedel, K. 1998. Microbial biosensors based on oxygen electrodes, p. 199-224. In A. Mulchandani and K. R. Rogers (ed.), Enzyme and microbial biosensors: techniques and protocols. Humana Press, Totowa, NJ.
- 41.Schaule, G., H. C. Flemming, and H. F. Ridgway. 1993. The use of CTC (5-cyano-2,3-ditolyl tetrazolium chloride) in the quantification of respiratory active bacteria in biofilms. Appl. Environ. Microbiol. 59:3850-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strunk, O., and W. Ludwig. 1995. ARB—a software environment for sequence data. Department of Microbiology, Technical University of Munich, Munich, Germany.
- 43.Thévenot, D. R., K. Toth, R. A. Durst, and G. S. Wilson. 2001. Electrochemical biosensors: recommended definitions and classification (technical report). Biosens. Bioelectron. 16:121-131. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, G., and G. G. Guilbault. 1994. Food biosensor analysis. Marcel Dekker, New York, NY.
- 45.Walsh, P. K., and D. M. Malone. 1995. Cell growth patterns in immobilization matrices. Biotechnol. Adv. 13:13-43. [DOI] [PubMed] [Google Scholar]
- 46.Wijffels, R. H. 2001. Biomass gradients, p. 101-122. In R. H. Wijffels (ed.), Immobilized cells. Springer-Verlag, Berlin, Germany.
- 47.Willardson, B. M., J. F. Wilkins, T. A. Rand, J. M. Schupp, K. K. Hill, P. Keim, and P. J. Jackson. 1998. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl. Environ. Microbiol. 64:1006-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf, B., M. Kraus, W. Baumann, M. Brischwein, R. Ehret, T. Henning, M. Lehmann, and A. Schwinde. 1997. Sensorik mit zellulären Systemen (Teil 1). Biotec 6:26-29. [Google Scholar]
- 49.ZoBell, C. E. 1941. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 4:42-75. [Google Scholar]