Abstract
As a means of investigating gene function, we developed a robust transcription fusion reporter vector to measure gene expression in bacteria. The vector, pTH1522, was used to construct a random insert library for the Sinorhizobium meliloti genome. pTH1522 replicates in Escherichia coli and can be transferred to, but cannot replicate in, S. meliloti. Homologous recombination of the DNA fragments cloned in pTH1522 into the S. meliloti genome generates transcriptional fusions to either the reporter genes gfp+ and lacZ or gusA and rfp, depending on the orientation of the cloned fragment. Over 12,000 fusion junctions in 6,298 clones were identified by DNA sequence analysis, and the plasmid clones were recombined into S. meliloti. Reporter enzyme activities following growth of these recombinants in complex medium (LBmc) and in minimal medium with glucose or succinate as the sole carbon source allowed the identification of genes highly expressed under one or more growth condition and those expressed at very low to background levels. In addition to generating reporter gene fusions, the vector allows Flp recombinase-directed deletion formation and gene disruption, depending on the nature of the cloned fragment. We report the identification of genes essential for growth on complex medium as deduced from an inability to recover recombinants from pTH1522 clones that carried fragments internal to gene or operon transcripts. A database containing all the gene expression activities together with a web interface showing the precise locations of reporter fusion junctions has been constructed (www.sinorhizobium.org).
Microbial genome sequences have revealed a multitude of genes for which functional information is almost completely lacking. One can infer that the functions of these genes are diverse, and hence, at a whole-genome level, it would seem prudent to adopt multiple approaches to the identification of gene functions. Several global approaches are being applied to the analysis of gene function in model organisms. These include in silico bioinformatic studies (2, 11, 19), systematic gene disruption (39), transcriptome microarray studies (1, 4, 6, 12), gene fusion and expression studies (3, 7, 21, 27, 44), promoter shutoff (35), the application of protein fusion tags (9, 43), proteomic analysis (16, 26), and two-hybrid screens to identify interacting proteins (13).
Genome-level studies of the legume symbiont Sinorhizobium meliloti have also been under way. This organism induces nitrogen-fixing root nodules on alfalfa. During the interaction between the eukaryotic host and prokaryotic bacterium, signals are exchanged which coordinate gene expression in both organisms to give rise to the N2-fixing nodule. The bacteria within the nodule are surrounded by a host-derived cell membrane and are supplied with a reduced carbon energy source(s) from the plant, and in turn they supply the plant with reduced nitrogen (30). As a free-living organism, the bacterium grows and survives in the soil environment and in the rhizosphere surrounding the plant root. In the laboratory, the free-living bacterium can grow in complex medium and in defined minimal media with glucose, succinate, or a broad range of carbon sources and can utilize ammonia, nitrate, or a range of other compounds as sources of nitrogen.
The S. meliloti genome contains three large replicons: the 3.65-Mb circular chromosome and the 1.36-Mb pSymA and 1.68-Mb pSymB megaplasmids (2, 19, 20). The sequence of the S. meliloti (strain Rm1021) genome revealed that this organism contains approximately 6,200 predicted protein-coding open reading frames (ORFs) (2, 19, 20). About 40% of the predicted genes are annotated as “unknown,” “hypothetical,” or “conserved hypothetical,” and with the exception of symbiotic and nitrogen fixation genes, very few of the remaining 60% have been functionally characterized. To begin to address this issue, analysis of gene expression in S. meliloti under various growth conditions, including in the root nodule, by using transcriptome microarrays has been reported (1, 4, 6, 12, 28), and an ORF library of the genome has been constructed (42).
As part of a functional genomic analysis of S. meliloti, we initiated a high-throughput study of the expression of many genes under various culture conditions. Here we describe the construction of a gene fusion library of the S. meliloti genome, in which the reporter fusions are integrated into the S. meliloti genome by homologous recombination. This avoids problems inherent in replicating plasmid systems which can disrupt regulation of expression due to copy number effects and rely on the presence of a promoter in the cloned DNA fragment. The gene fusion library was built to allow genome-scale expression analysis and detailed characterization of individual genes or groups of genes under various environmental conditions. To maximize screening efficiency, we constructed a new plasmid vector, pTH1522, which combines several features previously employed in other reporter vectors together with additional features that increase our ability to manipulate the plasmid and the host target genome. We report on the properties of the library, its gene coverage, identification of essential genes, and the results of initial screenings for reporter expression in complex and minimal media. Of the 2,480 genes annotated as encoding proteins of unknown function (PUFs), we have reporter fusions to 951. Of those, 16 appear to be essential for growth on complex media and around 40% show some expression under at least one of the three growth conditions tested. The potential of the library in assessing gene expression levels for large groups of genes under multiple growth conditions, including symbiosis, is discussed.
MATERIALS AND METHODS
Bacterial strains.
S. meliloti strain RmP110 is a derivative of Rm1021 in which a frameshift mutation in the pstC gene (pstC1021) is restored to the wild type (46). The S. meliloti genomic library in pTH1522 was stored as replicating plasmids in Escherichia coli DH5α and as cointegrant strains in S. meliloti RmP110. E. coli strain MT616 carries the mobilizing plasmid pRK600 (18). S. meliloti strains RmK990 and RmK991 carry pckA and chvI promoter fusions to gusA/tdimer2(12) and gfp+/lacZ, respectively, in pTH1522, and RmP319 and RmP320 carry fusions of the nifH promoter to gusA/tdimer2(12) and gfp+/lacZ, respectively, in pTH1703.
Construction of the reporter vector pTH1522.
The reporter plasmid pTH1522 was constructed as follows. The 1.7-kb replicon region together with the nic/bom site from pBR322 was PCR amplified using primers ML3340 and ML3341 (primer sequences are given in Table S1 in the supplemental material) and ligated with a 9.4-kb SpeI/EcoRI fragment carrying gfp+-lacY'Z-gusA-tdimer2(12)-Ω-aacC4-Ω from pTH1469 (43) (see Table 1 for details), yielding plasmid pTH1503. The Streptomyces phage attP50 site (25) was incorporated into primer ML3997. A 0.2-kb DNA fragment containing the Flp recombinase target (FRT)-trpA terminator-attP50 sites was PCR amplified from pTH1469 by using primers ML3997 and ML3397 and cloned into the SphI/SpeI sites in pTH1503, yielding pTH1504. The gfp+ gene with upstream stop codons in all three reading frames was obtained from pTH1469 by PCR using primers ML4277 and ML4274 and replaced the XhoI/BgIII fragment carrying lacZY′ in pTH1504, giving pTH1521. The lacZ gene was PCR amplified from pTH1469 by using primers ML4275 and ML4276 and inserted to replace the original copy of gfp+ adjacent to the attP50 as a SpeI/BgIII fragment in pTH1521, resulting in pTH1522. All constructs were confirmed by DNA sequencing.
TABLE 1.
Plasmids constructed during this work
| Plasmid | Description |
|---|---|
| pTH1469 | lacY′Z/gfp+, gusA/tdimer2(12), FRT site cloned in pUC19, Gmr |
| pTH1503 | lacY′Z/gfp+, gusA/tdimer2(12), ligated with pBR322 replicon region, Gmr |
| pTH1504 | pTH1503 with FRT-trpA terminator and attP50 site cloned into SphI/SpeI sites |
| pTH1521 | gfp+ with upstream stop codons replacing lacY′Z in pTH1504 |
| pTH1522 | lacZ gene replacing original copy of gfp+ in pTH1521 |
| pTH1591 | KpnI, SphI, PacI, ApaI, and NsiI sites removed from pTH1522 |
| pTH1596 | gfp+ gene from pTH1522 cloned into pUX19 |
| pTH1726 | MCS from primer ML6849 inserted into pTH1596 |
| pTH1728 | MCS from primer ML6851 inserted into pTH1596 |
| pTH1703 | EcoRV-XhoI fragment from pTH1726 inserted into pTH1591 |
| pTH1705 | EcoRV-XhoI fragment from pTH1728 inserted into pTH1591 |
| pTH1945 | Kmr/Nmr replacing Gmr in pTH1522 |
| pTH1946 | Kmr/Nmr replacing Gmr in pTH1703 |
| pTH1947 | Kmr/Nmr replacing Gmr in pTH1705 |
| pTH1565 | 2.5-kb pckA-chvI fragment inserted into pTH1522, pckA::gusA/tdimer2(12) orientation |
| pTH1566 | 2.5-kb pckA-chvI fragment inserted into pTH1522, pckA::gfp+/lacZ orientation |
| pTH1806 | 0.68-kb nifH fragment inserted into pTH1703, nifH::gusA-tdimer2(12) orientation |
| pTH1807 | 0.68-kb nifH fragment inserted into pTH1703, nifH::gfp+/lacZ orientation |
Construction of pTH1522 derivatives.
