Abstract
Biofilms, or surface-attached microbial communities, are both ubiquitous and resilient in the environment. Although much is known about how biofilms form, develop, and detach, very little is understood about how these events are related to metabolism and its dynamics. It is commonly thought that large subpopulations of cells within biofilms are not actively producing proteins or generating energy and are therefore dead. An alternative hypothesis is that within the growth-inactive domains of biofilms, significant populations of living cells persist and retain the capacity to dynamically regulate their metabolism. To test this, we employed unstable fluorescent reporters to measure growth activity and protein synthesis in vivo over the course of biofilm development and created a quantitative routine to compare domains of activity in independently grown biofilms. Here we report that Shewanella oneidensis biofilm structures reproducibly stratify with respect to growth activity and metabolism as a function of size. Within domains of growth-inactive cells, genes typically upregulated under anaerobic conditions are expressed well after growth has ceased. These findings reveal that, far from being dead, the majority of cells in mature S. oneidensis biofilms have actively turned-on metabolic programs appropriate to their local microenvironment and developmental stage.
Many bacteria in the environment are thought to grow as surface-attached microbial communities, or biofilms, and it has been suggested that this biofilm lifestyle may account for the remarkable persistence of bacterial populations in the face of changing environmental conditions (9). Biofilms are composed of many hundreds of cells, each of which experiences its own microenvironment due to strong chemical gradients that are established by metabolism and diffusion. Biofilm communities are therefore heterogeneous and spatially stratified, so that activity levels and biochemical processes occur differentially according to the location of a cell in a biofilm and the biofilm structure's biomass (42).
In previous studies workers have investigated the spatial organization of microbial biofilm communities. In mixed-species biofilms, by using measurements of metabolites, fluorescent in situ hybridization, and community analysis by PCR, it has been shown that species stratification occurs (7, 14, 15, 28, 31). For laboratory-based single-species biofilms, in several studies workers have investigated spatial patterns of cellular growth activity and metabolism using a variety of techniques (10, 20, 21, 24, 27, 30, 32, 38, 46-49). These studies demonstrated that there is heterogeneity with regard to cellular activity and metabolism and, in particular, that in biofilm communities there is decreased growth activity near the center of the biofilm, where it is expected that cells are nutrient limited. To our knowledge, however, no systematic studies have been done to characterize the specific metabolic processes that occur in biofilms and how they might change over the course of biofilm development. As a first step toward this end, we set out to create a system in which we could quantitatively and reproducibly measure spatiometabolic patterning in biofilms. In particular, we wanted to address the question of whether biofilm cells could decouple growth from metabolism. In numerous studies using a live/dead stain and other methods workers have concluded that cells in the middle of the biofilm are dead or not actively synthesizing proteins (19, 26, 43), but it is not clear how accurately these reporters reflect cell viability in a biofilm.
To explore the spatiometabolic stratification of developing and mature biofilms, we selected Shewanella oneidensis strain MR-1, a biofilm-forming facultative anaerobe with remarkable metabolic versatility. S. oneidensis can use oxygen and many other lower-potential substrates, including metals, as electron acceptors in respiration, making it an attractive experimental system with which to explore domains of metabolism within a biofilm (17, 40, 41, 44).
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
S. oneidensis strain MR-1 was grown planktonically using LML medium (pH 7.4), which contained 0.2 g/liter yeast extract, 0.1 g/liter peptone, 2.5 g/liter sodium HEPES, and 0.243 ml/liter 60% lactate syrup (5 mM lactate). Planktonic cells were grown with shaking at 250 rpm at 30°C. When required, antibiotics were added at the following final concentrations: 15 μg/ml tetracycline and 50 μg/ml kanamycin.
