Abstract
A multiplex terminal restriction fragment length polymorphism (M-TRFLP) fingerprinting method was developed and validated for simultaneous analysis of the diversity and community structure of two or more microbial taxa (up to four taxa). The reproducibility and robustness of the method were examined using soil samples collected from different habitats. DNA was PCR amplified separately from soil samples using individual taxon-specific primers for bacteria, archaea, and fungi. The same samples were also subjected to a multiplex PCR with the primers for all three taxa. The terminal restriction fragment length polymorphism profiles generated for the two sets of PCR products were almost identical not only in terms of the presence of peaks but also in terms of the relative peak intensity. The M-TRFLP method was then used to investigate rhizosphere bacterial, fungal, and rhizobial/agrobacterial communities associated with the dwarf shrub Calluna vulgaris growing in either open moorland, a mature pine forest, or a transition zone between these two habitats containing naturally regenerating pine trees. Rhizosphere microbial communities associated with Vaccinium myrtillus collected from the native pine forest were also investigated. In this study, individual PCR products from the three taxa were also pooled before restriction digestion and fragment size analysis. The terminal restriction fragment length polymorphism profiles obtained with PCR products amplified individually and with multiplexed and pooled PCR products were found to be consistent with each other in terms of the number, position, and relative intensity of peaks. The results presented here confirm that M-TRFLP analysis is a highly reproducible and robust molecular tool for simultaneous investigation of multiple taxa, which allows more complete and higher resolution of microbial communities to be obtained more rapidly and economically.
It is well established that microorganisms play an important role in the functioning of different ecosystems, but progress has been hindered by our inability to measure the vast diversity of microorganisms in environments. Molecular methods have been developed rapidly over the last decade, and they have overcome many of the problems associated with conventional culture methods. Use of these methods has shown that in many natural habitats the diversity and complexity of microbial communities (11, 12), as well as their spatial and temporal heterogeneity (18, 20, 28), are greater than initially anticipated. This has created additional problems for microbial ecologists as the great diversity of organisms is difficult to assess by any one approach and large numbers of samples must be analyzed to account for environmental variability.
A number of methods have been used to characterize microbial communities across a range of different ecosystems, but there are still important gaps in our knowledge which are vital for understanding ecological processes (33, 34). Often previous studies have focused on one or a few taxonomic groups of microorganisms. However, biotic interactions between components of the microbial community and with macroorganisms are extremely important in determining ecosystem processes in a given environment. For example, previous studies have described positive, negative, and neutral interactions between fungal and bacterial communities in several different ecosystems (4). It has also been suggested that a multitaxon approach would be more reliable for identifying bioindicators of environmental health (15, 32). Such an approach would help elucidate whether various microbial taxa are influenced by the same environmental factors and whether different taxa respond differently to environmental stresses.
Several culture-independent methods (31) have been used for studying microbial communities, such as analysis of phospholipid biomarkers and nucleic acid-based community analysis. Nucleic acid techniques have gained prominence recently because of their greater resolving power compared with methods such as phospholipid fatty acid analysis (33, 35). Most of these molecular methods are PCR based and target the rRNA gene cluster. PCR amplification of rRNA genes from soil DNA samples, combined with fingerprinting techniques such as denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphism (TRFLP), amplified ribosomal intergenic spacer analysis, and single-strand conformation polymorphism methods, provides detailed information about the microbial compositions of whole communities. Two of these techniques, DGGE and TRFLP analysis, are the methods that are most extensively used for studying changes in microbial community structure and diversity (3). While precise community information concerning several microbial taxa can be generated from the same sample by PCR amplification using individual taxon-specific primers, in practice the time and cost involved in performing multiple analyses with every experimental sample are a hindrance to more detailed investigations.
TRFLP analysis is an automated and sensitive fingerprinting method which uses fluorescently labeled primers for PCR, followed by restriction digestion and analysis of terminal fragments with a DNA sequencer. The sequencer recognizes only the fluorescently labeled terminal fragments, and therefore, in principle each fragment represents a unique operational taxonomic unit (OTU) in the sample (6, 23, 27, 30). The relative quantitative distribution within a profile can be determined, since the fluorescence intensity of each peak is proportional to the amount of genomic DNA present for each OTU in the sample (6). These advantages, together with the potential for high throughput, mean that TRFLP analysis has been widely used to study the structural and functional diversity of microbial communities and has recently been suggested to be the most appropriate molecular method for large-scale soil monitoring (5). However, like other molecular methods, so far it has been used to analyze only a single biomarker at a time.
The aim of the present work was to develop a rapid and sensitive method which could be used to simultaneously analyze communities of several microbial taxa with one PCR. Soil is one of the most complex natural environments, so we tested the robustness and reproducibility of the new method for soil samples by comparing data obtained by individual TRFLP analysis with data obtained by multiplex TRFLP (M-TRFLP) analysis. We then used the M-TRFLP approach to investigate rhizosphere microbial communities associated with the dwarf shrubs Calluna vulgaris and Vaccinium myrtillus across a moorland-Scots pine (Pinus sylvestris L.) vegetation gradient. C. vulgaris was present across the whole gradient, while V. myrtillus was found only under the pine trees. We carried out this experiment to investigate whether the rhizosphere microbial communities associated with C. vulgaris were similar irrespective of the habitat where it was found (mature pine forest, moorland, or a transition zone containing young naturally regenerating pine seedlings). Additionally, in the pine forest we also analyzed the rhizosphere microbial community of V. myrtillus to distinguish between the habitat and the selective influence of the plant species on the microbial communities. The potential applications and limitations of M-TRFLP assays are also discussed below.