To increase the versatility of the pTH1522 vector, multiple cloning sites (MCS) were engineered into the plasmid. Restriction sites KpnI, SphI, PacI, ApaI, and NsiI in pTH1522 were first removed sequentially by digestion with each restriction enzyme, treatment with Klenow and T4 DNA polymerases to create blunt ends, and then self-ligation, resulting in pTH1591. A BgIII/XhoI fragment carrying the gfp+ gene from pTH1522 was cloned into the BamHI/XhoI sites in pUX19 (29), yielding pTH1596. MCS were incorporated into primers ML6849 and ML6851. The gfp+ gene with the MCS were PCR amplified using primer pairs ML6849/ML4274 and ML6851/ML4274, restricted with XhoI/SstI, and then inserted into the same sites in pTH1596, resulting in plasmids pTH1726 and pTH1728. pTH1591 was digested with BgIII, treated with Klenow DNA polymerase, and then digested with XhoI and ligated with an EcoRV/XhoI fragment carrying MCS and gfp+ from pTH1726 or pTH1728, resulting in plasmids pTH1703 and pTH1705.
Plasmids pTH1945, pTH1946, and pTH1947 were constructed by in vivo replacement of the gentamicin resistance gene (aacC4) in the three reporter plasmids with the Neor/Kmr gene (ntpII) from pVO155 (36). Phage lambda red recombinase was employed for gene exchange (15). A 1.3-kb DNA fragment carrying the nptII gene and its promoter PnptII was PCR amplified using primers ML10244 and ML10245. The gel-purified fragment was transformed into electrocompetent E. coli MG1655 carrying pKD46 (15) and pTH1522, pTH1703, or pTH1705, permitting recombination between the 50-bp homologous sequences flanking PnptII-ntpII on the PCR product and PaacC4 and aacC4 in pTH1522, pTH1703, or pTH1705. New Kmr plasmids pTH1945, pTH1946, and pTH1947 were retransformed into E. coli DH5α and screened for Kmr Gms transformants.
A 2.5 kb SaII fragment carrying the partial S. meliloti chvI-pckA region was obtained from pTH137 (38) and inserted into the XhoI site in both orientations in pTH1522, yielding plasmids pTH1565 and pTH1566. A 680-bp DNA fragment including 0.42 kb upstream of nifH was PCR amplified from Rm1021 genomic DNA using primer pair ML2850 and NifH6 and primer pair NifH5 and ML2581. Following restriction with XhoI/BamHI, the DNA fragments were inserted into the XhoI and BglII sites in pTH1703, generating nifH transcriptional fusions to either gusA-tdimer2(12) (pTH1806) or gfp+-lacZ (pTH1807).
Construction of the S. meliloti genomic library.
Total genomic DNA was prepared from S. meliloti strain Rm1021 cells as follows. Ten micrograms of DNA was partially digested with Sau3AI (New England Biolabs) to generate fragments of between 500 bp and 20 kb. The ends of the fragments were filled in with dATP, dGTP, and Klenow DNA polymerase (New England Biolabs) to leave a two-nucleotide 5′ overhang. The digest was fractionated on a 1% low-melting-point agarose gel, and the 750-bp to 1.5-kb and 1.5- to 3-kb size fractions were excised, eluted (QIAGEN gel extraction kit), and quantified before ligation. Plasmid pTH1522 was isolated from a 1-liter culture of E. coli DH5α by using a QIAGEN Maxi Prep plasmid kit and then further purified by cesium chloride density gradient centrifugation. Ten micrograms of pTH1522 DNA was digested with XhoI and partially filled in with dTTP, dCTP, and Klenow DNA polymerase to leave a two-nucleotide 5′ overhang. Approximately 100 ng pTH1522 DNA was ligated with 100 to 200 ng Sau3AI-digested genomic DNA in a 20-μl reaction volume with Promega TA ligase at 4°C overnight (Promega). The ligation reaction mixture was diluted with 80 μl Tris-EDTA, and 5 μl was used to transform library efficiency E. coli DH5α competent cells (Invitrogen) and plated onto LB agar with 10 μg/ml gentamicin and 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The transformation efficiency was approximately 50 to 100 colonies/ng vector DNA. Colonies were grown overnight in 3 ml LB with 5 μg/ml gentamicin, and plasmid DNA was isolated from 2 ml of culture (QIAGEN spin prep kit) for DNA sequencing. Cultures were frozen in 96-well microtiter plates in LB plus 10% glycerol and stored at −70°C. Clones were numbered 1 to 6,596 with the prefix pFL for plasmid fusion library.
Plasmid transfer into Sinorhizobium meliloti.
Plasmids were transferred from E. coli to S. meliloti strain RmP110 in triparental matings with the helper strain MT616 as previously described (18). Transconjugants were selected on LB agar with 200 μg/ml streptomycin and 60 μg/ml gentamicin. Resultant strains were colony purified twice and then inoculated into 1.3 ml LBmc (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl, 2.5 mM MgSO4, 2.5 mM CaCl2) with 100 μg/ml streptomycin and 30 μg/ml gentamicin in 96-well deep-well plates (ABgene). The S. meliloti strains were given the designation SmFL (for S. meliloti fusion library) and the number corresponding to the E. coli clone and then were frozen in individual wells in 96-well microtiter plates in LB plus 15% glycerol and stored at −70°C.
DNA sequencing of S. meliloti cointegrant recombinants.
The identity of recombinant S. meliloti strains was confirmed by direct sequencing from genomic DNA with the primers ML4876 and ML4875. Genomic DNA was prepared from 5 ml of culture grown in LBmc with antibiotics (see above) to saturation. Bacteria were collected in 2-ml microcentrifuge tubes by centrifugation (4 ml total), washed once with 0.85% NaCl, and then resuspended in 750 μl 10 mM Tris, 25 mM EDTA (pH 8). Sodium dodecyl sulfate (SDS) was added to 1%, NaCl to 1 M, and proteinase K (Sigma) to 0.5 mg/ml. Samples were mixed gently and incubated at 65°C for 2 h. DNA was extracted once with buffer-saturated phenol, twice with 1:1 phenol-chloroform, and once with chloroform, and then ammonium acetate was added to 0.5 M and nucleic acids precipitated with an equal volume of isopropanol. The pellet was dissolved in 400 μl of 10 mM Tris, 1 mM EDTA with 20 μg/ml RNase A and incubated for 30 min at 37°C. DNA was extracted once with 1:1 phenol-chloroform and once with chloroform, ammonium acetate was added to 0.5 M, and DNA was precipitated with an equal volume of isopropanol. To avoid errors from pipetting, the optical density at 260 nm (OD260) of the whole sample was measured to determine the DNA concentration. The samples were then lyophilized and dissolved to a final concentration of 2 μg/ml for sequencing. Sequencing was done by the Mobixlab Central Facility, McMaster University.
Library database.
The library database consists of three modules: a raw data processing module, a MySQL database, and a PHP web interface. Raw data are taken from trace files generated from partial sequencing of each clone to determine the fusion junctions with the reporter genes by using primer ML4975 for the gfp+ gene boundary and primer ML4876 for the gusA gene boundary. Sequencing was done at the Mobixlab Facility at McMaster University and at the Genome Sciences Centre, BC Cancer Centre, Vancouver, British Columbia, Canada. Other sources of raw data come from the expression data from enzymatic assays of cointegrant strains.
Perl scripts have been written to process the raw DNA sequence data and to interface it with PHRED/PHRAP and BLASTN software. These scripts automated the process of base calling and produced FASTA-formatted sequence data for alignment with the S. meliloti genome. The scripts provided the following information about a cloned insert: the nucleotide positions of its 5′ and 3′ ends, the length, and the orientation of the insert with respect to the reporter genes in the plasmid. Expression data, in the form of spread sheets representing 96-well plates, were automatically transformed into text format and then the data from each well matched with its clone identification number before import into the database. The expression data were further refined by employing a background subtraction, specific activity calculation, and calculation of fold increase above values for wild-type controls.
