Abstract
The effect of the addition of synthetic sheep urine (SSU) and plant species on the bacterial community composition of upland acidic grasslands was studied using a microcosm approach. Low, medium, and high concentrations of SSU were applied to pots containing plant species typical of both unimproved (Agrostis capillaris) and agriculturally improved (Lolium perenne) grasslands, and harvests were carried out 10 days and 50 days after the addition of SSU. SSU application significantly increased both soil pH (P < 0.005), with pH values ranging from pH 5.4 (zero SSU) to pH 6.4 (high SSU), and microbial activity (P < 0.005), with treatment with medium and high levels of SSU displaying significantly higher microbial activity (triphenylformazan dehydrogenase activity) than treatment of soil with zero or low concentrations of SSU. Microbial biomass, however, was not significantly altered by any of the SSU applications. Plant species alone had no effect on microbial biomass or activity. Bacterial community structure was profiled using bacterial automated ribosomal intergenic spacer analysis. Multidimensional scaling plots indicated that applications of high concentrations of SSU significantly altered the bacterial community composition in the presence of plant species but at different times: 10 days after application of high concentrations of SSU, the bacterial community composition of L. perenne-planted soils differed significantly from those of any other soils, whereas in the case of A. capillaris-planted soils, the bacterial community composition was different 50 days after treatment with high concentrations of SSU. Canonical correspondence analysis also highlighted the importance of interactions between SSU addition, plant species, and time in the bacterial community structure. This study has shown that the response of plants and bacterial communities to sheep urine deposition in grasslands is dependent on both the grass species present and the concentration of SSU applied, which may have important ecological consequences for agricultural grasslands.
Intensification of upland pastures has been widespread in northwestern Europe, with formerly rough grazing areas undergoing improvement through fertilization, liming, and increased grazing (5). Intensification leads to shifts in the floristic composition in acidic upland grasslands, resulting in diminished floristic diversity (19). In Ireland and the United Kingdom, the predominant upland grassland formation on acidic soils is a species-rich but low-yielding plant community, with Agrostis capillaris as the predominant grass species and Nardus stricta, Holcus lanatus, Festuca rubra, and Festuca ovina typically at lower abundances (36). These seminatural grasslands can be improved to species-poor but high-yielding types dominated by Lolium perenne through intensive practices such as fertilization, grazing, and/or reseeding. A loss of diversity through intensification has been a cause for some concern (20, 23), particularly since little is known about impacts on below-ground biodiversity and the associated effects on biogeochemical and decompositional processes (40).
At present, there is no clear view as to what factors determine below-ground microbially diverse populations and activity. Several factors have been advanced, such as driving shifts in soil microbial community structure, including changes in physicochemical variables such as pH and nutrient levels (9, 26, 27), the influences of a floristic community change (16, 17), changes in soil physical structure resulting from tillage (24, 37), and grazing (4). Changes in plant species have been singled out as important drivers of microbial community composition during intensification in acidic upland grasslands (16, 17, 18), although in a microcosm study, Kennedy et al. (26) previously found that bacterial community composition was much more influenced by pH increase and nitrogen than by plant species.
Nitrogen supply and availability are important to the productivity of grazed pastures despite upland grazing areas rarely being fertilized (36). In the nutrient-poor acidic soils of temperate upland grasslands, low pH results in key dynamic processes within the nitrogen cycle, such as nitrification and net mineralization, operating only at low levels (46). In a study that compared soil properties along an ecocline between an improved mesotrophic grassland and an upland acidic grassland, Brodie et al. (6) found that nitrate levels in the acidic grassland were about half that of the mesotrophic grassland, whereas the levels of reduced N were approximately 36 times higher and probably held in organic matter. N supply in acidic grasslands is largely dependent on inputs from atmospheric N deposition, rainfall, and animal excretions such as urine and dung (21, 46).
Grazing can result in the deposition of large quantities of urine-N (400 to 1,200 kg N ha−1) within soils, resulting in perturbations of the grassland soil-plant system (14, 25). The main nitrogenous component of urine is urea, which is rapidly transformed by hydrolyzation to NH4+ (42). Only a small proportion is immediately captured by plant communities (13), with microbial processes such as nitrification and mineralization largely determining the subsequent fate of soil ammonium inputs (45). In a field experiment that examined the effects of urine deposition on Scottish grasslands, Williams et al. (47) showed that urine deposition substantially altered soil microbial communities in terms of bacterial and fungal counts, basal respiration rates, and community-level physiological profiles. Urine deposition in grazed pastures is spatially variable and leads to increases in soil pH directly below urine patches. This results in increased heterogeneity in soil chemistry and microbiology (30) and may be a major contributor to the extreme spatial variations characteristic of the microbiology of grazed upland pastures (35). In previous studies concerning urine deposition on pastures, microbial community changes have been assessed using conventional culture-based approaches (45, 46, 47); however, these approaches profile probably less than 1% of microbially diverse populations (43). Ritz et al. (35) used denaturing gradient gel electrophoresis to profile microspatial differences in grassland soil microbial communities; however, that study did not focus on urine deposition.