MATERIALS AND METHODS
Development of M-TRFLP method.
In the preliminary development work on the M-TRFLP method we used six soil samples obtained from a laboratory experiment. Lolium perenne was grown in pot soil from a grassland site (Cairnbrogie; National Grid reference NJ848 266; 10.2% organic matter) for 4 weeks prior to sampling. DNA was extracted from 0.5 g of each rhizosphere soil sample using an UltraClean soil DNA isolation kit according to the manufacturer's instructions (Mo Bio Laboratories, Carlsbad, CA).
(i) PCR conditions and optimization.
DNA samples were amplified with PCR primers (Table 1) specific for bacteria, fungi, and archaea individually. A multiplex PCR was also performed for each DNA sample, and the reaction mixture contained all three primer pairs for fungi, bacteria, and archaea. All PCR amplifications were performed using the same conditions. The PCR master mixture (50 μl) contained 1× NH4 reaction buffer, 2 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 250 μM, and 2.5 U of Biotaq DNA polymerase (all reagents obtained from Bioline, London, United Kingdom), as well as 20 μg bovine serum albumin (Roche Diagnostic, Lewes, United Kingdom) and 2 μl of template DNA. Initially, 10 pM of each primer was used for both individual and multiplex PCR, but the amplification of fungal internal transcribed spacer (ITS) regions in the multiplex PCR was poor (data not shown). Therefore, the concentration of fungal ITS primers was doubled to 20 pM for all further PCR amplifications. Both individual and multiplex PCRs were performed with a DYAD DNA Engine Peltier thermal cycler (MJ Research, Waltham, MA) using the same program. The program consisted of an initial step of 5 min at 95°C, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 1 min. The last cycle was followed by extension at 72°C for 10 min. PCR amplicons were stained with ethidium bromide and visualized on a 1% agarose gel using UV radiation.
TABLE 1.
Primers used in this study
| Primer | Fluorescent label | Sequence (5′ to 3′) | Target region | Specificity (reference) |
|---|---|---|---|---|
| 63f | None | AGGCCTAACACATGCAAGTC | 16S rRNA gene | Eubacteria (25) |
| 1087r | VIC (green) | CTCGTTGCGGGACTTACCCC | 16S rRNA gene | Eubacteria (19) |
| 1494r | VIC (green) | TACGGYTACCTTGTTACGAC | 16S rRNA gene | Eubacteria (22) |
| Ar3f | None | TTCCGGTTGATCCTGCCGGA | 16S rRNA gene | Archaea (16) |
| AR927r | NED (yellow) | CCCGCCAATTCCTTTAAGTTTC | 16S rRNA gene | Archaea (21) |
| ITS1f | FAMa (blue) | CTTGGTCATTTAGAGGAAGTAA | ITS | All fungi (14) |
| ITS4r | None | TCCTCCGCTTATTGATATGC | ITS | All fungi (39) |
| Rhiz-1244r | PET (red) | CTCGCTGCCCACTGTCAC | 16S rRNA gene | Rhizobia/agrobacteria (38) |
FAM, 6-carboxyfluorescein.
(ii) TRFLP analysis.
PCR products were purified using a GenElute PCR clean-up kit (Sigma-Aldrich, Dorset, United Kingdom) according to the manufacturer's instructions. Prior to digestion, purified PCR product concentrations were determined with a spectrophotometer (UV photometer; Eppendorf, Germany). All PCR products obtained from individual and multiplex PCRs were digested separately with the HaeIII, HhaI, MspI, and RsaI restriction enzymes in 20-μl reaction mixtures containing 500 ng of PCR products, 1× buffer, 0.1 μg μl−1 of acetylated bovine serum albumin, and 20 U of restriction enzyme (all reagents obtained from Promega, Southampton, United Kingdom). Samples were incubated at 37°C for 3 h, and this was followed by deactivation at 95°C for 15 min. TRFLP profiles were produced for amplicons generated with the primers for bacteria, fungi, and archaea individually, as well as in a multiplex PCR. After digestion, 2 μl of each sample was mixed with 0.3 μl of LIZ-labeled GS500 (−250) internal size standard and 12 μl of formamide (all reagents obtained from Applied Biosystems, Warrington, United Kingdom). Prior to fragment analysis, samples were denatured at 95°C for 5 min and then chilled on ice for 5 min. A fragment size analysis was carried out with an ABI PRISM 3130xl genetic analyzer (Applied Biosystems, Warrington, United Kingdom).
Data analysis.