The main web interface “SinoSeq” is found at http://www.sinorhizobium.org under the link to the Fusion Library home page and displays both a graphical representation of clone/genome relationship and the corresponding gene fusion expression data. It utilizes the sequence information from each clone along with the GenBank annotation designation of the S. meliloti genome sequence to produce a graphical representation depicting the alignment of an insert sequence against the genome. This provides the context of the transcriptional fusions upon integration of the reporter plasmid into the S. meliloti genome. Expression data associated with a clone are extracted from the database based on a unique clone identification and displayed as tables. Two other web interfaces, SinoExp and SinoFold, were developed for the user to query and display expression data as specific activities and as fold increases, respectively.
Growth of cultures and reporter enzyme assays.
Cultures were grown in 1.3 ml LBmc, with 100 μg/ml streptomycin and 30 μg/ml gentamicin, in 96-well deep-well plates for 24 h at 30°C with shaking and then were subcultured into 225 μl LBmc, M9-glucose, and M9-succinate in 96-well microtiter plates and incubated for a further 40 h at 30°C. All subsequent liquid handling steps were performed with a Perkin-Elmer Multiprobe II. 5× M9 salts was purchased from Difco and made up to a final concentration of 48 mM Na2HPO4, 17 mM KH2HPO4, 8.6 mM NaCl, and 18.6 mM NH4Cl. This was further supplemented with 2.5 mM Mg2SO4, 1.25 mM CaCl2, 1 μg/ml biotin, 10 ng/ml CoCl2, and 15 mM glucose or succinate as a carbon source. One hundred microliters of each culture was transferred to a fresh 96-well plate and culture turbidity (OD600) measured with a Tecan Safire microplate spectrophotometer. Fluorescence readings were also taken from this plate: GFP+ was read at wavelengths of 485 nm (excitation) and 510 nm (emission), and t-dimer2(12) was read at 552 nm (excitation) and 579 nm (emission). Relative fluorescence was determined as (fluorescence at 510/552 nm − background)/OD600.
β-Galactosidase activity was measured from 20 μl culture with 80 μl Z buffer (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) containing 0.8 mg/ml 2-nitrophenyl-β-d-galactopyranoside, 0.0125% SDS, and 40 mM β-mercaptoethanol. Reaction mixtures were incubated for 1 h at room temperature, and reactions were stopped with 100 μl 1 M Na2CO3. The end product was measured at 420 nm with a Tecan Safire plate reader. Specific activity was calculated as (1,000 × OD420)/(time × OD600 × volume of culture in reaction in ml). β-Glucuronidase activity was measured from 20 μl culture with 80 μl GUS buffer (50 mM sodium phosphate buffer [pH 7], 1 mM EDTA) containing 0.44 mg/ml p-nitrophenyl β-d-glucuronide, 0.0125% SDS, and 50 mM dithiothreitol. Reaction mixtures were incubated for 1 h at room temperature, and reactions were stopped with 100 μl 1 M Na2CO3. Activity was measured at 405 nm with a Tecan Safire microplate spectrophotometer. Specific activity was calculated as described above. All plates included cultures of Rm1021 and RmP110 as negative controls and RmK990 and RmK991 as positive controls. Expression results were entered into the database both as specific activity and as fold increase over background, where the values obtained from Rm1021 and RmP110 were used as the background level.
Plant and bacteroid assays.
Alfalfa (Medicago sativa) seedlings, growing in Leonard assemblies with Jensen's medium, were inoculated with approximately 106 S. meliloti cells in 10 ml sterile water as previously described (17). After 4 to 5 weeks, root nodules were picked from the plants (10 to 20 per strain), bacteroids were isolated (17), and enzyme assays were performed as described above; specific activity was calculated as (1,000 × OD420/405)/(time × mg protein in reaction). The stability of the integrated pTH1522 plasmids in viable bacteria from the nodule was checked for several randomly chosen recombinants by crushing of one nodule from each strain, serial dilution, and plating onto LB agar plates with and without the selective antibiotic gentamicin. Colony numbers on selective media were >95% those of nonselective media.
Nucleotide sequence accession number.
The complete sequence of plasmid pTH1522 is available from GenBank under accession number DQ316260.
RESULTS AND DISCUSSION
Properties of the reporter vector pTH1522.
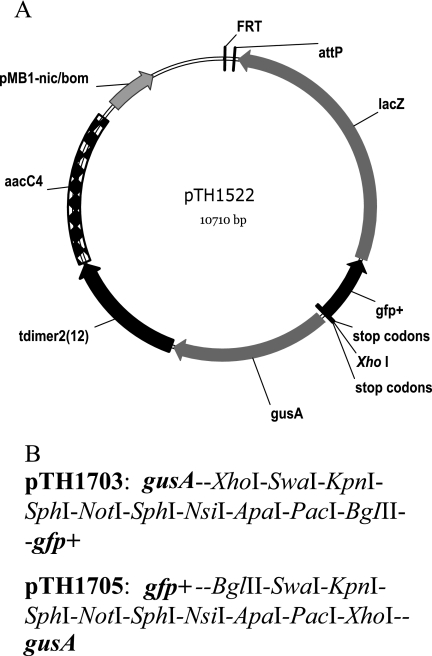
The reporter vector pTH1522 was constructed as a limited-host-range replicating plasmid with genes encoding both a fluorescent protein and an easily measured enzyme activity transcribed in either direction from a single unique XhoI cloning site. The plasmid carries the gentamicin resistance aacC4 gene flanked by transcription termination sites from pHPΩaac (8) and the Streptomyces coelicolor phage ΦC31 attP site for integrase-dependent recombination (25). Flanking the XhoI site are pairs of reporter genes encoding the enzyme β-glucuronidase (gusA) and a red fluorescent protein [tdimer2(12)] in one orientation and genes coding a green fluorescent protein (gfp+) and the enzyme β-galactosidase (lacZ) in the opposite orientation. Translation stop codons in three reading-frames are located upstream of the first reporter gene on either side of the XhoI site. An FRT site was included to allow Flp-mediated site-specific recombination from this site to other FRT sites in the host genome (33) (Fig. 1A). Plasmid pTH1522 can be transferred to a wide range of bacteria; however, while the plasmid replicates with a moderate copy number in E. coli, its ColE1 oriV host range is narrow, and it does not replicate in organisms such as Rhizobium and Pseudomonas. However, following nicking at the nic/bom site, it is mobilizable in trans, via the broad-host-range RK2 tra genes, into S. meliloti, where single-crossover, homologous recombination must occur between the cloned fragment and the S. meliloti genome in order to obtain gentamicin-resistant cointegrant colonies. The gfp+ variant used here is that described by Scholz et al. (41). It carries mutations that increase the folding efficiency and fluorescence yield of the encoded GFP protein. This GFP variant has fluorescence excitation and emission wavelength maxima of 491 nm and 512 nm, respectively. The red fluorescent protein (RFP), encoded by tdimer2(12), is a tandem genetic fusion in which two copies of dimer2 are joined with a 12-residue linker known to be resistant to protease (10). The dimer2 is derived from DsRed of Discosoma and carries 17 mutations relative to the DsRed protein. These mutations result in a protein that matures five times faster and has greater fluorescence brightness than wild-type DsRed. The tdimer2(12) derivative has excitation and emission wavelength maxima of 552 nm and 579 nm, respectively, and an extinction coefficient that is twice that of DsRed (10). Plasmids pTH1703 and pTH1705, derived from pTH1522, have been constructed with multiple cloning sites inserted at the position of the XhoI site (Fig. 1B). In addition, to allow greater flexibility in selection for the reporter plasmids in recipient strains, the gentamicin resistance gene, aacC4, in plasmids pTH1522, pTH1703, and pTH1705 has been replaced with the neomycin/kanamycin resistance gene (nptII) from Tn5. The resulting plasmids have been designated pTH1945, pTH1946, and pTH1947, respectively.
FIG. 1.
A. Plasmid pTH1522, with reporter genes gfp+, lacZ, gusA, and tdimer2(12) shown as divergent operons from the XhoI cloning site. Also indicated is the position of the FRT site, attP site, and gentamicin resistance gene and the locations of stop codons either side of the cloning site to prevent translational readthrough. B. Multiple restriction enzyme sites inserted in the pTH1522 derivatives, relative to the adjacent reporter genes.
