Abstract
In situ detection of methanogens within the family Methanobacteriaceae is sometimes known to be unsuccessful due to the difficulty in permeability of oligonucleotide probes. Pseudomurein endoisopeptidase (Pei), a lytic enzyme that specifically acts on their cell walls, was applied prior to 16S rRNA-targeting fluorescence in situ hybridization (FISH). For this purpose, pure cultured methanogens within this family, Methanobacterium bryantii, Methanobrevibacter ruminantium, Methanosphaera stadtmanae, and Methanothermobacter thermautotrophicus together with a Methanothermobacter thermautotrophicus-containing syntrophic acetate-oxidizing coculture, endosymbiotic Methanobrevibacter methanogens within an anaerobic ciliate, and an upflow anaerobic sludge blanket (UASB) granule were examined. Even without the Pei treatment, Methanobacterium bryantii and Methanothermobacter thermautotrophicus cells are relatively well hybridized with oligonucleotide probes. However, almost none of the cells of Methanobrevibacter ruminantium, Methanosphaera stadtmanae, cocultured Methanothermobacter thermautotrophicus, and the endosymbiotic methanogens and the cells within UASB granule were hybridized. Pei treatment was able to increase the probe hybridization ratio in every specimen, particularly in the specimen that had shown little hybridization. Interestingly, the hybridizing signal intensity of Methanothermobacter thermautotrophicus cells in coculture with an acetate-oxidizing H2-producing syntroph was significantly improved by Pei pretreatment, whereas the probe was well hybridized with the cells of pure culture of the same strain. We found that the difference is attributed to the differences in cell wall thicknesses between the two culture conditions. These results indicate that Pei treatment is effective for FISH analysis of methanogens that show impermeability to the probe.
Methanogenic archaea belonging to the family Methanobacteriaceae, comprising 23 species within four genera, have been characterized and isolated from various anoxic environments (3). In addition, recent studies based on 16S rRNA gene sequencing analyses revealed that a number of uncultured species within this family have been found in various anaerobic environments (8, 12, 17, 18, 21, 29, 31). Because of the ubiquity of methanogens within this family in the environment, they are thought to play important roles as one of the hydrogenotrophic methanogens. To precisely determine the abundance and localization of Methanobacteriaceae cells in environmental samples, fluorescence in situ hybridization (FISH) techniques targeting 16S rRNA genes have been frequently employed (7, 13, 17). However, some researchers reported difficulties in their detections (6, 9, 27, 30), probably because of the impermeability of oligonucleotide probes through the distinctive cell walls of this family, namely, pseudomurein. This could be, to some extent, solved by freeze-and-thaw (27) and proteinase K (9) treatments, but these treatments are laborious, and some taxonomic groups are very susceptible to these treatments.
The glycan strand of pseudomurein is composed of alternating β-(1→3)-linked N-acetyl-d-glucosamine and β-(1→3)-linked N-acetyl-l-talosaminuronic acid (instead of N-acetyl-d-muranosamine in murein) (14). Moreover, the constituents in the peptide strand cross-linking glycan strands are made up solely of l-amino acids. As far as we know, pseudomurein endoisopeptidase (Pei) is the only known specific enzyme that cleaves the specific site of the peptide moiety (15). Pei was originally found in (pro)phages in Methanothermobacter wolfeii and Methanothermobacter marburgensis (20, 23). The gene encoding the enzyme has been cloned (19).
Treatment with lysozyme and/or other murein-degrading enzymes is known to be effective for FISH analysis of some gram-positive bacteria, since thick cell walls of some gram-positive bacteria hinder oligonucleotide probes, which would otherwise permeate into cytoplasms (1, 2, 4). In this study, we attempted to apply pseudomurein endoisopeptidase to FISH analysis for clearly visualizing the methanogens in the family Methanobacteriaceae.
MATERIALS AND METHODS
Organisms, cultivation conditions, and FISH sample preparation.
The following representative methanogens of each genus in the family Methanobacteriaceae were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ): Methanobacterium bryantii DSM863T, Methanosphaera stadtmanae DSM3091T, Methanobrevibacter ruminantium DSM1093T, and Methanothermobacter thermautotrophicus DSM1053T (strain ΔH). These strains were cultivated on media recommended by the DSMZ until early or mid-exponential growth phase, and the cells were harvested by centrifugation. Cells were washed with filtered (0.2-mm pore size) phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde-PBS for 4 h at 4°C, followed by centrifugation and washing twice with filtered PBS.