TRFLP profiles were produced using the GeneMapper software (version 3.7; ABI, United Kingdom). Terminal restriction fragments (TRFs) were quantified using the advanced mode and second-order algorithm. Only peaks at positions between 50 and 500 bp were considered in order to avoid TRFs caused by primer-dimers and to obtain fragments within the linear range of the internal size standard. The relative abundance of a TRF in a TRFLP profile was calculated by dividing the peak height of the TRF by the total peak height of all TRFs in the profile. All peaks with heights that were less than 0.5% of the total peak height were not included in further analyses. This approach minimized the effect of variations in the TRFLP profiles caused by the quantity of DNA analyzed. A peak-by-peak comparison was carried out for TRFLP profiles produced from the same sample using individual PCR and multiplex PCR. TRFLP profiles (electropherograms) obtained using the two approaches were aligned with each other and compared, as were the tables of peaks generated by the GeneMapper software. Similarity matrices were also calculated for bacteria, fungi, and archaea for each individual DNA sample by comparing the TRFLP data generated using individual and multiplex PCRs. The similarity was expressed as a percentage determined by dividing the number of peaks present in both profiles by the total number of peaks present in the profile with the lower number of peaks.
Application of the M-TRFLP method. (i) Field site and sampling.
Samples of C. vulgaris and V. myrtillus rhizosphere soil were collected from a naturally regenerating Scots pine (P. sylvestris) forest at Abernethy, Cairngorm, Scotland (National Grid reference NJ027122) (9) which extends onto an open moorland. The area between the open moor and the forest has been partially colonized by Scots pine trees and is termed the transition zone. The colonizing trees are approximately 15 to 20 years old. Few seedlings are more than 1 to 1.3 m tall (9). Samples were taken along three parallel transects, one each in the forest, transition zone, and moorland regions. Each transect was 80 m long and was divided into 10 sections, so samples were obtained from a total of 11 points that were 8 m apart. A C. vulgaris plant and associated rhizosphere soil were collected from each point along the three transects. In addition, a V. myrtillus plant was also collected from each point along the forest transect. Rhizosphere-rhizoplane soil samples were taken from all plants collected (n = 44) (29). DNA was extracted from each sample as described above, but only 43 samples produced enough DNA for PCR amplification and were used in the analysis.
(ii) M-TRFLP analysis of field samples.
An M-TRFLP analysis was conducted as described above, except that the microbial taxa investigated were bacteria, fungi, and rhizobia. Rhizobia/agrobacteria were investigated instead of archaea to see if the presence of this group, which was expected to be present at low levels due to the low pH of the soil (9), could be detected using this approach. Initially, five randomly selected samples were used to test the efficacy and reliability of the approach since the complement of primers to be used in the multiplex PCR was different. As described above, TRFLP profiles were produced for PCR products generated for bacteria, fungi, and rhizobia individually and for products generated using a multiplex PCR performed with all three primer pairs. Additionally, for this experiment, the amplicons produced by using the primers for bacteria, fungi, and rhizobia/agrobacteria individually were pooled at a 2:2:1 ratio for each soil sample and were subjected to fragment size analysis. These samples were first digested with restriction enzymes HaeIII, HhaI, MspI, and RsaI. HhaI and MspI produced more peaks with an even distribution and higher consistency. The quantity of DNA used for restriction digestion was optimized with three different DNA concentrations (1,000, 500, and 200 ng) in 20-μl reaction mixtures. Samples containing 500 ng DNA gave the best results. Therefore, most subsequent restriction digestions were carried out with HhaI and MspI using 500 ng of DNA; the only exception was the individual rhizobial PCR product, for which 200 ng DNA was used.
All 43 soil DNA samples were amplified with three primer sets, one set for 16S rRNA genes specific for the bacterial community, one set for ITS specific for the fungal community, and one set for 16S rRNA genes specific for rhizobia/agrobacteria (63f and Rhiz-12444r), individually and in a multiplex PCR, resulting in 172 PCR products. TRFLP profiles were produced using two restriction enzymes (HhaI and MspI) as described above for (i) PCR products obtained using a single pair of primers (258 analyses), (ii) pooled PCR products generated using individual primers (86 analyses), and (iii) multiplex PCR products (86 analyses). The data were analyzed as described above, but the peaks generated using the rhizobial/agrobacterial primers and enzymes were also compared to the peaks predicted using the Microbial Community Analysis III database (http://mica.ibest.uidaho.edu/trflp.php).
Statistical analysis.
Fragment data obtained from TRFLP and M-TRFLP analyses were exported to Excel (Microsoft, Redmond, WA) and converted into a binary data table (presence or absence of individual peaks) and a relative abundance table (percentage of individual peaks in each profile). The relative abundance data were analyzed using principal-component (PC) analysis based on a correlation matrix appropriate for such data. A principal-coordinate (PCO) analysis was also performed with the binary data using GenStat (8th ed.; VSN International Ltd, Hempstead, United Kingdom) to identify patterns of microbial community structure in samples. PC and PCO analyses were carried out separately with the data obtained with each approach to test the consistency of the method.
RESULTS
Development and validation of the M-TRFLP method.