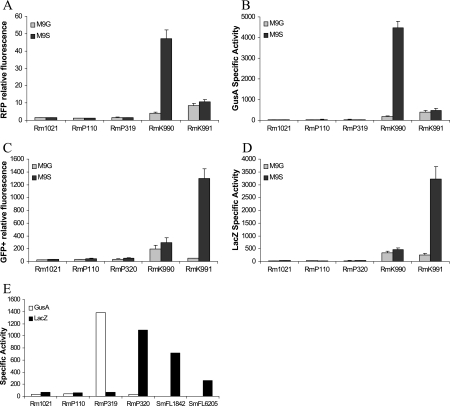
To evaluate the expression of the four reporter genes lacZ, gfp+, gusA, and tdimer2(12) in S. meliloti, we used two well-characterized S. meliloti promoters, pckA (phosphoenolpyruvate carboxykinase) and nifH (nitrogenase Fe protein), and in addition the chvI (transcription regulator) promoter, which is transcribed divergently from pckA. A 2.5-kb DNA fragment containing part of the pckA and chvI genes was cloned in both orientations into the XhoI site of the reporter plasmid pTH1522 and transferred into S. meliloti Rm1021 (strains RmK990 and RmK991). When RmK990 [pckA::gusA/tdimer2(12) and chvI::gfp+/lacZ] was grown in M9 medium, tdimer2(12) fluorescence and β-glucuronidase activities were high in M9-succinate and low in M9-glucose (Fig. 2A and B). In the reverse orientation, when pckA is fused to gfp+/lacZ in strain RmK991, GFP+ fluorescence and β-galactosidase activity were much higher in succinate-grown than in glucose-grown cells (Fig. 2C and D). These results are consistent with previous work on the induction of expression of pckA by succinate (38). The chvI promoter is not regulated by glucose or succinate and shows similar levels of expression in both media (RmK990 in Fig. 2C and RmK991 in Fig. 2A). We note that when the pckA promoter is driving tdimer2(12) transcription (Fig. 2A), the RFP fluorescence values are much lower than the GFP fluorescence values obtained when the pckA promoter is driving gfp+ transcription (Fig. 2C). The reduced sensitivity of RFP [tdimer2(12)] as a reporter compared to GFP+ (and LacZ and GusA) was also reflected in the data obtained with the fusion library (see below).
FIG. 2.
Expression of reporter genes from the pckA, chvI, and nifH promoters in free living cells and bacteroids. A. RFP relative fluorescence from strains grown in M9 with glucose (M9G) or with succinate (M9S) as the sole carbon source. B. GusA specific activity (in Miller units) from strains grown in M9 with glucose or succinate as the sole carbon source. C. GFP relative fluorescence from strains grown in M9 with glucose or succinate as the sole carbon source. D. LacZ specific activity from strains grown in M9 with glucose or succinate as the sole carbon source. E. GusA and LacZ specific activity from bacteroid protein extracts isolated from alfalfa root nodules. Rm1021 and RmP110, wild-type controls; RmP319 and RmP320, nifH promoter in either orientation; RmK990 and RmK991, chvI and pckA promoters; SmFL1842, lacZ fusion to the dme promoter; SmFL6205, lacZ fusion to the tme promoter. Error bars indicate standard deviations.
To assess expression of the reporter genes in the symbiotic state, a DNA fragment containing the nifH promoter was cloned into pTH1703 in both orientations and transferred to S. meliloti. Resultant cointegrant strains were then inoculated onto 2-day alfalfa seedlings and root nodules were harvested after 4 weeks, as described in Materials and Methods. When nifH was fused to gusA/tdimer2(12) in RmP319 or to gfp+/lacZ in RmP320, no expression of reporter genes could be detected in M9-glucose or M9-succinate medium (Fig. 2A to D). However, as expected, gusA was highly expressed in nodules inoculated with RmP319, and lacZ expression was high in nodules inoculated with RmP320 (Fig. 2E). In addition, two strains selected from the gene fusion library, SmFL1842 and SmFL6205, which carry gfp+/lacZ fusions to the dme and tme genes, respectively, were also included. Expression results from those fusions also agree with previously published work (17).
Properties of the S. meliloti genome library.
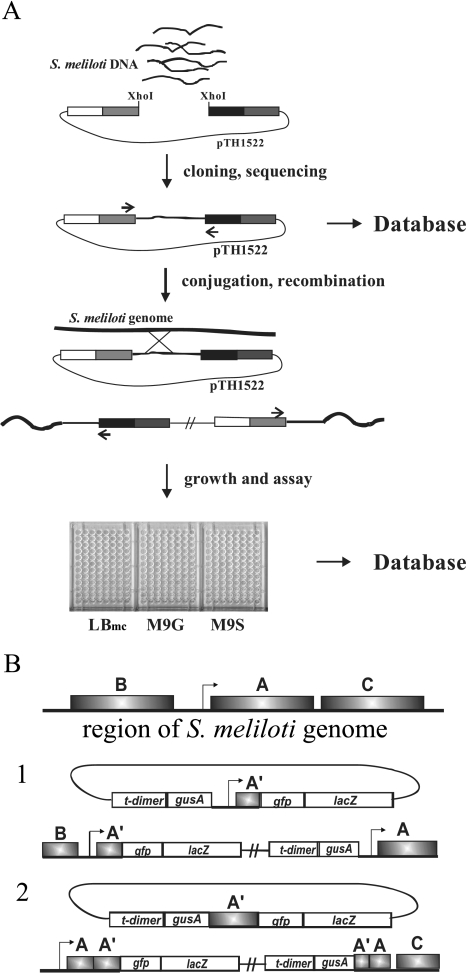
We have constructed a library containing 0.75- to 3-kb random DNA fragments from the S. meliloti genome in the reporter vector pTH1522. The strategy for constructing, assaying, and reporting the data from the library is outlined in Fig. 3A. To ensure a high frequency of insertion and minimize self-ligation of the vector, both the XhoI site of the pTH1522 DNA and the Sau3AI-digested S. meliloti genomic DNA were partially filled in, as described in Materials and Methods, to create complementary two-base overhangs. DNA sequence data from over 6,000 E. coli colonies revealed a cloning efficiency of >95%. The bidirectional reporter genes transcribed from the unique XhoI cloning site of pTH1522 effectively double the number of gene fusions generated relative to conventional unidirectional reporter fusion plasmids. Upon transfer to S. meliloti, pTH1522 clones carrying S. meliloti insert DNA recombine into the genome via single-crossover homologous recombination to yield gentamicin-resistant recombinants in which the cloned region is duplicated (Fig. 3B). Depending on the boundaries of the fragment present in pTH1522, single-crossover recombination can restore completely functional genes or operon transcripts or disrupt the gene or operon transcript (Fig. 3B). Each pTH1522 clone was transferred to S. meliloti strain RmP110 by conjugation, and of the 6,298 E. coli clones examined, a total of 5,795 recombinant strains were generated. In addition to the approximately 400 clones that had no S. meliloti DNA insert, an additional 101 clones that did carry S. meliloti gene fragments failed to yield recombinants, and these clones are discussed further below.
FIG. 3.
Schematic of the pTH1522 reporter gene fusion library construction and analysis. A. S. meliloti DNA fragments were cloned into pTH1522 and sequenced (primers are shown as small arrows), and the sequence information was entered into the database. Following conjugation and recombination, some strains were resequenced to confirm integrity (primers are indicated by small arrows), and then all strains were assayed in LBmc, M9 plus glucose (M9G), and M9 plus succinate (M9S) and the results were entered into the database. B. Diagrammatical representation of the genome organization following homologous recombination into the S. meliloti genome when the cloned fragment includes the promoter region of a gene (1) or an internal gene fragment (2).