M. thermautotrophicus strain ΔH was also cocultured with a syntrophic acetate-oxidizing bacterium, Thermacetogenium phaeum DSM12270T, as previously described (10). Cocultured cells were harvested by centrifugation at the mid-exponential growth phase, and the cells of M. thermautotrophicus were fractionated from mixed cells as previously described, with some modifications (11). Briefly, precipitated cells were washed twice with T-DTE buffer (10 mM Tris-HCl, pH 7.5, and 2.5 mM dithioerithritol) and resuspended in T-DTE buffer. For selective lysis of Thermacetogenium phaeum, mutanolysin (500 units; Sigma) and lysozyme (2.5 mg; Wako Pure Chemical Industries Ltd., Osaka, Japan) were added into the cell suspension and incubated at 55°C with vigorous shaking for 15 min. The cell suspension was ultracentrifuged with 10 parts Percoll at 45,000 × g at 4°C for 2 h in order to completely fractionate M. thermautotrophicus cells from unlysed cells of Thermacetogenium phaeum. Fractionated cells of M. thermautotrophicus were recovered using a syringe from the centrifugal tube and washed at least twice with T-DTE buffer. The cells were fixed with 4% paraformaldehyde-PBS in a manner similar to that for pure cultured methanogens as described above.
Endosymbiotic methanogen cells in ciliates were examined. An anaerobic, free-living ciliate, Trimyema compressum, which is maintained in our laboratory, was used. This ciliate harbors methanogenic endosymbionts belonging to the family Methanobacteriaceae (unpublished data). The ciliate cells were grown for 2 weeks on the medium with a food bacterium, Lactococcus lactis, as previously described (32). T. compressum cells were harvested by centrifugation at a minimum centrifugal force for 1 min with a multipurpose swing bucket centrifuge (Swingman; Tanahashi Kikai Co., Ltd., Tokyo, Japan). Cells were washed twice with filtered PBS, suspended, and fixed with 4% paraformaldehyde-PBS for 1 h at 4°C, followed by centrifugation and washing with filtered PBS twice. All of the fixed cells were resuspended in ethanol-PBS (1:1, vol/vol) and incubated at 4°C overnight. Fixed samples were stored at −20°C before use.
A granular sludge from a laboratory-scale upflow anaerobic sludge blanket (UASB) reactor was also examined. The granule sample was kindly provided by the Nagaoka University of Technology. The reactor was operated as previously described (28). The granules taken from the reactor were washed with PBS and fixed with 4% paraformaldehyde-PBS solution for 4 h at 4°C, followed by centrifugation and washing with filtered PBS several times. Fixed granules were resuspended in ethanol-PBS (1:1, vol/vol) and incubated at 4°C overnight. The granules were homogenized with a plastic pestle and stored at −20°C before use.
Cloning, heterogeneous expression, and purification of pseudomurein endoisopeptidase.
The pseudomurein endoisopeptidase gene from M. wolfeii DSM2970 (peiW) was cloned and expressed with the pET expression system to obtain the recombinant enzyme (rPeiW). Frozen cells of M. wolfeii DSM2970 were a kind gift from H. Morii (University of Occupational and Environmental Health, Japan). Genomic DNA was extracted from the frozen cells by using the benzyl chloride method (34). PCR amplification of peiW and the construction of the expression vector were performed as described in previous reports (19), except for the following modifications. Primers PeiWF2 (5′-AGGTGATCATATGGAAGTGGGGCTAAATG-3′ [the NdeI site is underlined]) and PeiWR2 (5′-AACAACTCGAGCATGTCTCTGCCACAAAC-3′ [the XhoI site is underlined]) were used for amplification. The PCR product of peiW was cloned into the TOPO TA cloning vector (Invitrogen) to create plasmid pPeiW2-4. After the sequence of cloned peiW was verified, pPeiW2-4 was digested with NdeI and XhoI. The digested fragment was ligated into pET-19b digested with NdeI and XhoI to create an expression vector, pPETPeiW, and this construct was transformed into Escherichia coli BL21(DE3). The transformant was grown in the LB broth with 50 μg/ml ampicillin at 30°C until the optical density at 600 nm (OD600) reached to 0.5, followed by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration, 250 μM) and incubation for another 5 h. Cells were harvested by centrifugation and resuspended in binding buffer (20 mM phosphate-0.5 M NaCl-20 mM imidazole). To disrupt cells, the suspension was passed through a French press at 1,000 lb/in2 several times until the suspension became transparent. Cell-free extract was obtained after centrifugation and sequential filtration with a 0.45-μm-pore-size membrane filter. Recombinant PeiW (rPeiW) was purified from the cell-free extract with a His Trap HP kit (Amersham Biosciences) as recommended by the manufacturer. The protein concentration of purified rPeiW was determined with a Bio-Rad protein assay (Bio-Rad) according to the manufacturer's instructions. Homogeneity of the purified rPeiW (approximately 3 mg) was confirmed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis by Coomassie brilliant blue staining. Purified rPeiW was stored under anoxic conditions at 4°C. rPeiW activity was measured based on a procedure previously described by Morii and Koga (22). M. thermautotrophicus strain ΔH cells were used as a substrate for the enzymatic reaction. Preparation of the substrate was performed in an anaerobic chamber (Don Whitely Scientific Limited, West Yorkshire, England) under an N2-H2-CO2 atmosphere (80:10:10, vol/vol), except for cultivation and centrifugation steps. M. thermautotrophicus cells were harvested by centrifugation at the late exponential growth state, washed twice with H buffer (50 mM HEPES [pH 7.0]-5 mM dithiothreitol-20.8 mM Na2S), and suspended in H buffer to give an OD600 value of 0.8. The cell suspension (3 ml) was transferred into a screw-cap test tube, and the tube was sealed with a butyl-rubber stopper. The headspace was replaced with N2 gas (approximately 200 kPa). The tube was preincubated at 55°C for 10 min, followed by the addition of the purified rPeiW (50 μl) into the test tube with a syringe. This mixture was incubated at 55°C with agitation at 180 rpm for 1 h. One unit of the rPeiW activity was defined as a 0.1-unit decrease of the OD600 for 1 h. Specific activity of the purified rPeiW preparation was 1,053 units/mg protein. rPeiW is available from the authors upon request.