S. oneidensis MR-1 derivatives constitutively expressing the GFPmut3* fluorescent protein (green) and enhanced cyan fluorescent protein (ECFP) were generated using mini-Tn7-KSGFP and mini-Tn7-KSCFP to insert the gfpmut3* or ecfp gene at the unique attTn7 site in the MR-1 genome, creating strains DKN308 and DKN309, respectively (the mini-Tn7 system is described in the supplemental material) (2, 6, 8, 12, 22, 29, 45). To allow detection of growth activity in S. oneidensis, the reporter system described by Sternberg et al. was used (38). The NotI fragment of pSM1606 (38) containing the growth rate-regulated Escherichia coli rrnB P1 promoter (3) fused to the gfp(AAV) gene encoding an unstable green fluorescent protein (GFP) was cloned into the highly stable broad-host-range plasmid pME6031 (16) to create plasmid pTK4. GFP(AAV) is unstable because it has a specific C-terminal oligopeptide extension that makes it susceptible to relatively fast degradation by intracellular tail-specific proteases, thereby enabling real-time expression imaging (1). pTK4 was transferred by conjugation into the ECFP-expressing strain S. oneidensis DKN309 to obtain strain DKN310. The mtrB reporter strain, DKN311, was made by homologous recombination. Primers were designed to amplify flanking 1-kb regions up- and downstream of the end of the mtrB gene from S. oneidensis MR-1 (upstream region primers 5′ CGGGATCCGCGGCCGCATAATACCCAAGTAGAAGAA and 3′ ATCAATCAACTAGTTCTAGAGCGATTAGAGTTTGTAACTCATGCT; downstream region primers 5′ GCAGCAGTTTAATGCTAGCGAACATTTGCCTCATATGCTCAAAAG and 3′ ATAAGAATGCGGCCGCTGTTGAATTGAATCCCCTGTT) and insert eyfp(AAV) between them. eyfp(AAV) encodes an unstable yellow fluorescent protein and was constructed by amplifying eyfp from pMP4658 (5) using primers that included the AAV sequence to fuse the AAV C-terminal peptide tag sequence to eyfp. Using NotI restriction sites, the upstream region, the sequence for eyfp(AAV), and the downstream region were inserted into the kanamycin resistance suicide plasmid pSMV10 such that eyfp(AAV) was between the upstream and downstream regions of the end of the mtrB gene. This plasmid was cloned into E. coli WM3064 (36) and mated with S. oneidensis MR-1, selecting for strains with kanamycin resistance. Such transconjugants were plated on plates containing LB medium supplemented with 5% sucrose to select for secondary recombinants lacking the integrated plasmid, which encodes sucrose sensitivity. Colonies from these plates were tested by PCR for the presence of the eyfp(AAV) gene. Plac-ecfp from miniTn7(Gm)PA1/04/03 ecfp-a (23) was inserted into pME6031 using KpnI and MluI to create plasmid pTK5, constitutively expressing ecfp. This plasmid was introduced into the MR-1 mtrB-yfp(AAV) strain DKN311 by conjugation to create strain DKN312, which constitutively expressed ecfp and expressed eyfp(AAV) from the end of the mtr operon.
Determination of plasmid stability.
To determine the stability of pTK4 and pTK5 in S. oneidensis MR-1 biofilms, S. oneidensis with pTK4 was grown for 5 days in 96-well polyvinyl chloride plates at 30°C in LML medium. As biofilm cells were attached to the walls of the well, the medium was exchanged every day to eliminate planktonic cells. After 5 days, the biofilm cells that were still attached to the well walls were removed and resuspended. Dilutions were plated on LB medium plates and incubated at 30°C overnight. A total of 309 colonies from the plates were picked, and each colony was streaked on plates containing LB medium and plates containing LB medium supplemented with tetracycline that were incubated at 30°C overnight. Of the 309 colonies, 308 grew on both types of plates, indicating that 99.7% of MR-1 cells were tetracycline resistant and therefore contained pTK4.
Measurement of GFP expression and stability.
Cells were grown overnight in LML medium and allowed to reach stationary phase. Cells were then transferred into fresh medium to obtain an optical density at 600 nm (OD600) of 0.0143 and were grown aerobically for 20 h. Samples for measurement of growth and fluorescence were taken using a Bio-Tek Synergy HT. To measure growth, the optical density at 600 nm was used. For green fluorescence, measurements were taken using a 485/20 excitation filter and a 528/20 emission filter.
Determination of respiration rates.
S. oneidensis overnight cultures were diluted into fresh LML medium and allowed to reach stationary phase. Cells were then transferred to a bottle with a stopper into which no oxygen could be introduced. The oxygen concentration was determined over time using an oxygen microelectrode until the concentration reached zero. The slope of the data was calculated, and this slope was the oxygen consumption rate for the number of cells in the flask. CFU counts were obtained for each flask, and the slope was divided by the number of cells in the flask to determine the rate of oxygen consumption per cell.
Biofilm experiments.
A flow cell system was constructed so that biofilms could be grown under constant conditions and images could be obtained over time. The flow cells had four channels machined from polyurethane with coverslips attached with epoxy. Each channel was 40.6 mm long, 11.4 mm wide, and 0.203 mm deep. LML medium (0.2 g/liter yeast extract, 0.1 g/liter peptone, 2.5 g/liter sodium HEPES, 0.043 ml/liter 60% lactate syrup; pH 7.4) containing 0.5 mM lactate was run through the flow cell system. Each flow cell was inoculated with 300 μl of a culture in the exponential phase at an OD600 of approximately 0.075 using a sterile syringe. Flow was not started immediately in order to allow cells to attach to the surface. After 1 h the flow was started at a rate of 4.1 μl/s, or 1.5 rpm, with a Watson-Marlow peristaltic pump. The flow continued at this rate for the length of the experiment. Using this technique, biofilms were grown for up to 7 days.
Confocal fluorescence microscopy was used to obtain images of bacterial biofilms grown in the flow cells. A Zeiss LSM 510 inverted microscope with a 63× Achroplan water immersion lens at the Caltech Beckman Institute Biological Imaging Center was used. Z-series images were acquired for multiple fields of view at multiple times during the experiment. For imaging egfp or eyfp, the excitation wavelength was 488 nm and the emission filter used was a BP500-550 filter. For imaging ecfp the excitation wavelength was 420 nm and the emission filter used was a BP465-485 filter. Images were processed using the Imaris software.