The end points for each cloned DNA insert were identified by DNA sequencing, and the precise location of the cloned fragment within the annotated S. meliloti genome was determined. A graphical representation of each cloned fragment aligned with a map of the S. meliloti genome and showing the fusion junctions with the reporter genes can be found on the library database website (http://www.sinorhizobium.org; follow links to the Fusion Library home page). Sequences of clones which did not carry an insert were deleted from the database, as were any clones where the two end sequences mapped on different replicons or were further than 3.5 kb apart, hence eliminating any clones that might carry two or more different DNA fragments. Summary statistics, including the number of gfp+/lacZ and gusA/tdimer2(12) fusions to S. meliloti genes, together with the numbers of unique gene fusions, are given in Table 2. The bidirectional nature of the reporter vector ensures that clones that carry an insert spanning an intergenic region with divergently transcribed promoters will generate strains with fusions to both genes. Some clones carry internal gene fragments that generate a transcriptional fusion but also cause a loss-of-function mutation upon recombination into the genome. In addition, there are a number of genes that have independent fusions to both gfp+/lacZ and gusA/tdimer2 (12) (some examples can be found in Tables S2 and S3 in the supplemental material). Of the 6,204 annotated protein-coding genes in the S. meliloti genome, the library contains fusions to about 45% of the predicted ORFs.
TABLE 2.
Statistics for the S. meliloti reporter gene fusion library
| Parameter | Value |
|---|---|
| Total no. of pTH1522 clones in E. coli | 6,298 |
| Total no. of cloned DNA fragments in database | 5,896 |
| Total no. of fusion junctions sequenced | 11,519 |
| Total no. of S. meliloti recombinant strains with transcriptional fusionsa | 4,392 |
| Total no. of unique gene fusions (no duplicates) | 2,874 |
| Total no. of unique gusA/tdimer2(12) fusions (no duplicates) | 1,834 |
| Total no. of unique gfp+/lacZ fusions (no duplicates) | 1,814 |
| No. of gfp+/lacZ fusions to genes with unknown functionb | 1,250 |
| No. of gusA/tdimer2(12) fusions to genes with unknown functionb | 1,254 |
| Total no. of cloned intergenic sequences with both gfp+/lacZ and gusA/tdimer2(12) transcriptional fusions | 670 |
| Total no. of cloned internal gene fragments (potential loss-of-function mutations) | 512 |
A transcriptional fusion is defined as a reporter gene fusion within an identified ORF.
Genes with unknown function are defined as genes annotated as “hypothetical,” “conserved hypothetical,” “putative,” or “unknown” and given an Sm designation only (20).
Many genes are transcribed as operons, with from 2 to as many as 12 ORFs encoded on one RNA transcript. We queried the S. meliloti genome to calculate the number of potential operons, defined as those ORFs transcribed from the same DNA strand and with less than 75 base pairs of separation. Statistics regarding the number of these predicted operons to which the library contained at least one transcriptional fusion are shown in Table 3. Whereas coverage of single genes is approximately 45%, for predicted operons containing two or more ORFs, coverage is 76%, and for predicted operons of three or more genes, we have representation of 90 to 100%. It is of interest that the number of gene fusions in each replicon is proportional to the replicon size: 24% for pSymA, 27% for pSymB, and 48% for the chromosome. This represents direct evidence that the megaplasmids and chromosome copy number are the same.
TABLE 3.
Operon representation in the S. meliloti reporter gene fusion library
| Genes/operona | No. of operons | % Operons with at least one fusion |
|---|---|---|
| 1 | 2,673 | 46 |
| 2 | 709 | 64 |
| 3 | 253 | 90 |
| 4 | 120 | 94 |
| 5 | 62 | 100 |
| 6 | 38 | 100 |
| 7 | 26 | 94 |
| 8 | 5 | 100 |
| 9 | 6 | 100 |
| 10 | 4 | 100 |
| 11 | 0 | NAb |
| 12 | 2 | 100 |
Putative operons selected as genes transcribed from the same DNA strand and with an intergenic region of <75 bp.
NA, not applicable.
High-throughput expression screening.
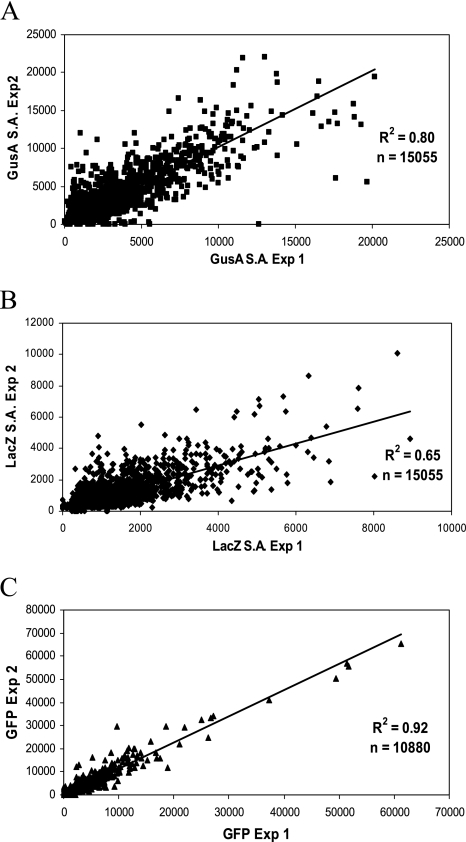
The S. meliloti strains derived from the library clones were assayed for all four reporter protein activities: green fluorescent protein (GFP+), red fluorescent protein [tdimer2(12)], β-galactosidase (LacZ), and β-glucuronidase (GusA). To assay the two enzyme activities in a high-throughput format using microtiter plates, several modifications were made to the original, published assay procedures (34). Because the enzyme assays were performed in polystyrene 96-well microtiter plates, it was not possible to use chloroform or toluene to permeabilize the cells, as these organic compounds react with the plastic. To avoid using a separate step for permeabilization as described by Griffith and Wolf (24) and to reduce the number of liquid-handling steps required, we developed a single-step addition assay in which 80 μl of assay buffer containing enzyme substrate (2-nitrophenyl-β-d-galactopyranoside or p-nitrophenyl-β-d-glucuronide) and 0.0125% SDS as a cell-permeabilizing agent is added to 20 μl of bacterial culture. We found that a final concentration of 0.01% SDS was sufficient to efficiently permeabilize the cells and gave enzyme activity results comparable to those obtained by previous methods. Cultures were grown in complex medium (LBmc) and in M9 minimal medium with either glucose or succinate as the sole carbon source then assayed as described in Materials and Methods. The complete S. meliloti fusion library was screened twice in two separate experiments with an interval of several months between them. As an indication of the reproducibility of the screening procedure, all the data points from the three growth conditions were plotted against each other (Fig. 4). The correlation between the two sets of data is shown for GusA activity (Fig. 4A), LacZ activity (Fig. 4B), and GFP+ relative fluorescence (only the values from M9 medium) (Fig. 4C); as stated before, the RFP fluorescence readings were too low to be meaningful and are not included. The correlation coefficients for the GusA activity and GFP+ fluorescence are good, at 0.8 and 0.92, respectively. However, the correlation coefficient for the LacZ activities was less satisfactory, at only 0.65. Most of the LacZ values fell below 3,000 units, and greater variability was seen with the highly expressed strains. Some of this variability may be due to the length of the RNA transcript required to produce full-length protein, as the correlation for GFP+ fluorescence, the product of the first gene in the pTH1522 reporter vector operon, is much higher. The GusA activity values show a larger spread from low to high values, although as expected, the majority of fusions show low expression values. The expression data were entered into the database containing the sequence information, and the interactive web interface allows access to the data for any specified clone. For each clone, the website shows the end points of the DNA fragment cloned in pTH1522 aligned with an annotated map of the genome region, the positions of the fusion junctions, the expression data for all four reporter genes as the raw data (including cell optical densities), the enzyme activity, the specific activity/relative fluorescence, and the fold increase over the RmP110 background control value.
FIG. 4.
Correlation between the two data sets from the expression screening. A. GusA specific activities obtained from library screening experiment 1 plotted against GusA specific activities obtained from library screening experiment 2. Values for all strains from the three test media are used (n = 15,055). B. LacZ specific activities from library screen experiment 1 plotted against LacZ specific activities obtained from library screen experiment 2. Values for all strains from the three test media are used (n = 15,055). C. GFP relative fluorescence values from experiment 1 plotted against GFP relative fluorescence values from experiment 2. Values for all strains from M9-glucose and M9-succinate media only are used (n = 10,880). r2 values are shown on the graphs.
Verification of the S. meliloti strains.