rPeiW treatment of methanogen cells for FISH.
Fixed cells were spotted onto Vectabond (Vector Laboratories)-coated eight-well glass slides. After samples were dried at room temperature, the glass slides were immersed into 50%, 70%, and 99.5% ethanol for 3 min each to dehydrate the specimens. The amount of rPeiW applied was optimized for each species of methanogen to prevent excessive reaction (lysis) and maintain the morphologies of the cells. The rPeiW solution was appropriately diluted with H buffer (6 to 140 units/ml), applied (50 μl) to each specimen, and incubated at 55°C for 5 min in a preincubated polypropylene tube (50 ml) containing a filter paper (17Chr; Whatman) soaked in H buffer. The total amount of rPeiW applied to each specimen was as follows: 0.4 units for cells of Methanobacterium bryantii, M. thermautotrophicus, and Methanosphaera stadtmanae; 2.3 units for cells of Methanobrevibacter ruminantium; 0.3 units for cocultured cells of M. thermautotrophicus; 7.0 units for anaerobic ciliates; and 0.35 units for homogenized UASB granules. The glass slide was washed with distilled and deionized water (ddW) to stop the enzymatic reaction. The samples were dried at room temperature and dehydrated with ethanol as described above. H buffer was also applied in the same manner as the untreated control.
Freeze-and-thaw and proteinase K treatments.
Freeze-and-thaw treatment was performed as previously reported (27). Proteinase K treatment was performed as described previously by Hara et al. (9).
Whole-cell FISH and microscopic observation.
FISH was performed according to a procedure described previously (27), except that prehybridization with 1% blocking reagent (Roche Diagnostics) in a hybridization buffer was done at 46°C for 1 h after treatment with rPeiW. The specific oligonucleotide probe for the family Methanobacteriaceae, MB1174 (24), labeled with rhodamine, was employed for visualizing the cell. Hybridization was performed with hybridization buffer containing 35% formamide at 46°C for 2 h. Unhybridized probe was removed by washing with the same hybridization buffer without probe at 48°C for 30 min. With respect to the granular sludge sample, a mixed oligonucleotide probe for the domain Bacteria (EUB338, EUB338 II, EUB338 III, and EUB338 IV) (5, 26), labeled with Cy5, was also employed to visualize bacterial cells. After washing of the unhybridized MB1174 probe, the glass slide was thoroughly rinsed with ddW and dried in the dark, hybridization was subsequently performed with hybridization buffer containing 5% formamide at 46°C overnight, and the unhybridized probe was washed off in the same manner as described above. The glass slide was thoroughly rinsed with ddW and dried in the dark at room temperature. Specimens, except for the ciliate and the granular sample, were examined under a phase-contrast and fluorescence microscope (AX-80; Olympus, Tokyo, Japan) equipped with a WIG cube (excitation, 520 to 550 nm; emission, >580 nm) (Olympus, Tokyo, Japan). Phase-contrast micrographs and fluorescent micrographs in the same visual fields were captured by an 8-bit-scale digital camera (ORCA-ER; Hamamatsu Photonics K. K., Shizuoka, Japan) and saved as TIF files by using digital image analysis software (Aqua-Lite; Hamamatsu Photonics K. K., Shizuoka, Japan). Both micrographs were superimposed by Paint Shop Pro 9 (Corel Japan Ltd., Tokyo, Japan) to differentiate probe-hybridized and unhybridized cells. The ratio of probe-hybridized cells to total cells for Methanobacterium bryantii, M. thermautotrophicus, Methanosphaera stadtmanae, and Methanobrevibacter ruminantium was determined by counting over 500 cells. Sections of hybridized probe in the ciliate specimen and homogenized granular sludge sample were obtained with a confocal laser scanning microscope (LSM5 Pa; Zeiss). Differential interference micrographs (the ciliate sample) and fluorescent micrographs of sections were also superimposed as described above.