LIVE/DEAD staining.
S. oneidensis biofilms were grown in flow cells. At the final time point, the flow was stopped, and 1.5 μl of propidium iodide and 1.5 μl of Syto 9 from a Molecular Probes BacLight LIVE/DEAD L7012 stain kit in 1 ml of LML medium containing 0.5 mM lactate were injected into the tubing upstream of the biofilm. The flow was started briefly to deliver the stain to the biofilm. The flow was then stopped for 1 min to allow staining to occur, and then it was resumed to wash away residual stain. Images were obtained using an excitation wavelength of 488 nm and the BP500-550 emission filter for green fluorescence. The excitation wavelength was 543 nm and the emission filter was an LP605 filter to obtain images for red fluorescence.
mtrB expression with various oxygen levels.
LB medium containing 20 mM lactate was prepared. Ten milliliters of medium was added to 250-ml anaerobic Balch tubes. The tubes were closed with stoppers and flushed with the appropriate oxygen concentrations; 0%, 2%, and 10% oxygen tanks contained the appropriate concentrations of oxygen, and the remainder was nitrogen. Tubes containing 21% oxygen were flushed with filtered air. Fumarate (20 mM) was added to 0% oxygen cultures. Cells were grown overnight in aerobic cultures. Cells were then diluted to obtain an OD600 of 0.075. Cultures were set up for each oxygen concentration in duplicate. Cells were allowed to grow for 2 h or less until the OD600 was 0.150. Cells were then collected and treated with QIAGEN RNAprotect, and the RNA was immediately extracted using a QIAGEN RNAqueous Micro kit. The RNA was treated with DNase using the QIAGEN DNase I treatment and DNase inactivation protocol.
To synthesize cDNA, reverse transcriptase PCR (RT-PCR) was done using an Applied Biosystems TaqMan kit with a 50-μl reaction mixture. To quantify the cDNA, quantitative RT-PCR was performed. Twenty-one-base-pair primers flanking a 100-bp region for the mtrB, gfp, envZ, and recA genes were diluted to obtain a concentration of 5 pmol/μl. For each reaction, 1 μl cDNA from the RT-PCR, 6.5 μl of diethyl pyrocarbonate-treated water, 10 μl of 2× SYBR green mixture, and 2.5 μl of a primer pair mixture were added to one well of a 96-well quantitative PCR plate. A no-cDNA control was also included for each primer set. Quantitative RT-PCR analysis was performed using an Applied Biosystems Gene Amp 7500 sequence detector. A fluorescence value of 0.5 on a log scale was selected as the threshold for comparison of cycle numbers. The envZ and recA genes were used for normalization.
Quantitative analysis of biofilm images.
Biofilms images were obtained at multiple times in multiple fields of view. For each image, the fluorescence intensity profile for the reporter was mapped through the center x axis of the structure using NIH ImageJ. The fluorescence values were exported as a text file of x coordinates and fluorescence intensity for each pixel. We wrote an analysis program to analyze and process the data. The brightest pixel was defined as 100% fluorescence, and all fluorescence values were determined relative to this value. To automatically determine the edges of a structure, an edge detection algorithm was used, where the region of greatest contrast between the empty space surrounding the biofilm structure and the fluorescence of the structure was defined to be the edge. Using the defined edges, the center could then be specified. Then the intensity of each 9-pixel bin was determined in relation to its distance from the center of the structure, and a plot of fluorescence intensity versus distance from the center of the structure was constructed automatically for each image. This analysis generated a fluorescence profile for each image. Twelve biofilms for the growth reporter strain DKN310 and eight biofilms for the anaerobic reporter strain DKN312, representing images from all stages in biofilm development, were analyzed in this way in order to determine the size of the structure and its fluorescence profile. Biofilms were binned into representative sizes, and the average fluorescence profile for all the structures of a size, with a minimum of six structures in each category, was calculated.
RESULTS
Development of S. oneidensis biofilms.
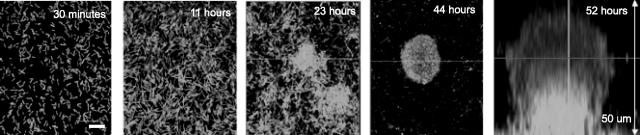
S. oneidensis was labeled with a constitutively expressing GFP from a lac basal promoter. Cells were grown in a flow cell system with LML medium containing 0.5 mM lactate, and images were obtained using confocal laser scanning microscopy. As shown in Fig. 1, S. oneidensis first formed a monolayer; after 23 h, the first structures became three-dimensional mounds; and at about 44 h, these structures began to become mushroom-like with a distinct stalk and cap. This basic mushroom morphology was then maintained until approximately 100 h, when the biofilms began to dissolve at both the cap and the stalk. Although S. oneidensis forms single-layer biofilms under some conditions (41), in our flow cell system biofilms developed as mushroom-shaped structures very similar to those described for other bacteria, including Pseudomonas aeruginosa, Pseudomonas fluorescens, and E. coli (37).