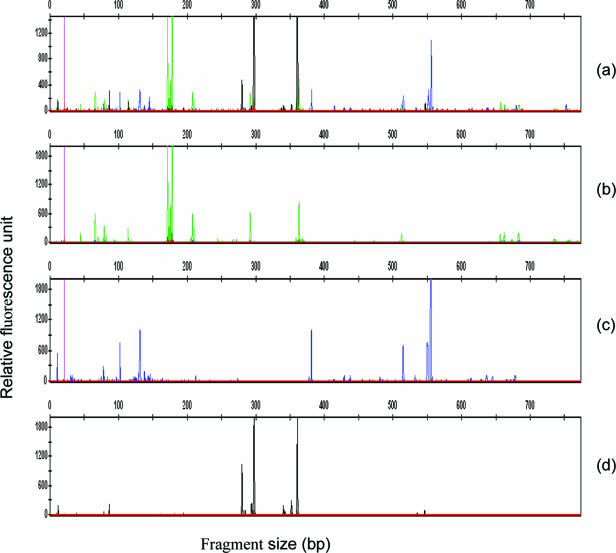
A summary of the rationale behind the M-TRFLP method is shown in Fig. 1. The two different approaches, single-TRFLP and M-TRFLP analyses, produced almost identical profiles (Fig. 2) for each taxon primer pair. There were similarities between profiles not only in terms of the presence and absence of peaks but also in terms of the relative dominance of particular peaks for any single taxon. Most of the samples analyzed by the single-TRFLP and M-TRFLP methods were >90% similar to each other for all taxa regardless of the restriction enzyme used. Between 6 and 10 peaks (TRFs) were produced for archaea using the archaon-specific primers in all samples. A peak-by-peak comparison suggested that TRFLP and M-TRFLP analyses produced identical profiles for three samples (100% similarity). Lower levels of similarity (>86%) between the results of the single and multiplex approaches were obtained for samples 4, 5, and 6. This was due in part to the fact that the level of one TRF was below the threshold value of selection (i.e., <0.5% based on the total peak fluorescence intensity). For bacterial communities, 20 to 31 TRFs were used for comparison with the same selection criteria. Again, the level of similarity between the profiles produced using the two different approaches was high (>90%). Manual comparison of the data revealed that there were only minor differences between the profiles generated using the two approaches, with the majority of the differences resulting from one to three TRFs in one profile whose levels were below the 0.5% total fluorescence cutoff limit. The same trend was found for fungi, although the number of TRFs for fungi (24 to 49 TRFs) was higher than the number of TRFs for bacteria.
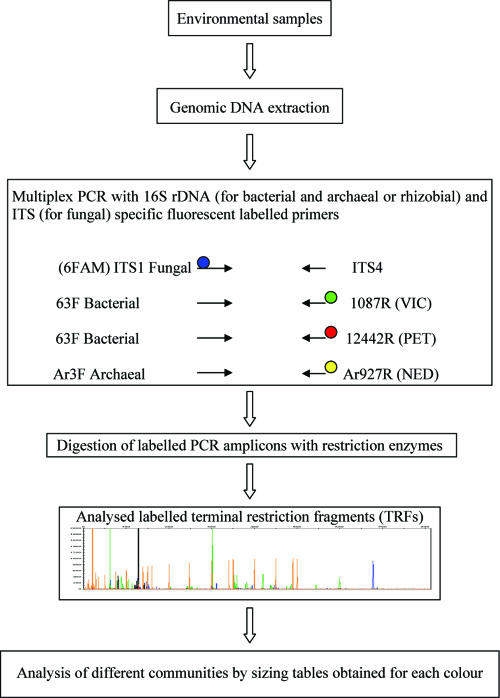
FIG. 1.
Outline of the procedure for M-TRFLP analysis. Different colors at the primer end represent different fluorescent labels. The fluorescent labels used in this study were 6-carboyfluorescein (FAM) (blue), VIC (green), NED (yellow), and PET (red).
FIG. 2.
Profiles obtained for archaeal, bacterial, and fungal communities by M-TRFLP and individual TRFLP analyses for a single sample. (a) Profile generated by M-TRFLP analysis for bacterial (green), fungal (blue), and archaeal (yellow but appears black on GeneMapper) communities together using multiplex PCR. (b to d) TRFLP profiles generated for the same sample from PCR products obtained using only bacterium-specific (b), fungus-specific (c), and archaeon-specific (d) primers in separate PCRs.
Use of the M-TRFLP method for analysis of field soils.