Transmission electron microscopy.
Pure cultured and cocultured M. thermautotrophicus cells were grown under the conditions described above until mid-log phase. Cells were harvested by centrifugation, fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at 4°C for 6 h, and then postfixed in 2% osmium tetroxide for 60 min at 4°C. The fixed cells were suspended in 1% aqueous uranyl acetate for 1 h at room temperature. Suspended cells were embedded in 1.5% agarose before dehydration with a graded ethanol series. The dehydrated specimens were embedded in Spurr resin. Ultrathin sections were cut with an ultramicrotome (Ultracut-N; Leichert-Nissei), mounted onto copper grids, and stained with uranyl acetate and lead citrate. Images of sections were obtained by using a transmission electron microscope (75 kV) (H-7000; Hitachi, Japan).
RESULTS AND DISCUSSION
FISH of pure cultures with rPeiW treatment.
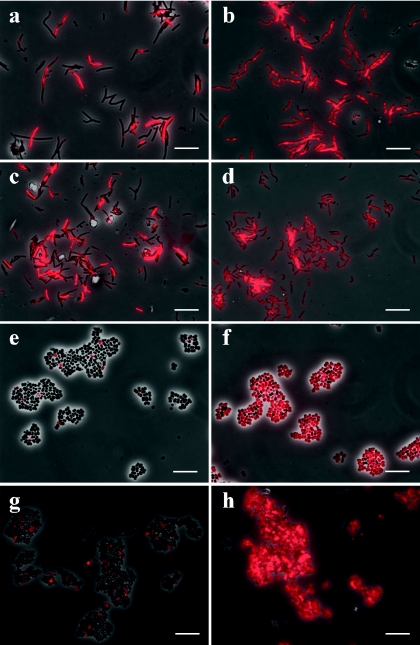
To verify the effect of rPeiW, four representative methanogens within the family Methanobacteriaceae were tested. Their FISH images are shown in Fig. 1. Without rPeiW treatment, the ratio of probe-hybridized cell counts (red color) to total cell counts varied in each specimen: whereas a relatively large percentage of cells of Methanobacterium bryantii (45%) and M. thermautotrophicus (34%) were hybridized without treatment (Fig. 1a and c), a very small number of the Methanosphaera stadtmanae (6%) and Methanobrevibacter ruminantium (7%) cells were hybridized without treatment (Fig. 1e and g). The notable difference in probe-hybridizing ratios between cells of the four species not treated with rPeiW of the four species (Fig. 1a, c, e, and g) can be explained by the difference in their cell wall thicknesses (16). The cell wall of Methanobrevibacter ruminantium has been reported to be about twice as thick as that of Methanobacterium bryantii and M. thermautotrophicus. We then applied rPeiW to these cells. The rPeiW treatment remarkably improved the probe-hybridizing ratio (more than 95%) in all samples (Fig. 1b, d, f, and h). In particular, probe hybridization of Methanosphaera stadtmanae and Methanobrevibacter ruminantium cells was significantly altered (Fig. 1f and h). Cells of Methanobrevibacter ruminantium required a relatively higher concentration of rPeiW (2.3 units) for detection by FISH analysis than the other strains (0.4 units). These results indicate that rPeiW facilitates the probe permeability to the cytoplasm, although the optimum concentration of rPeiW depends on individual strains (species).
FIG. 1.
FISH images of pure cultured methanogens within the family Methanobacteriaceae: Methanobacterium bryantii DSM863T (a and b), M. thermautotrophicus DSM1053T (c and d), Methanosphaera stadtmanae DSM3091T (e and f), and Methanobrevibacter ruminantium DSM1093T (g and h). The left panels (a, c, e, and g) show untreated controls, and the right panels (b, d, f, and h) show rPeiW-treated cells. Bars, 10 μm.
FISH of cocultured cells of M. thermautotrophicus.