FIG. 1.
Time course of S. oneidensis biofilm development. Cells expressed a constitutive GFP. The first four panels (30 min and 11, 23, and 44 h) show x-y slices at the base of the developing biofilm structure. The final panel (52 h) shows a z profile through the structure. Scale bar = 10 μm.
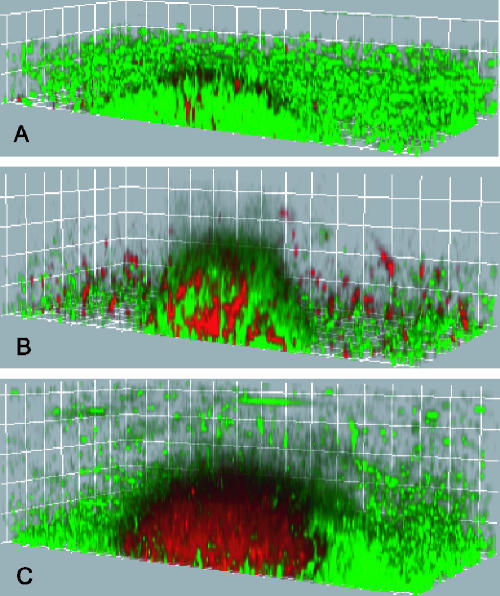
LIVE/DEAD stain for S. oneidensis biofilms.
To establish the live/dead patterns within an S. oneidensis biofilm, as typically determined with the Molecular Probes LIVE/DEAD BacLight stain, biofilms grown in the flow cell system were stained with the LIVE/DEAD reagent at different developmental points (Fig. 2). At early points or in structures with little biomass, most of the biofilm stained “live.” As the biomass of biofilms increased, cells throughout the structure stained “dead,” until the biomasses of the structures reached their maximum values when almost all cells in the middle of the structure stained “dead.” These patterns match the “live” and “dead” profiles reported for similar biofilms (11, 26). Yet in some studies workers have observed a discrepancy between LIVE/DEAD staining and other measures of viability, indicating that there may be a potential for bias with this method (35). The LIVE/DEAD stain uses two fluorescent nucleic acid stains, Syto9 and propidium iodide. Syto9 is used to quantify “live” cells because it can permeate cells under all conditions; propidium iodide is used to quantify “dead” cells because, as a highly charged molecule, it is unable to permeate cells with a strong electrochemical gradient across the membrane (i.e., the chemiosmotic potential). Although the results obtained with the LIVE/DEAD stain typically correlate with the results of growth assays performed in liquid or solid nutrient medium, as described in the Molecular Probes manual, little is known about how the membrane properties of cells in biofilms change with time. Given this uncertainty and that the LIVE/DEAD stain assay is a terminal assay, we reasoned that an in vivo method for assessing growth might give a more accurate representation of the metabolic state of the biofilm.
FIG. 2.
Live/dead staining in S. oneidensis biofilms. Green indicates the “live” stain, Syto9, and red indicates the “dead” stain, propidium iodide. The grid lines are 10 μm apart. (A) When the diameter is 60 μm, most cells continue to stain “live.” (B) When the diameter is ∼80 μm, cells throughout the biofilm start to stain “dead.” (C) When the diameter is ∼140 μm, almost all cells in the middle of the biofilm stain “dead.”
Patterns of growth activity in biofilms determined using the rrnB P1 reporter.
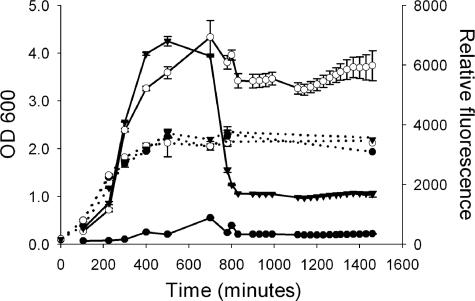
Derivatives of S. oneidensis MR-1 with in vivo fluorescent reporters of growth activity were constructed to spatially monitor cell growth in situ during the development and maturation of a biofilm. The growth reporter plasmid pTK4 was made using the system designed by Sternberg et al. (38), in which GFP is expressed from the E. coli rrnB P1 ribosomal promoter, whose activity correlates with ribosome production (3). S. oneidensis MR-1 containing pTK4 fluoresces green with an unstable GFP, GFP(AAV), when cells are producing ribosomes, a process associated with growth and protein synthesis capacity. To determine if this reporter functions as a good indicator of growth state in S. oneidensis, time course experiments were performed with well-characterized planktonic cultures. As shown in Fig. 3, for both the constitutively green fluorescent strain, DKN308, and the strain containing the ribosomal green fluorescent reporter, DKN310, fluorescence increased with OD600 until the concentration reached 1.2 ×108 cells/ml, where the OD600 plateaued. The level of fluorescence then stayed high in strain DKN308 but decreased in strain DKN310, reaching zero 240 min after the cell density stopped increasing. Green fluorescence from the ribosomal reporter increased during the exponential phase, as expected for a marker of growth, and then decreased due to the combined effects of RNA downregulation and prompt protein turnover during the stationary phase. These results verify that this GFP reporter is a sensitive, time-resolved indicator of growth activity in S. oneidensis.