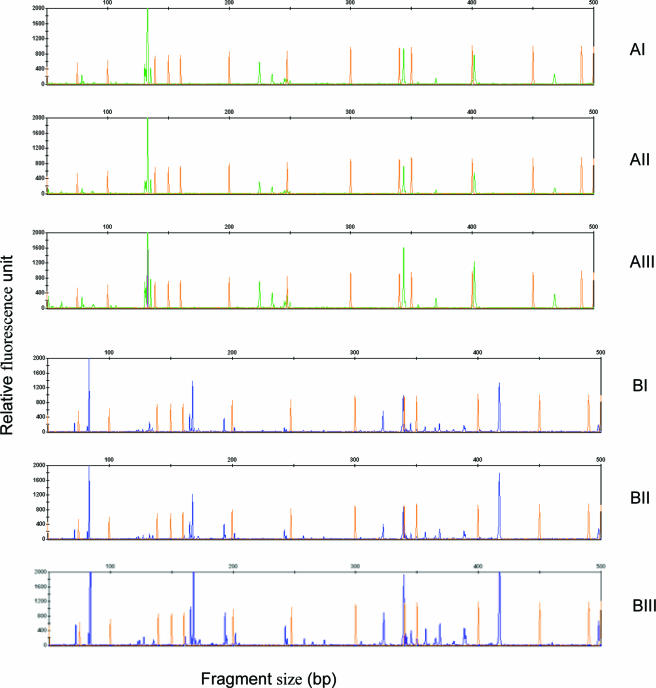
In the analysis of field soils experiment, three different sets of data were generated for each sample. TRFLP profiles were generated for (i) PCR products for individual taxa (bacteria, fungi, and rhizobia/agrobacteria), (ii) pooled PCR products generated using individual PCR primers, and (iii) multiplex PCR products (i.e., M-TRFLP). Again, when the data for the bacterial, fungal, and rhizobial communities were analyzed, the profiles generated for the same samples by TRFLP analysis of individual, pooled PCR products and by M-TRFLP analysis were consistent with each other (Fig. 3). A peak-by-peak comparison for each microbial taxon revealed a very high level of similarity for the data produced using the three different approaches (data not shown). The minor variation again was due to the loss of minor peaks whose levels were below the 0.5% cutoff limit. Again, the raw data produced nearly identical peak profiles for the TRFLP and M-TRFLP obtained for each taxon of a given sample.
FIG. 3.
(AI to AIII) Bacterial community profiles generated by three different TRFLP approaches. (AI) Bacterial community profile for one sample in M-TRFLP analysis, generated from multiplex PCR. (AII) TRFLP profile generated from pooled PCR products from individual PCRs. (AIII) TRFLP profile for the bacterial community of the same sample obtained from individual PCR using eubacterium-specific primers. For comparison, only results for LIZ (orange; marker) and VIC (green; linked to bacterial primer) are shown in panels AI and AII. (BI to BIII) Comparison of profiles for fungal communities in the same sample generated by three TRFLP approaches: TRFLP profiles of the fungal community obtained from M-TRFLP (BI), pooled PCR (BII), and individual PCR (BIII) products. In panels BI and BII, only results for LIZ and 6-carboyfluorescein (blue; linked to fungal primer) are shown in order to compare profiles obtained with the different TRFLP approaches.
The database search of the Microbial Community Analysis III website predicted the presence of only two TRFs for rhizobia/agrobacteria when samples were digested with HhaI (153 and 154 bp) and one TRF when samples were digested with MspI (90 bp) for the primer combination used in this study. However, in our samples, 156- and 158-bp TRFs were detected in samples digested with HhaI. The signal intensities for these two TRFs in the M-TRFLP analysis were significantly lower than the signal intensities for TRFs generated from samples amplified with the individual rhizobial/agrobacterial primers or the pooled PCR products. Samples digested with MspI produced two TRFs (86 and 88 bp) in both individual TRFLP and M-TRFLP analyses instead of the one predicted peak at 90 bp. The level of similarity of the TRFLP profiles obtained for rhizobia/agrobacteria was lower (85%) than the level of similarity for the other taxa when the two techniques were compared. However, when the raw data were analyzed with a reduced cutoff value for the signal intensity, the same TRFs were detected in all samples. However, in addition to the predicted TRFs, there were also peaks detected in several samples which were inconsistent across the samples and had very low signal intensities. In a later experiment, to increase the fluorescence intensity, we amplified a number of these samples with the reverse bacterial primer 1494R instead of 1087R. This approach led to a severalfold increase in the fluorescence intensity of rhizobial/agrobacterial peaks in M-TRFLP analyses. A further increase in the fluorescence in M-TRFLP analyses was observed when both primers were targeted exclusively at the rhizobial/agrobacterial group (data not shown).
Statistical analysis of field sample data.
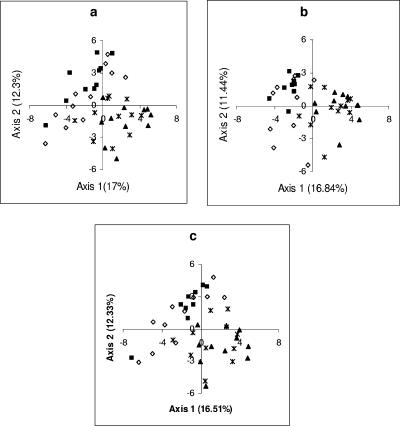
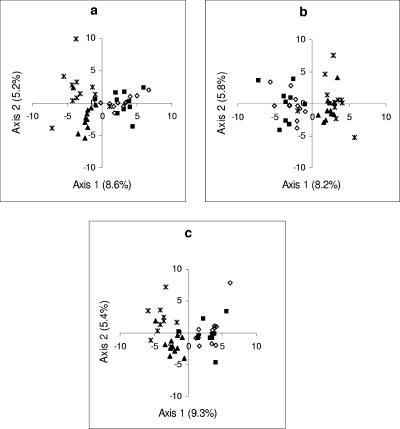
Relative abundance data obtained using the three TRFLP approaches (individual PCR, pooled PCR products, and multiplex PCR) for the field-collected samples were analyzed separately and compared with each other to examine both the bacterial and fungal communities for the two enzymes used. Figure 4 shows the clustering of samples based on the PC scores of dimensions 1 and 2. The PCO scores were obtained from the analysis of bacterial 16S rRNA genes digested with the HhaI enzyme. The ordination of the data showed that the samples from the forest could be discriminated from the samples from the moorland and transition zone. However, the bacterial communities associated with forest V. myrtillus and C. vulgaris could not be discriminated. This pattern was consistent for all three TRFLP approaches. The results obtained from the PC analysis of the fungal ITS regions digested with the HhaI enzyme were similar to the bacterial 16S rRNA data, but there was slightly stronger clustering according to the habitat type and all the samples from the forest, irrespective of the plant type, were clustered together. However, fungal communities were also discriminated on the basis of habitat type (i.e., moorland, transition zone, and forest). This pattern was consistent in the individual TRFLP and M-TRFLP analyses (Fig. 5). Similar results were also obtained for bacterial and fungal communities when the samples were digested with MspI. PCO analysis of binary data for these samples produced similar results, although the distinction between the transition zone fungal community and the moorland fungal community was less evident (data not shown).