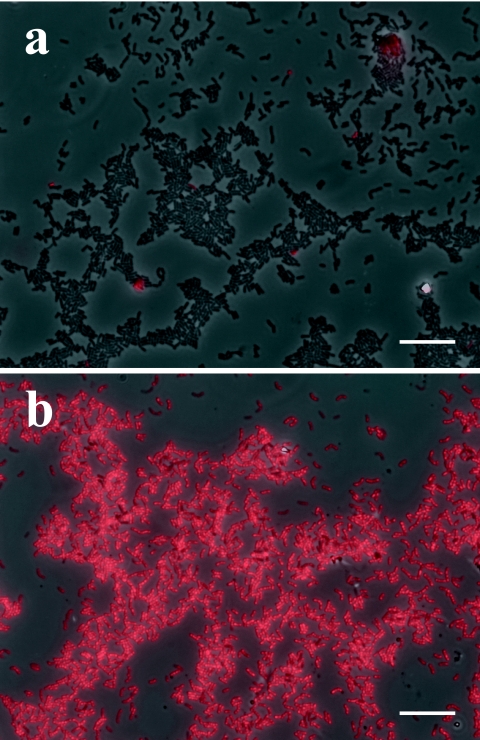
Figure 2 shows FISH images of cocultured cells of M. thermautotrophicus. Cocultured M. thermautotrophicus cells were collected from the two-membered coculture by selective lysis of the partner cells (syntroph) and by density gradient ultracentrifugation with Percoll. Very interestingly, almost none of the hybridization signals were observed in the cocultured cells without treatment (Fig. 2a), whereas more than 30% of the pure cultured cells (identical strain to that in the coculture) were hybridized with the probe even without rPeiW treatment (Fig. 1c). However, once rPeiW was applied, nearly all of the cells showed hybridization signals (Fig. 2b). These results strongly indicate that rPeiW treatment gives a remarkable improvement in probe hybridization for cocultured M. thermautotrophicus cells.
FIG. 2.
FISH images of cultured cells of M. thermautotrophicus. (a) Untreated control; (b) rPeiW-treated cells. Bars, 10 μm.
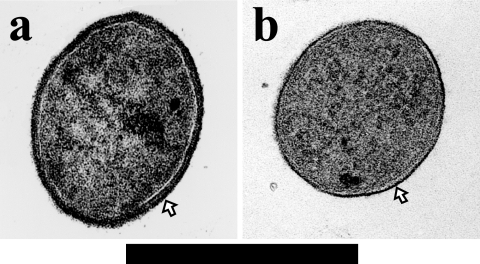
One of the interesting morphological differences between pure culture and coculture of M. thermautotrophicus cells was that the length of cocultured cells was one-third to one-quarter as long as those of pure cultured cells. Moreover, transmission electron microscopy observation revealed that cocultured cells had a cell wall that was at least three times thicker than that of cells from pure culture (Fig. 3a and b). Similar morphological alterations of cells depending on growth temperature were also reported previously (33). The most likely cause of these morphological differences is the H2 concentration between the two culture conditions; i.e., under coculture conditions, the partial H2 pressure is kept very low (less than 100 Pa) as a result of interspecies H2 transfer between the acetate-oxidizing H2-producing syntroph and M. thermautotrophicus. Moreover, the growth of M. thermautotrophicus in coculture is much slower than that in pure culture (approximate doubling time based on CH4 production rate of 3 h and 7 days, respectively) (data not shown). We also confirmed that the increase in the cell wall thickness of the M. thermautotrophicus strain ΔH cells occurred in coculture with a syntrophic butyrate-oxidizing bacterium, Syntrophothermus lipocalidus. These results indicate that the thickness of the cell wall is affected by the concentration of substrate (H2) but probably not by partner microorganisms.
FIG. 3.
Ultrathin-section (vertical direction) micrographs of transmission electron microscopy of M. thermautotrophicus cells grown in coculture (a) and pure culture (b). Bar, 0.5 μm. The outermost layers indicated by arrows are the cell wall (pseudomurein).
Endosymbiotic methanogens.