FIG. 3.
Fluorescence levels and OD600 for S. oneidensis MR-1 (filled circles), S. oneidensis DKN308 constitutively expressing GFP (open circles), and S. oneidensis DKN310 expressing GFP(AAV) from a ribosomal promoter, representing growth activity (filled inverted triangles). OD600 is indicated by dashed lines, and fluorescence is indicated by solid lines. As cells grow through the exponential phase, the fluorescence levels increase, but when the stationary phase is reached, the fluorescence levels from the constitutively expressed GFP remain high whereas the fluorescence from the growth-active version decreases rapidly. The error bars indicate standard deviations of triplicate cultures; in some cases the error is less than the size of the symbol.
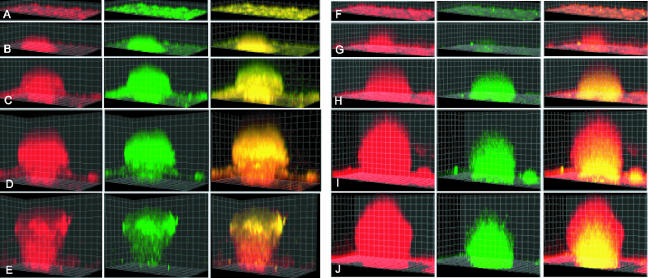
To monitor growth activity for the entire developing biofilm over time, pTK4 was used as one part of a two-color fluorescent system imaged by confocal microscopy. The pTK4 plasmid was introduced into S. oneidensis DKN309, a strain of S. oneidensis that also expresses ECFP constitutively from a basal lac promoter. The resulting new strain, DKN310, fluoresced cyan at all times to mark cellularity and also fluoresced green to mark cells actively engaged in growth, as indicated by active synthesis of the ribosomal machinery. To determine if there was a defined developmental progression and what parameters (size, shape, age, etc.) correlated most directly with growth activity, S. oneidensis DKN310 biofilms were grown in flow cells for up to 100 h. Individual biofilms were imaged longitudinally at multiple times and in multiple fields of view, using dual-channel fluorescence imaging of cyan and green with a Zeiss 510 confocal laser scanning microscope. A representative developmental course is shown in Fig. 4A to E. The growth activity, indicated by pTK4 reporter fluorescence, was uniform throughout the biofilm at early stages in development in structures up to 60 μm high that did not have a mushroom cap. When the structures developed a mushroom cap, they could then get up to 120 μm high and there could still be active growth in all regions of the biofilm. As structures got larger and reached the final mature state, up to 140 μm high, the growth activity in the cap was maintained, but the growth activity in the stalk was maintained only in the outer ∼25 μm.
FIG. 4.
Development of a mushroom structure in an S. oneidensis biofilm with a ribosomal (growth) reporter (DKN310) (A to E) and an anaerobic reporter gene (mtrB) (DKN312) (F to J). The grid squares are 10 μm by 10 μm. In the first column the cells are constitutively expressing ecfp and the fluorescence from ecfp is false red. In the second column of panels A to E, the cells expressing the growth-active GFP(AAV) are green. In the second column of panels F to J, the cells expressing the mtrB reporter are green. The third column is an overlay of the red and green channels. (A) 18 h and 8 μm high; (B) 28 h and 18 μm high; (C) 41 h and 52 μm high; (D) 65 h and 92 μm high; (E) 77 h and 112 μm high; (F) 17 h and 8 μm high; (G) 29 h and 31 μm high; (H) 47 h and 58 μm high; (I) 71 h and 104 μm high; (J) 85 h and 118 μm high.
Patterns of metabolism under anaerobic conditions in biofilms determined using the mtrB reporter.
To determine if metabolism (i.e., protein synthesis and energy generation) might occur in regions where cells are not actively growing, a reporter that is expressed under these conditions was required. It is known that as biofilm biomass increases, molecular oxygen (O2) levels in the inner regions decrease (34, 39), and these regions might correspond to domains where the rrnB P1 growth reporter is not active. To test whether this might be relevant for our system, we determined the O2 consumption rate for stationary-phase S. oneidensis cells in LML medium. Using this rate (55.3 mg O2/liter/s) and a biofilm diffusion model, as described by Stewart (39), we estimated that the concentration of O2 required to sustain growth was depleted approximately 30 μm into the biofilm. This is consistent with O2 profiles measured for many other biofilm systems (7, 34, 39). Therefore, if cells are still capable of active metabolism in regions where there is low growth activity, it is expected that genes expressed under low-O2 conditions would most likely be upregulated in these regions.