FIG. 4.
PC analysis for bacterial community obtained with three different TRFLP approaches (using the HhaI restriction enzyme) for 43 soil samples from the Abernethy forest. The values in parentheses on the x and y axes indicate the amount of variation accounted for by each axis. The plots are ordination plots of the first two dimensions of PC scores from individual TRFLP data (a), from M-TRFLP data (b), and from TRFLP data for pooled PCR products (c). Symbols: ▪, forest, Calluna; ⋄, forest, Vaccinium; ▴, transition zone, Calluna; *, moorland, Calluna.
FIG. 5.
PC analysis for fungal community obtained with three different TRFLP approaches (using the HhaI restriction enzyme) for 43 soil samples from the Abernethy forest region. The values in parentheses on the x and y axes indicate the amount of variation accounted for by each axis. The plots are ordination plots of the first two dimensions of PC scores from individual TRFLP data (a), from M-TRFLP data (b), and from TRFLP data for pooled PCR products (c). Symbols: ▪, forest, Calluna; ⋄, forest, Vaccinium; ▴, transition zone, Calluna; *, moorland, Calluna.
DISCUSSION
Our results confirm that M-TRFLP analysis is as reproducible and consistent as TRFLP analysis but is a cheaper, less labor-intensive, and more rapid method for analysis of microbial communities when information for more than one taxon is required. The TRFLP method has been one of the most widely used methods for analysis of microbial communities since its first use with soil samples (30). It has proved to be more consistent and reproducible than other fingerprinting methods because of its automated analysis mode and because an internal size standard is included in every sample. In addition, it can provide semiquantitative data which can be used for generating information on the relative abundance of operational taxonomic units (6, 27). The M-TRFLP method is a significant advance compared with the TRFLP method and also with other molecular methods used in microbial ecology as it allows simultaneous characterization of up to four different targets in one reaction mixture. This is unique in the sense that no other fingerprinting method can do this at the present time, and the M-TRFLP approach is likely to be a very useful intermediate approach between single-taxon and phylogenetic microarray methods. However, M-TRFLP analysis is subject to systematic biases like any PCR-based method (40) and has the same limitations as TRFLP analysis (13, 24). Nonetheless, because of its reproducibility and consistency with the individual TRFLP method, the M-TRFLP method has considerable advantages and should be a valuable tool in microbial ecology research.
In our experiment, PCR optimization was required, and we had to double the concentration of the fungal primers compared with the concentration of the bacterial primers. This probably reflected the sizes of the genomes of different taxa or primer specificity. Profiles produced for the same sample by individual TRFLP and M-TRFLP analyses were very similar (>90%). Whatever differences were observed were mainly due to the mode of data analysis. The loss of small TRFs, which are often artifacts (6), is well documented for TRFLP analysis. However, detection of these TRFs can be optimized if necessary by lowering the fluorescence threshold or by manual correction (30). When we compared raw data without this analysis, a peak-by-peak comparison resulted in almost 100% similarity between data produced using the two approaches. We took this approach to data analysis because it is a well-established method for avoiding very small peaks which may have appeared as artifacts (6). Another reason for this variation may have been minor shifts in the positions of a few peaks during electrophoresis of the samples which were not taken into account during data analysis. Even so, the high level of similarity between individual TRFLP and M-TRFLP profiles shows the robustness of the new method. When products from individual PCR were pooled before restriction digestion, the resulting profiles were almost identical to those obtained from the corresponding individual TRFLP and M-TRFLP samples, which suggested that at least in this experiment, it was not essential to use individual PCR when M-TRFLP analysis is performed.
It is known that greater discrimination in TRFLP profiles for bacterial communities can be obtained by labeling the forward primer because of length heterogeneities at the 5′ end of the gene (30). To take this fact into account, we also tested other approaches in which forward bacterial primers and reverse fungal primers or only forward primers for both bacterial and fungal communities were labeled to investigate the impact on M-TRFLP profiles. We also tested six samples where four different primer pairs (Cytophagales-specific primers in addition to bacterial, fungal, and archaeal primers) were tested together for M-TRFLP analysis. These approaches had no effect on the robustness of the method, and the data were reproducible and consistent with the other data presented here (data not shown). However, in the present study, the number of OTUs obtained for the bacterial community with the labeled reverse primer was not significantly different from number of OTUs obtained with the labeled forward primer.