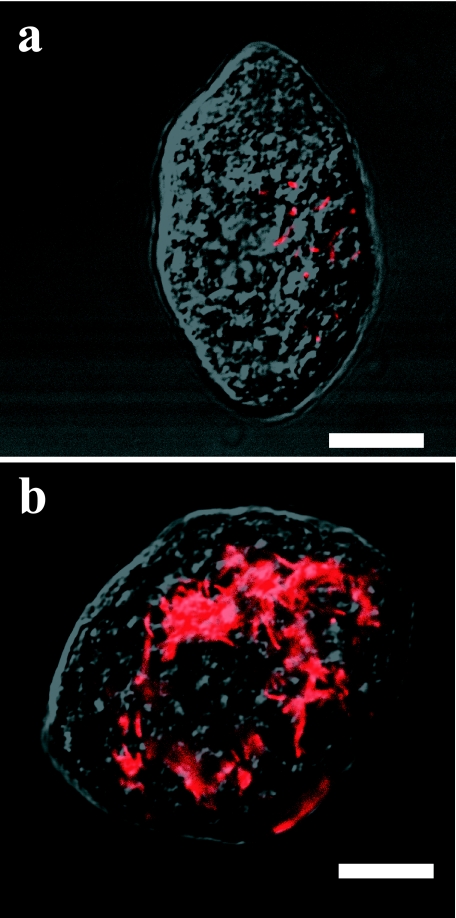
We also applied rPeiW to endosymbiotic methanogens in the ciliate Trimyema compressum. The host ciliates and endosymbiotic methanogens are shown in Fig. 4. Endosymbionts were hardly observed in the host ciliate without rPeiW treatment (Fig. 4a). However, rPeiW treatment enabled the visualization of a large number of endosymbiotic methanogens within the ciliate (Fig. 4b). The effectiveness was verified by examining another dozen of the protist cells with rPeiW treatment. Our previous study indicated that endosymbionts within T. compressum cells are closely related to the genus Methanobrevibacter from the 16S rRNA gene cloning library analysis (unpublished data). Without rPeiW treatment, probe-hybridizing ratios were less than 5% in the cells of other species of the genus Methanobrevibacter, such as Methanobrevibacter arboriphilicus DSM1125T and Methanobrevibacter curvatus DSM11111T (unpublished data). These results strongly suggest that Methanobrevibacter species cells intrinsically have thick cell walls regardless of their ways of living (symbiosis and free-living).
FIG. 4.
FISH images of endosymbiotic methanogens within a free-living anaerobic ciliate, Trimyema compressum. (a) Untreated control; (b) rPeiW-treated cell. Bars, 10 μm.
UASB granules.
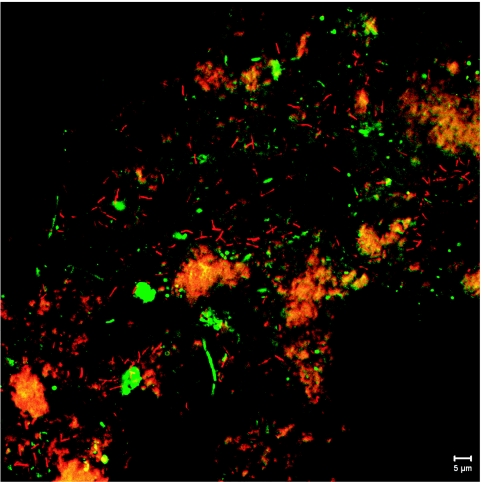
rPeiW treatment of granular sludge from the laboratory-scale UASB reactor was examined. The microbial community structure in the granule has been analyzed previously (28). We applied rPeiW to the granular sludge to evaluate its effectiveness. Figure 5 shows a sectioned micrograph of the rPeiW-treated homogenized granule taken by the confocal laser scanning microscope. Rod-shaped cells (red) resembling Methanothermobacter cells, which had been detected by the clone library analysis (28), were detected by the rPeiW treatment, whereas they were rarely viewed in the untreated sample (data not shown). These cells were distributed unevenly within the granule. Moreover, abiotic substances with strong autofluorescence (yellow-orange in Fig. 5) sometimes hampered the observation so that a quantitative comparison with the untreated sample was impossible. However, apparent increases in the numbers of cells and cell clumps detected by FISH analysis were observed when rPeiW was applied. Bacterial cells were not influenced by rPeiW treatment, and they were detected by the EUB338 probe mixture in the same field (green in Fig. 5). Moreover, detection of other methanogens such as Methanosaeta spp. seemed not to be affected by rPeiW treatment (unpublished data). These results strongly show that rPeiW specifically reacts with cells of Methanobacteriaceae and enables us to visualize them within complex microbial communities such as a UASB granular sludge.
FIG. 5.
Sectioned micrograph of a homogenized UASB granule taken by a confocal laser scanning microscope. The sample was simultaneously hybridized with rhodamine-labeled MB1174 (probe for the family Methanobacteriaceae) (red) and Cy5-labeled EUB338mix (probe for the domain Bacteria) (green). Blocks with a strong yellow-orange color were abiotic substances. Bar, 5 μm.
Instead of rPeiW pretreatment, freeze-and-thaw and proteinase K treatments were also applied to all samples examined in this study. However, these treatments did not significantly improve hybridization (data not shown). They even damaged the morphologies of specimens and caused a loss of methanogen cells during the pretreatment processes. The ciliate specimen was notably susceptible to these treatments. It is important to conserve morphological details of a host when attempting to observe the spatial distributions of endosymbionts within the host cell. rPeiW worked well on endosymbiotic methanogens and allowed us to examine their distributions within the host while conserving its intact morphology. We also successfully detected endosymbionts within the ciliates of a termite with rPeiW treatment (unpublished data).