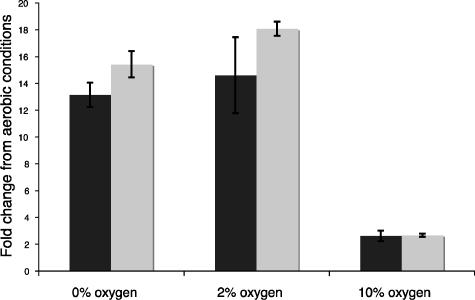
Previous work in our lab and the labs of other workers suggested that mtrB might be such a gene. mtrB is expressed under low-O2 conditions by Shewanella, and its product, an outer membrane β-barrel protein, facilitates electron transfer under anaerobic conditions (4, 25). Therefore, a reporter construct for mtrB was made, and a gene encoding an unstable yellow fluorescent protein, eyfp(AAV), was inserted into the chromosome after the mtr operon, creating strain DKN311. To establish whether yellow fluorescent protein fluorescence from the mtr operon in DKN311 is an accurate indicator of metabolism under anaerobic conditions, DKN311 was grown planktonically under aerobic, microaerobic, and anaerobic conditions. Quantitative real-time PCR experiments conducted with different O2 concentrations (0%, 2%, 10%, and 21% O2) showed that mtrB and eyfp expressed from the mtr operon were upregulated 12-fold only with 0% and 2% O2 (Fig. 5), validating the conclusion that eyfp(AAV) expression in the mtr operon can be used as a marker of new metabolism in low-oxygen domains. DKN312, the reporter strain used for imaging, was created by inserting the pTK5 plasmid that constitutively expresses ECFP from the lac promoter into strain DKN311; DKN312 constitutively expresses ECFP and expresses EYFP only under anaerobic or microaerobic conditions. Because EYFP and ECFP have an absolute requirement for O2 to fold properly, either color of fluorescence implies that at least trace amounts of oxygen are present in a biofilm (13).
FIG. 5.
Fold changes in expression, as measured by quantitative RT-PCR, of mtrB and eyfp in S. oneidensis DKN312 relative to aerobic conditions (21% O2). Solid bars, mtrB; gray bars, eyfp. mtrB and eyfp expression is upregulated under anaerobic and microaerobic conditions. The error bars indicate the ranges for duplicate cultures.
Biofilms of S. oneidensis DKN312 were grown exactly as described above for the growth activity reporter strain, DKN310. Figures 4F to J show that mtrB was not expressed at early stages of biofilm development, which is consistent with full O2 availability in structures less than 60 μm in diameter. mtrB expression appeared in the interior spatial domains of biofilms only at late developmental stages in structures more than 100 μm in diameter, when the interior cells were likely O2 limited.
Quantitative analysis of expression patterns.
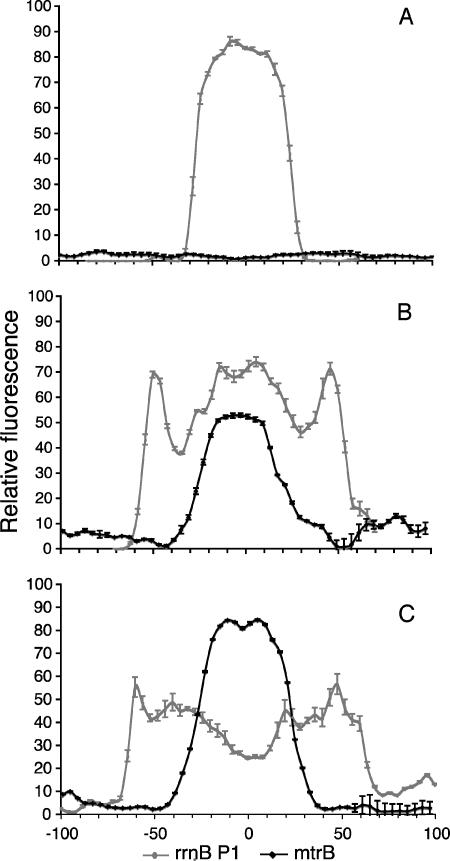
To assess the reproducibility of these results for multiple individual biofilms and in replicate experiments, we developed a quantitative analysis system to map reporter expression profiles for multiple biofilms. Using this system, a plot of fluorescence intensity versus distance from the center of the structure was constructed automatically for each image. For the different biofilm developmental stages, averages for S. oneidensis DKN310 (or growth activity) and S. oneidensis DKN312 (the mtrB strain) were calculated. The patterns of the reporters were remarkably consistent relative to structure size and shape. The most important factor in determining these patterns was not the time that the biofilm had developed, but its size, which is a far better and more consistent correlate of reporter gene activity domain. This quantitative analysis also revealed that the mtrB expression profiles were the inverse of the profiles for the growth activity GFP marker (Fig. 6).
FIG. 6.
Quantitative analysis of patterns of growth activity and metabolism in S. oneidensis biofilms. The gray lines show growth activity profiles for strain DKN310 (rrnB P1), and the black lines show mtrB expression of strain DKN312. (A) Biofilm structures approximately 60 μm in diameter. (B) Structures approximately 110 μm in diameter. (C) Structures approximately 140 μm in diameter. Each line represents an average of a minimum of six different structures. The error bars indicate the standard deviations of the binned pixel intensity values for all the images included in the plot. For panels B and C, local minima at the edges of the colonies are regions with no cells, thought to be extracellular polysaccharide. The patterns of expression relative to the size of the biofilm structure are remarkably consistent across multiple structures, and mtrB continues to be expressed in regions where growth activity has decreased.