In the initial experiment using primers 63F and 1087R for bacteria and primers 63F and 1244R for rhizobia/agrobacteria, the peak heights for the rhizobial/agrobacterial TRFs were very low, although the peaks were detectable. However in the subsequent experiment, we increased the peak height severalfold for the rhizobial/agrobacterial community by using the 63F and 1494R primers instead of 1087R for the bacterial community in the multiplex PCR. This result might be explained by the fact that in the first experiment, rhizobial PCR products from the first cycle had binding sites for both bacterial and rhizobial reverse primers, which led to competition between them in the second cycle and every subsequent cycle. This hypothesis is supported by the observation that a further increase in the peak height and intensity was obtained when the samples were amplified in a multiplex PCR using forward and reverse primers exclusively targeting the rhizobial/agrobacterial group (primers 63F and Rhiz1244R but no eubacterial reverse primers). Thus, complementarity of primers should be taken into account during selection of primer sets for M-TRFLP analysis, and for the best results, both primers used should probably be as specific as possible for a target taxon.
The PC and PCO analyses of the data generated by the individual and M-TRFLP methods for each taxon confirmed that the two methods gave similar results (Fig. 4). The bacterial communities were separated on the basis of habitat type, and the communities associated with C. vulgaris from the moorland, the transition zone, and the forest were clearly separated from each other. There was no separation between the communities associated with C. vulgaris or V. myrtillus within the forest. A similar pattern was obtained for the fungal communities, with even tighter clustering based on habitat type, which confirmed that both the bacterial and fungal communities in the ecosystems tested were influenced by the habitat type. Similar results have been obtained previously for the same site for the fungal community using DGGE (2). In addition, several previous studies produced similar results, where habitat or soil type had a stronger impact on microbial communities than the individual plant species had (1, 7, 8, 17, 26, 35). Again, a slight difference between the PCO plots generated from individual and M-TRFLP analyses may be attributed to the method used to identify TRFs. However, the overall trends of the results were very similar, emphasizing the reproducibility of the new method. The better discrimination of the fungal community is due in part to the greater resolution obtained using ITS primers (3). Combining the data for the different taxa gives even better discrimination of habitat type.
M-TRFLP analysis has numerous potential applications in addition to studying biotic interactions. It may be useful in situations where information on multiple taxa or functional genes is required but the sample size is limited, as is often the case when environmental samples are studied at the small scales (e.g., root tips or biofilms). TRFLP analysis (10) and multiplex PCR (36, 37) have been used previously for identification of pathogens. The M-TRFLP method combines the features from multiplex PCR and TRFLP together and provides a rapid and economic way to detect important organisms, such as pathogens. M-TRFLP analysis can also be used to increase the phylogenetic resolution of individual OTUs in a community profile by using four primers for the same marker gene that are labeled with different dyes. This allows generation of four TRFs for the same OTU which can be used to identify the OTU to the genus level and in some cases to the species level.
Acknowledgments
We thank Fiona Moore and Rebekka Artz for critically reviewing the manuscript.
This research was supported by the Scottish Executive Environment and Rural Affairs Department (SEERAD). A patent application has been filed for the commercial application of the M-TRFLP (reference number, UK 0525523.7).
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Alvey, S., C. H. Yang, A. Buerkert, and D. E. Crowley. 2003. Cereal/legume rotation effects on rhizosphere bacterial community structure in West African soils. Biol. Fertil. Soils 37:73-82. [Google Scholar]
- 2.Anderson, I. C., C. D. Campbell, and J. I. Prosser. 2003. Diversity of fungi in organic soils under a moorland—Scots pine (Pinus sylvestris L.) gradient. Environ. Microbiol. 5:1121-1132. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, I. C., and J. W. G. Cairney. 2004. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ. Microbiol. 6:769-779. [DOI] [PubMed] [Google Scholar]
- 4.Artursoon, V., R. D. Finlay, and J. K. Jansson. 2006. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 8:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Black, H. I. J., C. Wood, N. Parekh, P. M. Chamberlain, A. Scott, K. Ritz, J. A. Harris, C. D. Campbell, and W. Towers. 2006. SQID phase I: prioritising biological indicators of soil quality for deployment in a national-scale soil monitoring scheme. Project no. SP0529. Scoping biological indicators of soil quality, in press. [Online.] http://www.defra.gov.uk/. Defra, London, United Kingdom.