Having generated the fusion library in S. meliloti, it was important to confirm that the fusions in the S. meliloti cointegrant strains were, as predicted, from the junction sequences obtained from plasmid DNA isolated from E. coli. Accordingly, we employed direct sequencing from total DNA extracted from the S. meliloti transconjugants. Genomic DNA was isolated from approximately 450 S. meliloti::pTH1522 strains and sequenced with primer ML4876 (see Table S1 in the supplemental material). Over 96% of the strains gave the expected sequence, and of the few that did not, the genomic sequence revealed that the error resulted from misnumbering of the strain. All such errors were corrected in the database. Further confirmation of the integrity of the library comes from consistency in expression results where several fusions within the same gene exist, all giving the same expression profile. Examples of this can be found in Tables S2 and S3 in the supplemental material.
Gene expression in minimal medium versus complex medium.
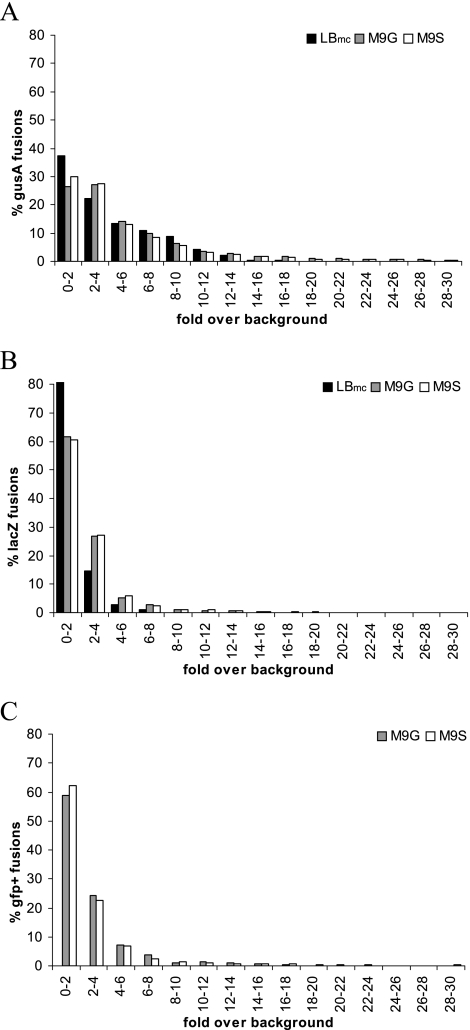
To gain insight into global gene expression levels in S. meliloti, as determined from the reporter fusions, the expression data for bona fide fusions were plotted as the numbers of clones (percentage of total) versus the expression level (given as fold above background) (Fig. 5). The expression frequency distribution, as revealed by these plots, demonstrates that >60% of the genes with gfp+/lacZ fusions are expressed at levels less than twofold above background in all three medium types (Fig. 5B and C), whereas the number is closer to 50% expressed at less than threefold over background for genes with gusA/tdimer2(12) fusions (Fig. 5A). The numbers of unique gene fusions expressed at low to background levels under the three growth conditions used are summarized in Table 4. A smaller percentage of genes are not expressed in M9 minimal medium compared to in LBmc, suggesting that more genes are expressed in minimal media, perhaps because more diverse biosynthesis is required for growth. In comparison, some microarray studies have estimated the number of genes expressed above background in S. meliloti grown in rich medium to be about two-thirds (4), whereas proteomic analysis of S. meliloti grown in defined medium detected about 2,000 gene products, corresponding to approximately 30% (26).
FIG. 5.
Distribution of expression levels for the gene fusion library. A. Number of gusA/tdimer2(12) gene fusions as a percentage of the total number of gusA/tdimer2(12) fusions with different expression levels of β-glucuronidase, measured as fold over background, in the three different test media (LBmc, M9-glucose [M9G], and M9-succinate [M9S]). B. Number of gfp+/lacZ gene fusions as a percentage of the total number of gfp+/lacZ fusions with different expression levels of β-galactosidase, measured as fold over background, in the three different test media (LBmc, M9-glucose, and M9-succinate). C. Number of gfp+/lacZ gene fusions as a percentage of the total number of gfp+/lacZ fusions with different expression levels of GFP+ fluorescence, measured as fold over background, in two different test media (M9-glucose and M9-succinate).
TABLE 4.
Numbers of gene fusions not expressed in complex or minimal media
| Fusion type | No. (%c) expressed in:
|
|
|---|---|---|
| LBmc only | M9-glucose and M9-succinate | |
| Unique gfp+/lacZ fusionsa | 1,469 (80) | 1,004 (55) |
| Unique gusA/tdimer2(12) fusionsb | 954 (53) | 764 (42) |
Unique lacZ transcriptional fusions expressed <2-fold above background.
Unique gusA transcriptional fusions expressed at <3-fold above background.
Percentages calculated from the number of unique fusions for each type of fusion.
Identification of genes highly expressed in complex medium.
The expression data were also analyzed to identify the gene fusions that are highly expressed (lacZ >4-fold above background and gusA >5-fold above background) in either complex medium (LBmc) or minimal media (M9 glucose and M9 succinate). The values are summarized in Table 5, and the list of genes identified can be found in Tables S2 and S3 in the supplemental material, respectively. Of the genes found to be highly expressed only in LBmc medium, many were similarly reported in microarray studies (4, 6). These include the genes involved in chemotaxis and motility, such as cheA, cheR, cheWI, flaB, fliG, and flgK; genes involved in metabolism of miscellaneous carbon compounds, such as agaL2, glpD, paaD, paaE, SMc00981, and SMc00982; and genes involved in nitrogen reduction, such as napA, napB, and norQ.
TABLE 5.
Numbers of gene fusions highly expressed in either complex or minimal media
| Fusion type | No. (%a) expressed in:
|
|
|---|---|---|
| LBmcb | M9-glucose and M9-succinatec | |
| Unique gfp+/lacZ fusions | 25 (1.3) | 173 (9.6) |
| Unique gusA/tdimer2(12) fusions | 62 (3.5) | 141 (7.8) |
Percentages calculated from the number of unique fusions for each type.
Number of gene fusions showing >4-fold over background expression of lacZ and >5-fold over background expression of gusA in LBmc (complex medium).
Number of gene fusions showing >4-fold over background expression of lacZ and >5-fold over background expression of gusA in M9 medium with either glucose or succinate as the sole carbon source.
Identification of genes highly expressed in minimal media.
Among the genes highly expressed in M9 medium with glucose or succinate as the sole carbon source are many genes involved in exopolysaccharide production, including exoF1, exoF3, exoH, exoY, and mrcA2, and in β-1,2-glucan synthesis, including ndvA and ndvB, as has been reported from microarray experiments (4, 6). The genes for thiamine biosynthesis, i.e., thiC, thiG, and thiO, are very highly expressed, reflecting the requirement of thiamine for growth in minimal medium (18, 40). The genes for synthesis of the siderophore rhizobactin, i.e., rhbB, rhbC, rhbF, rhlE2, and rhtA, are also very highly expressed (31). Interestingly we also find high expression of SMa2337, the gene immediately upstream of rhbA, confirming its role in rhizobactin synthesis as shown recently by Cuiv et al. (14). The high expression of the siderophore biosynthesis genes suggests that S. meliloti is iron limited in M9 medium. This is also reflected in the high expression of genes involved in hemin transport, i.e., hmuS, hmuT, and SMc02726 (shmR). These genes were also shown to be induced under iron-limiting conditions by microarray analysis (12). Interestingly, the conserved hypothetical protein SMc01514, which lies adjacent to the hmu gene cluster, is also highly expressed, suggesting that it may also be involved in hemin uptake or utilization. Expression of SMc01514 was also shown to be increased in response to iron limitation by microarray analysis (12). In addition, we also observe elevated expression, in minimal medium, of genes annotated as being involved in amino acid, purine, and pyrimidine biosynthesis.
Expression of PUFs.