DISCUSSION
Although various single-species biofilms have been well studied with respect to what controls their physical development, far less attention has been paid to their metabolic dynamics. A standard interpretation of the LIVE/DEAD stain results (Fig. 1) might be that large subpopulations of cells within mature biofilms do not actively generate energy and are therefore dead. However, the results obtained with our in vivo metabolic activity reporter system do not support this interpretation. Although the patterns of cell viability at early stages in biofilm development (diameter, <60 mm) correlate with the results obtained with the LIVE/DEAD stain, beginning in the middle developmental stages (60 mm < diameter < 140 mm), the patterns of metabolic activity diverge from the LIVE/DEAD stain patterns. Larger regions of growth activity (as measured with the rrnB P1 reporter) are observed, and subpopulations where metabolism decouples from growth (as measured by the mtrB reporter) also appear. In mature biofilms (diameter, >140 mm), although growth activity is restricted to the outer ∼25 mm, strong fluorescent signatures of mtrB expression dominate the interior; this is in sharp contrast to what is seen in the LIVE/DEAD stain, where all cells in the interior stain “dead.”
The patterns of growth activity and mtrB expression are remarkably consistent and correlate specifically with the size of the biofilm structure. This demonstrates that growth-inactive regions of the biofilm are nevertheless metabolically active. At a minimum, they generate sufficient energy to synthesize recently induced new proteins. The involvement of the mtr operon further implies that there is competence for key aspects of cellular metabolism, including electron transport and anaerobic metabolism that is enabled by mtr gene products. Thus, there appear to be major interior domains of biofilms where cells generate energy although they are not actively growing. The state of these interior cells may be akin to the stationary phase of planktonic cultures or might represent biofilm-specific metabolism. What is clear from the time-resolved imaging studies is that the vast majority of cells in all regions of a biofilm are physiologically active, even though they eventually run different programs of activity that depend on the spatial domain and, presumably, the associated microenvironment.
In conclusion, we observed remarkably reproducible spatiometabolic stratification in S. oneidensis biofilms. Our findings are leading us to rethink previous interpretations of what it means to be a “dead” cell in a biofilm and have implications for understanding how cells in a biofilm react to antibiotics, toxins, or other changes in environmental conditions. Cells that are maintained in a nongrowing state yet are still capable of synthesizing proteins may respond to introduced agents in unexpected ways and have the ability to act as a reservoir of survival. Indeed, in many natural systems where microorganisms are present at high cell densities, such as infections, microbial mats, and engineered bioreactors, it is likely that a significant fraction of the population is not actively growing (9, 42). A better understanding of what defines and controls the capabilities and activities of this growth-inactive state not only is essential for understanding basic aspects of biofilm biology but also is relevant for applications that aim to exploit the metabolic activity of biofilms for energy conversion and other purposes (18, 33).
Supplementary Material
Acknowledgments
We thank William Berelson for help with O2 respiration measurements, Søren Molin for plasmids, and the Caltech Biological Imaging Center for use of the facility.
This work was supported by grants from the Office of Naval Research, the Packard Foundation, and the Howard Hughes Medical Institute to D.K.N.
Footnotes
Published ahead of print on 25 August 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, M. S., and R. L. Gourse. 1994. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J. Bacteriol. 176:5560-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beliaev, A. S., D. K. Thompson, T. Khare, H. Lim, C. C. Brandt, G. Li, A. E. Murray, J. F. Heidelberg, C. S. Giometti, J. Yates 3rd, K. H. Nealson, J. M. Tiedje, and J. Zhoui. 2002. Gene and protein expression profiles of Shewanella oneidensis during anaerobic growth with different electron acceptors. OMICS J. Integr. Biol. 6:39-60. [DOI] [PubMed] [Google Scholar]
- 5.Bloemberg, G. V., A. H. Wijfjes, G. E. Lamers, N. Stuurman, and B. J. Lugtenberg. 2000. Simultaneous imaging of Pseudomonas fluorescens WCS365 populations expressing three different autofluorescent proteins in the rhizosphere: new perspectives for studying microbial communities. Mol. Plant-Microbe Interact. 13:1170-1176. [DOI] [PubMed] [Google Scholar]
- 6.Choi, K. H., J. B. Gaynor, K. G. White, C. Lopez, C. M. Bosio, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443-448. [DOI] [PubMed] [Google Scholar]
- 7.Cole, A. C., M. J. Semmens, and T. M. LaPara. 2004. Stratification of activity and bacterial community structure in biofilms grown on membranes transferring oxygen. Appl. Environ. Microbiol. 70:1982-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, N. L. 1989. Transposon Tn7. American Society for Microbiology, Washington, D.C.