- 6.Blackwood, C. B., T. Marsh, S. H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossio, D. A., K. M. Scow, N. Gunapala, and K. J. Graham. 1998. Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb. Ecol. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 8.Brodie, E., S. Edwards, and N. Clipson. 2002. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 44:260-270. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, S. J., C. D. Campbell, and G. Puri. 2003. Native woodland expansion: soil chemical and microbiological indicators of change. Soil Biol. Biochem. 35:753-764. [Google Scholar]
- 10.Christensen, J. E., J. A. Stencil, and K. D. Reed. 2003. Rapid identification of bacteria from positive blood cultures by terminal restriction fragment length polymorphism profile analysis of the 16S rRNA gene. J. Clin. Microbiol. 41:3790-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis, T. P., and W. T. Sloan. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221-226. [DOI] [PubMed] [Google Scholar]
- 12.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhiza and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 15.Gillison, A. N., and N. Liswanti. 2004. Assessing biodiversity at landscape level in northern Thailand and Sumatra (Indonesia): the importance of environmental context. Agric. Ecosyst. Environ. 104:75-86. [Google Scholar]
- 16.Giovannoni, S. J., F. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligonucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girvan, M. S., J. Bullimore, A. S. Ball, J. N. Pretty, and A. M. Osborn. 2004. Responses of active bacterial and fungal communities in soils under winter wheat to different fertilizer and pesticide regimens. Appl. Environ. Microbiol. 70:2692-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green, J. L., A. J. Holmes, M. Westoby, I. Oliver, D. Briscoe, M. Dangerfield, et al. 2004. Spatial scaling of microbial eukaryote diversity. Nature 432:747-750. [DOI] [PubMed] [Google Scholar]
- 19.Hauben. L., C. Vauterin, J. Swings, and E. R. B. Moore. 1997. Comparison of 16S ribosomal DNA sequence of all Xanthomonas species. Int. J. Syst. Bacteriol. 47:328-335. [DOI] [PubMed] [Google Scholar]
- 20.Horner-Devine, M. C., M. Lage, J. B. Hughes, and B. J. M. Bohannan. 2004. A taxa-area relationship for bacteria. Nature 432:750-753. [DOI] [PubMed] [Google Scholar]
- 21.Jurgens, G., K. Lindstrom, and A. Saano. 1997. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl. Environ. Microbiol. 63:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 23.Liu, W. T., T. L. Marsh, H. Cheng, and F. J. Forney. 1997. Characterization of microbial diversity by determining restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marschner, P., C. H. Yang, R. Lieberei, and D. E. Crowley. 2001. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437-1445. [Google Scholar]
- 27.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (TRFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 28.Nicol, G. W., L. A. Glover, and J. I. Prosser. 2003. Spatial analysis of archaeal community structure in grassland soil. Appl. Environ. Microbiol. 69:7420-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nunan, N., T. J. Daniell, B. K. Singh, A. Papert, J. W. McNicol, and J. I. Prosser. 2005. Links between plant and rhizoplane bacterial communities in grassland soils, characterized using molecular techniques. Appl. Environ. Microbiol. 71:6784-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (TRFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 31.Prosser, J. I. 2002. Molecular and functional diversity in soil micro-organisms. Plant Soil 244:9-17. [Google Scholar]
- 32.Sauberer, N., K. P. Zulka, M. Bensperg-Traun, H. M. Berg, G. Bieringer, and N. Milasowszky. 2004. Surrogate taxa for biodiversity in agricultural landscapes of eastern Austria. Biol. Conserv. 117:181-190. [Google Scholar]
- 33.Singh, B. K., P. Millard, A. S. Whiteley, and J. C. Murrell. 2004. Unravelling rhizosphere-microbial interactions: opportunities and limitations. Trends Microbiol. 12:386-393. [DOI] [PubMed] [Google Scholar]
- 34.Singh, B. K. 2005. Microbial diversity: progress and important knowledge gaps. ASM News 71:210-211. [Google Scholar]
- 35.Singh, B. K., S. Munro, E. Reid, B. Ord, J. M. Potts, E. Paterson, and P. Millard. 2006. Investigating microbial community structure in soil by physiological, biochemical and molecular fingerprinting methods. Eur. J. Soil Sci. 57:72-82. [Google Scholar]
- 36.Song, Y., N. Kato, C. Liu, Y. Matsumiya, H. Kato, and K. Watanabe, K. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 37.Stankovic, M., L. Rakicevic, D. Mikovic, G. Jankovic, and A. Nikolic. 2005. Indirect diagnosis of haemophilia B by multiplex PCR/RFLP. Clin. Lab. Haematol. 27:145-146. [DOI] [PubMed] [Google Scholar]
- 38.Tom-Petersen, A., T. D. Leser, T. L. Marsh, and O. Nybroe. 2003. Effect of copper amendment on the bacterial community in agricultural soil analysed by T-RFLP. FEMS Microbiol. Ecol. 46:53-62. [DOI] [PubMed] [Google Scholar]
- 39.White, T. J., T. D. Bruns, S. Lee, and J. Taylor. 1990. Analysis of phylogenetic relationship by amplification and direct sequencing of ribosomal RNA genes, p. 315-322. In M. A. Innis, D. H. Gelfond, J. J. Sainsky, and T. J. White (ed.), PCR protocol: a guide to method and applications. Academic Press, New York, N.Y.
- 40.Yu, C. P., R. Ahuja, G. Sayler, and K. H. Chu. 2005. Quantitative molecular assay for fingerprinting microbial communities of wastewater and estrogen-degrading consortia. Appl. Environ. Microbiol. 71:1433-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]