Concluding remarks.
Here, we show that rPeiW could partially degrade cell walls of methanogens within the family Methanobacteriaceae and facilitate the permeation of oligonucleotide probes into their cytoplasms, resulting in significant increases in the number of cells detectable by FISH analysis. In addition, rPeiW acts on the cell wall of Methanobacteriaceae, creating a more effective method than conventional treatments, such as freeze-and-thaw and proteinase K treatments, in terms of the specificity and conservation of morphologies of entire communities during FISH analysis.
The other important finding of our study is that cells of Methanothermobacter, one of the cosmopolitan methanogens in anaerobic environments, change their cell wall thickness depending upon their way of living, and rPeiW treatment is very effective for them when they are grown syntrophically. Considering that methanogens in nature exist in extremely low partial H2 pressure (∼10 Pa) provided by syntrophic bacteria in the degrading process of complex organic matters (25, 35), Methanobacteriaceae may increase their cell wall thickness under such conditions, although the reason for this still remains unclear. The results from the UASB granule could support this phenomenon. Without any appropriate pretreatments, those methanogens in their natural habitats would hardly be detected by FISH, resulting in an imprecise estimation of their abundance and spatial distribution.
rPeiW may be a powerful tool for other in situ analysis techniques for methanogens belonging to the family Methanobacteriaceae in natural environments. Several techniques have recently been developed for in situ visualization of microbes in natural habitats, such as tyramide signal amplification-FISH, catalyzed reporter deposition-FISH, and in situ PCR (for a recent review, see reference 36). These techniques would allow us to analyze in situ information on microbes with low rRNA contents or with a certain functional activity, which we are hardly able to speculate on using conventional FISH analysis. However, these techniques involve the permeation of catalytic molecules into specimen cytoplasm for their reactions, such as horseradish peroxidase and DNA polymerase, which have higher molecular weights than oligonucleotide probes. Our method could make a significant contribution to these new techniques.
Acknowledgments
We gratefully acknowledge Hiroyuki Morii of the University of Occupational and Environmental Health, Satoshi Hattori and Moriya Ohkuma of RIKEN, and Masaaki Kawashima, Akinori Iguchi, and Kengo Kubota of the Nagaoka University of Technology with deepest appreciation for giving us the cells of M. wolfeii, for helpful advice on the treatment of ciliates, and for operation of the UASB reactor, a preparation of granular samples, and a helpful suggestion for preparation of the granular sample, respectively.
K.N. is financially supported by JSPS research fellowships for young scientists. This work was supported by the New Energy and Industrial Technology Development Organization.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beimfohr, C., A. Krause, R. Amann, W. Ludwig, and K. H. Schleifer. 1993. In situ identification of lactococci, enterococci and streptococci. Syst. Appl. Microbiol. 16:450-456. [Google Scholar]
- 3.Boone, D. R., W. B. Whitman, and Y. Koga. 2001. Family I. Methanobacteriaceae, p. 214-233. In G. M. Garrity et al. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer Verlag, New York, N.Y. [Google Scholar]
- 4.Carr, E. L., K. Eales, J. Soddell, and R. J. Seviour. 2005. Improved permeabilization protocols for fluorescence in situ hybridization (FISH) of mycolic-acid-containing bacteria found in foams. J. Microbiol. Methods 61:47-54. [DOI] [PubMed] [Google Scholar]
- 5.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 6.Egert, M., U. Stingl, L. D. Bruun, B. Pommerenke, A. Brune, and M. W. Friedrich. 2005. Structure and topology of microbial communities in the major gut compartments of Melolontha melolontha larvae (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 71:4556-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Embley, T. M., B. J. Finlay, R. H. Thomas, and P. L. Dyal. 1992. The use of rRNA sequences and fluorescent probes to investigate the phylogenetic positions of the anaerobic ciliate Metopus palaeformis and its archaeobacterial endosymbiont. J. Gen. Microbiol. 138:1479-1487. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez, A., S. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, and J. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara, K., N. Shinzato, T. Oshima, and A. Yamagishi. 2004. Endosymbiotic Methanobrevibacter species living in symbiotic protists of the termite Reticulitermes speratus detected by fluorescent in situ hybridization. Microbes Environ. 19:120-127. [Google Scholar]
- 10.Hattori, S., Y. Kamagata, S. Hanada, and H. Shoun. 2000. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 50:1601-1609. [DOI] [PubMed] [Google Scholar]
- 11.Hattori, S., H. Luo, H. Shoun, and Y. Kamagata. 2001. Involvement of formate as an interspecies electron carrier in a syntrophic acetate-oxidizing anaerobic microorganism in coculture with methanogens. J. Biosci. Bioeng. 91:294-298. [DOI] [PubMed] [Google Scholar]
- 12.Horn, M. A., C. Matthies, K. Kusel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Ohashi, and H. Harada. 2000. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl. Environ. Microbiol. 66:3608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandler, O., and H. König. 1993. Cell envelopes of archaea: structure and chemistry, p. 223-259. In M. Kates et al. (ed.), The biochemistry of Archaea (Archaebacteria). Elsevier Science Publishers B.V., Amsterdam, The Netherlands.