- 9.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finelli, A., C. V. Gallant, K. Jarvi, and L. L. Burrows. 2003. Use of in-biofilm expression technology to identify genes involved in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:2700-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groh, J. L., Q. Luo, J. D. Ballard, and L. R. Krumholz. 2005. A method adapting microarray technology for signature-tagged mutagenesis of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl. Environ. Microbiol. 71:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 14.Harmsen, H. J., A. D. Akkermans, A. J. Stams, and W. M. de Vos. 1996. Population dynamics of propionate-oxidizing bacteria under methanogenic and sulfidogenic conditions in anaerobic granular sludge. Appl. Environ. Microbiol. 62:2163-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmsen, H. J., H. M. Kengen, A. D. Akkermans, A. J. Stams, and W. M. de Vos. 1996. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl. Environ. Microbiol. 62:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 17.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1093-1094. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, D. E., D. R. Bond, R. A. O'Neil, C. E. Reimers, L. R. Tender, and D. R. Lovley. 2004. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb. Ecol. 48:178-190. [DOI] [PubMed] [Google Scholar]
- 19.Hope, C. K., D. Clements, and M. Wilson. 2002. Determining the spatial distribution of viable and nonviable bacteria in hydrated microcosm dental plaques by viability profiling. J. Appl. Microbiol. 93:448-455. [DOI] [PubMed] [Google Scholar]
- 20.Huang, C. T., K. D. Xu, G. A. McFeters, and P. S. Stewart. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 64:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, C. T., F. P. Yu, G. A. McFeters, and P. S. Stewart. 1995. Nonuniform spatial patterns of respiratory activity within biofilms during disinfection. Appl. Environ. Microbiol. 61:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch, B., L. E. Jensen, and O. Nybroe. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187-195. [DOI] [PubMed] [Google Scholar]
- 23.Lambertsen, L., C. Sternberg, and S. Molin. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6:726-732. [DOI] [PubMed] [Google Scholar]
- 24.Lequette, Y., and E. P. Greenberg. 2005. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lies, D. P., M. E. Hernandez, A. Kappler, R. E. Mielke, J. A. Gralnick, and D. K. Newman. 2005. Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for biofilms. Appl. Environ. Microbiol. 71:4414-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 27.Majors, P. D., J. S. McLean, G. E. Pinchuk, J. K. Fredrickson, Y. A. Gorby, K. R. Minard, and R. A. Wind. 2005. NMR methods for in situ biofilm metabolism studies. J. Microbiol. Methods 62:337-344. [DOI] [PubMed] [Google Scholar]
- 28.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers, C. R., and J. M. Myers. 1997. Replication of plasmids with the p15A origin in Shewanella putrefaciens MR-1. Lett. Appl. Microbiol. 24:221-225. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen, A. T., T. Tolker-Nielsen, K. B. Barken, and S. Molin. 2000. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 31.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabaey, K., N. Boon, S. D. Siciliano, M. Verhaege, and W. Verstraete. 2004. Biofuel cells select for microbial consortia that self-mediate electron transfer. Appl. Environ. Microbiol. 70:5373-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen, K., and Z. Lewandowski. 1998. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng. 59:302-309. [DOI] [PubMed] [Google Scholar]
- 35.Renye, J. A., Jr., P. J. Piggot, L. Daneo-Moore, and B. A. Buttaro. 2004. Persistence of Streptococcus mutans in stationary-phase batch cultures and biofilms. Appl. Environ. Microbiol. 70:6181-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saltikov, C. W., and D. K. Newman. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. USA 100:10983-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternberg, C., B. B. Christensen, T. Johansen, A. T. Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart, P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thormann, K. M., R. M. Saville, S. Shukla, and A. M. Spormann. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thormann, K. M., R. M. Saville, S. Shukla, D. A. Pelletier, and A. M. Spormann. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J Bacteriol. 186:8096-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolker-Nielsen, T., and S. Molin. 2000. Spatial organization of microbial biofilm communities. Microb. Ecol. 40:75-84. [DOI] [PubMed] [Google Scholar]
- 43.van der Mei, H. C., D. J. White, J. Atema-Smit, E. van de Belt-Gritter, and H. J. Busscher. 2006. A method to study sustained antimicrobial activity of rinse and dentifrice components on biofilm viability in vivo. J. Clin. Periodontol. 33:14-20. [DOI] [PubMed] [Google Scholar]
- 44.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49 Pt. 2:705-724. [DOI] [PubMed] [Google Scholar]
- 45.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 46.Wentland, E. J., P. S. Stewart, C. T. Huang, and G. A. McFeters. 1996. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12:316-321. [DOI] [PubMed] [Google Scholar]
- 47.Werner, E., F. Roe, A. Bugnicourt, M. J. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. S. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, K. D., P. S. Stewart, F. Xia, C. T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida, A., and H. K. Kuramitsu. 2002. Streptococcus mutans biofilm formation: utilization of a gtfB promoter-green fluorescent protein (PgtfB:gfp) construct to monitor development. Microbiology 148:3385-3394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.