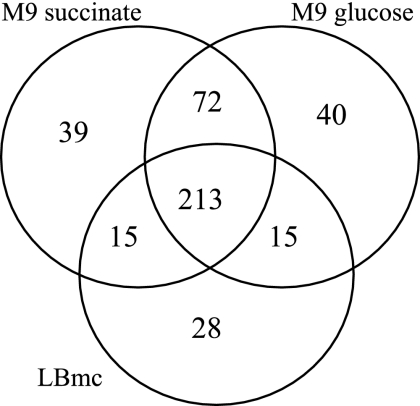
Of the 6,204 predicted ORFs in the S. meliloti genome, 2,480 are annotated as “conserved hypothetical,” “hypothetical,” or “unknown.” The S. meliloti::pTH1522 reporter gene fusion library contains transcriptional fusions to 951 of these proteins of unknown function. Analysis of the expression data for strains with fusions to just these genes reveals that approximately 60% are not expressed in complex medium (LBmc) or in minimal medium with either glucose or succinate as the sole carbon source. The remaining 40% show various degrees of expression in the different test media. Many are constitutively expressed in all three media, others in only one medium type, and some in different combinations of two medium types. These results are illustrated as a Venn diagram in Fig. 6, and a list of the genes identified, with data from one representative clone, is given in Table S4 in the supplemental material. For the genes that showed little or no expression in minimal medium, we are currently screening them with a battery of carbon and nitrogen compounds to find inducers of expression, with the aim of further elucidating function.
FIG. 6.
Venn diagram of PUF expression in LBmc, M9-glucose, and M9-succinate. The numbers of genes annotated as “conserved hypothetical,” “hypothetical,” and “unknown” expressed in the three test media analyzed are shown. Numbers represent lacZ gene fusions with >3-fold over background expression and gusA gene fusions with >4-fold over background expression.
Opposite-orientation expression.
In a very few cases activity was detected for the reporter gene that was not an in-frame transcriptional fusion as predicted from the S. meliloti sequence. After verification of the DNA sequence (as described in Materials and Methods) and reassaying of the reporter gene activity, four clones were identified as giving greater than threefold above background activity of the antisense reporter gene, and these are listed in Table 6. While the reason for the aberrant expression of these gene fusions has not been experimentally investigated, in each case the fusion junction was near the 5′ end of the ORF but transcribed in the antisense direction. One possible explanation could be that expression is occurring due to transcription of an antisense RNA involved in regulating expression of the gene, a phenomenon now recognized as being important in prokaryotic gene regulation as well as eukaryotic regulation (23, 32). We have analyzed the sequences upstream of the antisense fusion for putative promoters by using neural network promoter prediction at www.fruitfly.org, and antisense promoters are predicted in SMb20842 (score of 0.87, where 1.0 is the highest value) and SMb21465 (score of 0.80). Interestingly, in a recent publication similar findings of antisense transcription for two other S. meliloti genes expressed in early symbiosis were reported (47). It will be of interest to pursue these findings further.
TABLE 6.
S. meliloti strains with reporter gene expression in the antisense orientation
| SmFL no. | Gene | Annotation | Protein description |
|---|---|---|---|
| 310 | SMb20842 | Putative TonB-dependent receptor protein | |
| 2372 | SMa1561 | cheB2 | Probable CheB2 chemotaxis methylesterase |
| 4196 | SMc01023 | tpiA1 | Triosephosphate isomerase |
| 5433 | SMb21465 | prsE | Protein secretion protein, HlyD family |
Identification of essential genes.
A major advantage of the approach we have taken to the functional analysis of the S. meliloti genome is that it allows the identification of essential genes. As outlined above and shown in Fig. 3B, when the S. meliloti DNA gene fragment cloned in pTH1522 is internal to a gene or operon, recombination will disrupt the genomic copy and lead to loss-of-function mutation. If that occurs in a gene essential for growth in rich medium, then recombinant colonies will not be recovered. During transfer of the pTH1522 library clones to S. meliloti, we found 101 clones that did not generate recombinant strains. In several cases, multiple independent clones from the same gene failed to generate recombinants. For other genes where only one pTH1522 clone existed, the mating was repeated to verify the failure to obtain recombinants. Lists of the gene disruptions which failed to yield recombinants are given in Table 7 (for annotated genes) and Table 8 (for genes encoding proteins of unknown function).
TABLE 7.
Potential essential genes (annotated)
| No. of clones and gene name | Description |
|---|---|
| Multiple clones | |
| aidB | Acyl-CoA dehydrogenase |
| atpD | ATP synthase beta chain |
| cobT | Cobalamin biosynthesis |
| dxs | 1-Deoxyxylulose-5-phosphate synthase |
| fabI2 | Enoyl-acyl carrier protein reductase NADH |
| ftsY | Cell division protein |
| leuS | Leucyl-tRNA synthetase |
| murD | UDP-N-acetylmuramoylalanine-d-glutamate ligase |
| nrd | Ribonucleotide reductase |
| nuoN | NADH dehydrogenase I chain N |
| nusA | N utilization substance protein A |
| pbp | Penicillin binding protein |
| pheS | Phenylalanine-tRNA synthetase alpha chain |
| pheT | Phenylalanine-tRNA synthetase beta chain |
| rpsL | Ribosomal protein |
| rplL | Ribosomal protein |
| rpoC | RNA polymerase beta chain |
| pdhB | Dihydrolipoamide S-acetyltransferase |
| Single clone | |
| accC | Biotin carboxylase |
| argS | Arginyl-tRNA synthetase |
| aspS | Aspartyl-tRNA synthetase |
| cysS | Cysteinyl-tRNA synthetase |
| dnaB | Replicative DNA helicase |
| dnaG | DNA primase |
| dnaX | DNA polymerase III subunit Tau |
| exsC | Putative 6-pyruvoyl tetrahydropterin synthase |
| lysS | Lysyl-tRNA synthetase |
| hsdS | Type I restriction enzyme, modification enzyme |
| ileS | Isoleucyl-tRNA synthetase |
| ligA | DNA ligase |
| lppB | Lipoprotein precursor |
| lpxB | Lipid-A-disaccharide synthase |
| lytB | Penicillin tolerance protein |
| parB | Chromosome partitioning protein |
| parC | Topoisomerase IV subunit A |
| rpsE | 50S ribosomal protein L5 |
| rpmD | 50S ribosomal protein L30 |
| pyrG | CTP synthase |
| secA | Preprotein translocation subunit |
| thrS | Threonyl-tRNA synthetase |
| tolA | Hypothetical signal peptide protein |
| tolB | Periplasmic component of Tol biopolymer transporter |
| tsf | Elongation factor TS |
TABLE 8.
Potential essential genes (unknown function)
| No. of clones and gene name | Description |
|---|---|
| Multiple clones | |
| SMc00722 | Transmembrane protein |
| SMa2253 | Conserved hypothetical |
| Single clone | |
| SMc00074 | Hypothetical transmembrane signal peptide protein |
| SMc00471 | Putative sensor histidine kinase |
| SMc00532 | Conserved hypothetical protein |
| SMc00611 | Hypothetical transmembrane protein |
| SMc00951 | Transmembrane protein |
| SMc01721 | Inner membrane protein |
| SMc02090 | Conserved hypothetical protein |
| SMc02756 | Putative sensor histidine kinase |
| SMc02802 | Conserved hypothetical protein |
| SMc03872 | Hypothetical protein |
| SMc04010 | Hypothetical protein |
| SMc04347 | Conserved hypothetical protein |
| SMb20057 | ABC-type transporter permease |
| SMb21182 | Conserved membrane anchor protein |
Many of the genes identified as essential are as expected, such as genes for the amino-acyl tRNA synthetases, DNA and RNA polymerase subunits, and ribosomal proteins and genes involved in replication, transcription and translation, cell division, and protein translocation (Table 7), and these genes all map to the S. meliloti chromosome. In addition, there were 19 clones representing 16 genes encoding PUFs. Most of these were also located on the S. meliloti chromosome. SMc00074 is probably transcribed in an operon with rimJ and so may be involved in the synthesis or modification of ribosomal proteins. SMc01721 lies in an operon with rnpA, encoding the protein component of RNase P. The gene for the RNA component of RNase P has not been annotated for S. meliloti, but based on homology with rnpB from A. tumefaciens C58 (22, 45), it may lie between SMc01857 and SMc01856. Also of interest is SMc02090, which lies in an operon with lpxD, -A, and -B and so may be involved in lipopolysaccharide biosynthesis; we also show that lpxB is essential (Table 7). SMc02756 is the first gene in a long operon encoding mostly hypothetical proteins, but it includes SMc02760, which is annotated as encoding a nuclease/helicase. Finally, SMc02802 lies in an operon with leuS but also adjacent to the parA and -B genes, which are also shown to be essential. Interestingly, there was one gene on the pSymA megaplasmid, SMa2253, for which several different clones existed but each failed to produce recombinants. This result was unexpected, since the whole of pSymA has been deleted from S. meliloti strain Rm2011 without loss of viability (37). Whether this is because the gene is essential or because changes in expression are lethal, such as in a toxin-antitoxin system, remains to be determined, as these clones contained the 5′ upstream region of this gene and so should not have caused loss of function when recombined into the genome but may have resulted in loss of regulation.