- 15.Kiener, A., H. König, J. Winter, and T. Leisinger. 1987. Purification and use of Methanobacterium wolfei pseudomurein endopeptidase for lysis of Methanobacterium thermoautotrophicum. J. Bacteriol. 169:1010-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langenberg, K. F., M. P. Bryant, and R. S. Wolfe. 1968. Hydrogen-oxidizing methane bacteria. II. Electron microscopy. J. Bacteriol. 95:1124-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lepp, P. W., M. M. Brinig, C. C. Ouverney, K. Palm, G. C. Armitage, and D. A. Relman. 2004. Methanogenic Archaea and human periodontal disease. Proc. Natl. Acad. Sci. USA 101:6176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 19.Luo, Y., P. Pfister, T. Leisinger, and A. Wasserfallen. 2002. Pseudomurein endoisopeptidases PeiW and PeiP, two moderately related members of a novel family of proteases produced in Methanothermobacter strains. FEMS Microbiol. Lett. 208:47-51. [DOI] [PubMed] [Google Scholar]
- 20.Luo, Y., P. Pfister, T. Leisinger, and A. Wasserfallen. 2001. The genome of archaeal prophage ψM100 encodes the lytic enzyme responsible for autolysis of Methanothermobacter wolfeii. J. Bacteriol. 183:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 22.Morii, H., and Y. Koga. 1992. An improved assay-method for a pseudomurein-degrading enzyme of Methanobacterium wolfei and the protoplast formation of Methanobacterium thermoautotrophicum by the enzyme. J. Ferm. Bioeng. 73:6-10. [Google Scholar]
- 23.Pfister, P., A. Wasserfallen, R. Stettler, and T. Leisinger. 1998. Molecular analysis of Methanobacterium phage ΨM2. Mol. Microbiol. 30:233-244. [DOI] [PubMed] [Google Scholar]
- 24.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schink, B., and M. Friedrich. 1994. Energetics of syntrophic fatty acid degradation. FEMS Microbiol. Rev. 15:85-94. [Google Scholar]
- 26.Schmid, M. C., B. Maas, A. Dapena, K. van de Pas-Schoonen, J. van de Vossenberg, B. Kartal, L. van Niftrik, I. Schmidt, I. Cirpus, J. G. Kuenen, M. Wagner, J. S. S. Damste, M. Kuypers, N. P. Revsbech, R. Mendez, M. S. Jetten, and M. Strous. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 1999. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekiguchi, Y., Y. Kamagata, K. Syutsubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 29.Shinzato, N., T. Matsumoto, I. Yamaoka, T. Oshima, and A. Yamagishi. 1999. Phylogenetic diversity of symbiotic methanogens living in the hindgut of the lower termite Reticulitermes speratus analyzed by PCR and in situ hybridization. Appl. Environ. Microbiol. 65:837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokura, M., M. Ohkuma, and T. Kudo. 2000. Molecular phylogeny of methanogens associated with flagellated protists in the gut and with the gut epithelium of termites. FEMS Microbiol. Ecol. 33:233-240. [DOI] [PubMed] [Google Scholar]
- 31.Wright, A. D., A. J. Williams, B. Winder, C. T. Christophersen, S. L. Rodgers, and K. D. Smith. 2004. Molecular diversity of rumen methanogens from sheep in Western Australia. Appl. Environ. Microbiol. 70:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada, K., Y. Kamagata, K. Nakamura, Y. Inamori, and I. Nakamura. 1994. Selectivity of food bacteria for the growth of anaerobic ciliate Trimyema compressum. Arch. Microbiol. 161:229-233. [Google Scholar]
- 33.Zeikus, J. G., and R. S. Wolfe. 1973. Fine structure of Methanobacterium thermoautotrophicum: effect of growth temperature on morphology and ultrastructure. J. Bacteriol. 113:461-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu, H., F. Qu, and L. H. Zhu. 1993. Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res. 21:5279-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis: ecology, physiology, biochemistry & genetics. Chapman & Hall, New York, N.Y.
- 36.Zwirglmaier, K. 2005. Fluorescence in situ hybridisation (FISH)—the next generation. FEMS Microbiol. Lett. 246:151-158. [DOI] [PubMed] [Google Scholar]