Clones representing two genes on the pSymB megaplasmid also failed to produce recombinants. SMb21182 is represented by only one clone that contains an internal gene fragment, so recombination would lead to loss of function. The predicted gene product shows homology to acyl coenzyme A (acyl-CoA) transferases and lies in an operon with SMb21181, annotated as encoding a putative glutaryl-CoA dehydrogenase, but little else is known about this gene. In parallel experiments using the FRT/Flp recombinase system to delete defined regions of pSymB, this gene was deleted without loss of viability (B. Poduska and T. M. Finan, unpublished results). SMb20057 encodes an ABC transporter permease protein, and the clone representing this gene also contains an internal fragment which would cause loss of function of the transporter following recombination. We have confirmed that this gene is essential for growth on complex medium, and a more detailed analysis of this transport system is currently under way (J. Cheng, unpublished data).
Conclusions.
Since the sequencing of the S. meliloti genome, there have been several approaches taken towards understanding the roles of the many genes with both assigned and unassigned functions. Gene expression under specific conditions such as osmotic stress, phosphate limitation, and symbiosis has been analyzed by transcriptome microarray analysis (1, 4, 6), an ORF library of the genome has been constructed (42), and recently a transposon insertion library has been generated (39). Proteomic analysis by two-dimensional protein gel comparisons (16, 26, 37) and metabolite profiling (5) have also added to our knowledge of S. meliloti metabolism under various conditions.
We have taken a different approach. The S. meliloti reporter library is proving to be a versatile tool, allowing rapid analyses for gene expression under many growth conditions together with in situ staining for analysis of gene expression in the symbiotic state in root nodules. The pTH1522 library allows us to identify gene (and operon) knockout mutants. The reporter activity data nicely complement microarray data and readily allow more detailed analysis. We are now using the library to analyze classes of genes such as those induced upon phosphate starvation, all of the solute transport systems annotated in the S. meliloti genome (T. H. Mauchline et al., submitted for publication), and fusions to genes annotated as being involved in amino acid, purine, and pyrimidine biosynthesis, to further elucidate function both in the free-living state and in symbiosis with alfalfa. We are also exploiting the FRT site located in pTH1522 to direct deletion of defined regions of the S. meliloti genome. The versatility of the pTH1522 vector thus allows the integration of several approaches to the analysis of gene function and should be applicable to many other organisms.
Supplementary Material
Acknowledgments
We thank Bridget Kelly, Kris Knorr, Jihoon Hyun, Jordan Wronzberg, and Michelle Anstey for technical assistance during the construction of the library.
This work was funded by Genome Canada through the Ontario Genomics Institute. We gratefully acknowledge support from an NSERC Genomics award, which funded the initial phase of this work.
Footnotes
Published ahead of print on 8 September 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ampe, F., E. Kiss, F. Sabourdy, and J. Batut. 2003. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 4:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. BioTechniques 29:240-245. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barsch, A., T. Patschkowski, and K. Niehaus. 2004. Comprehensive metabolite profiling of Sinorhizobium meliloti using gas chromatography-mass spectrometry. Funct. Integr. Genomics 4:219-230. [DOI] [PubMed] [Google Scholar]
- 6.Becker, A., H. Berges, E. Krol, C. Bruand, S. Ruberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Kuster, C. Liebe, A. Puhler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant Microbe Interact. 17:292-303. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnason, J., C. M. Southward, and M. G. Surette. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 9.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao, T. C., J. Buhrmester, N. Hansmeier, A. Puhler, and S. Weidner. 2005. Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl. Environ. Microbiol. 71:5969-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke, P., P. O. Cuiv, and M. O'Connell. 2005. Novel mobilizable prokaryotic two-hybrid system vectors for high-throughput protein interaction mapping in Escherichia coli by bacterial conjugation. Nucleic Acids Res. 33:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuiv, P. O., P. Clarke, D. Lynch, and M. O'Connell. 2004. Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. J. Bacteriol. 186:2996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djordjevic, M. A. 2004. Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4:1859-1872. [DOI] [PubMed] [Google Scholar]
- 17.Driscoll, B. T., and T. M. Finan. 1997. Properties of NAD(+)- and NADP(+)-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti and differential expression of their genes in nitrogen-fixing bacteroids. Microbiology 143:489-498. [DOI] [PubMed] [Google Scholar]
- 18.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 21.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399-404. [DOI] [PubMed] [Google Scholar]
- 24.Griffith, K. L. and R. E. Wolf, Jr. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem. Biophys. Res. Commun. 290:397-402. [DOI] [PubMed] [Google Scholar]
- 25.Groth, A. C., E. C. Olivares, B. Thyagarajan, and M. P. Calos. 2000. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. USA 97:5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerreiro, N., M. A. Djordjevic, and B. G. Rolfe. 1999. Proteome analysis of the model microsymbiont Sinorhizobium meliloti: isolation and characterisation of novel proteins. Electrophoresis 20:818-825. [DOI] [PubMed] [Google Scholar]
- 27.House, B. L., M. W. Mortimer, and M. L. Kahn. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krol, E., and A. Becker. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics 272:1-17. [DOI] [PubMed] [Google Scholar]
- 29.Lies, D. P. 1994. Genetic manipulation and the overexpression analysis of posttranslational nitrogen fixation regulation in Rhodospirillum rubrum. Ph.D. thesis. University of Wisconsin, Madison.
- 30.Long, S. R. 2001. Genes and signals in the rhizobium-legume symbiosis. Plant Physiol. 125:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O. Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacLellan, S. R., L. A. Smallbone, C. D. Sibley, and T. M. Finan. 2005. The expression of a novel antisense gene mediates incompatibility within the large repABC family of alpha-proteobacterial plasmids. Mol. Microbiol. 55:611-623. [DOI] [PubMed] [Google Scholar]
- 33.McLeod, M., S. Craft, and J. R. Broach. 1986. Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2μm circle. Mol. Cell. Biol. 6:3357-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118:31-44. [DOI] [PubMed] [Google Scholar]
- 36.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 37.Oresnik, I. J., S. L. Liu, C. K. Yost, and M. F. Hynes. 2000. Megaplasmid pRme2011a of Sinorhizobium meliloti is not required for viability. J. Bacteriol. 182:3582-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osteras, M., B. T. Driscoll, and T. M. Finan. 1995. Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J. Bacteriol. 177:1452-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pobigaylo, N., D. Wetter, S. Szymczak, U. Schiller, S. Kurtz, F. Meyer, T. W. Nattkemper, and A. Becker. 2006. Construction of a large signature-tagged mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl. Environ. Microbiol. 72:4329-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Randhawa, G. S., and R. Hassani. 2002. Role of rhizobial biosynthetic pathways of amino acids, nucleotide bases and vitamins in symbiosis. Indian J. Exp. Biol. 40:755-764. [PubMed] [Google Scholar]
- 41.Scholz, O., A. Thiel, W. Hillen, and M. Niederweis. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein, p6. Eur. J. Biochem. 267:1565-1570. [DOI] [PubMed] [Google Scholar]
- 42.Schroeder, B. K., B. L. House, M. W. Mortimer, S. N. Yurgel, S. C. Maloney, K. L. Ward, and M. L. Kahn. 2005. Development of a functional genomics platform for Sinorhizobium meliloti: construction of an ORFeome. Appl. Environ. Microbiol. 71:5858-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661-670. [DOI] [PubMed] [Google Scholar]
- 44.Van Dyk, T. K., Y. Wei, M. K. Hanafey, M. Dolan, M. J. Reeve, J. A. Rafalski, L. B. Rothman-Denes, and R. A. LaRossa. 2001. A genomic approach to gene fusion technology. Proc. Natl. Acad. Sci. USA 98:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 46.Yuan, Z. C., R. Zaheer, and T. M. Finan. 2005. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium melitoti. J. Bacteriol. 188:1089-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, X. S., and H. P. Cheng. 2006. Identification of Sinorhizobium meliloti early symbiotic genes by use of a positive functional screen. Appl. Environ. Microbiol. 72:2738-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.