The work reported herein tests the hypothesis that urine deposition is a major influence on microbial community structural changes in upland grassland soils and that this process may be influenced by the species of grass present. To examine these effects, a sensitive molecular community profiling approach (automated ribosomal intergenic spacer analysis [ARISA]) was used to assess bacterial community structures.
MATERIALS AND METHODS
Soil.
Soil was collected in July 2004 from an area of unimproved Nardo-Galion grassland at Longhill, Kilmacanogue, County Wicklow, Ireland (United Kingdom national grid reference O 218 124), as described previously (26).
Microcosms.
Microcosms were prepared by weighing 80 g (dry mass) soil into black polyvinyl chloride pots (40-mm diameter, 110-mm height), which had been pierced to allow free drainage of water. Pots were planted with 20 to 25 surface-sterilized (2% sodium hypochlorite for 5 min) seeds (Emorsgate Seeds, Kings Lynn, United Kingdom) of either Agrostis capillaris or Lolium perenne. A set of control pots was left unplanted (bare soil). Forty-eight pots of each of the two plant species and bare soil were prepared, and microcosms were incubated in a greenhouse in a randomized block design for 75 days from seed germination, from 4 August to 17 October 2004. Water content was maintained at 35% (of overall pot weight) by the addition of distilled water as necessary. Plants remained untrimmed throughout the experiment and exhibited healthy growth throughout the initial 25 days of the experiment, prior to treatment. A synthetic sheep's urine (SSU) solution was prepared according to a method described previously by Clough (10) and was applied at three concentrations equivalent to 200, 500, and 800 kg N ha−1, representing low, medium, and high concentrations, respectively. Urea was mixed with the inorganic salts solution immediately before application. After 25 days, both planted and unplanted pots were treated as follows: (i) no addition of SSU (zero SSU), (ii) addition of SSU equivalent to 200 kg N ha−1 (low SSU), (iii) addition of SSU equivalent to 500 kg N ha−1 (medium SSU), and (iv) addition of SSU equivalent to 800 kg N ha−1 (high SSU). Microcosms were destructively sampled after 35 days (10 days after SSU treatment) and after 75 days (50 days after SSU treatment) to investigate whether short-term SSU-induced effects were still evident after an extended time period. All soil within microcosm pots was separated from plant matter and was considered rhizosphere soil. Soil was sieved to <4 mm and stored at 4°C for 48 h for pH, microbial activity, and microbial biomass analysis and at −20°C for molecular analysis.
Total microbial biomass, microbial activity, and pH.
Total microbial activity was measured as triphenylformazan dehydrogenase activity (1) and was determined based on a modification of a method described previously by Thalmann (41). Soil microbial biomass carbon was measured by substrate-induced respiration using a modification of methods described previously by Anderson and Domsch (2) and West and Sparling (44). Both methods were described previously by Brodie et al. (6). Microcosm soil samples were analyzed for pH according to a method described previously by Sparks et al. (39).
Total soil DNA extraction and purification.
Total soil DNA was extracted as described by previously by Brodie et al. (6). Briefly, 0.5 g of soil was added to tubes containing glass and zirconia beads to which hexadecyltrimethylammonium bromide extraction buffer was added. DNA was eluted in a final volume of 50 μl and was suitable for PCR amplification.
Bacterial community fingerprinting using ARISA.
The 16S-23S intergenic spacer region from the bacterial rRNA operon was amplified using forward primer S-D-Bact-1522-b-S-20 (eubacterial rRNA small subunit [5′-TGCGGCTGGATCCCCTCCTT-3′]) and reverse primer L-D-Bact-132-a-A-18 (eubacterial rRNA large subunit [5′-CCGGGTTTCCCCATTCGG-3′]) (32). Amplified sequences contained the intergenic spacer region plus approximately 150 bp corresponding to the 20 nucleotides of the forward primer and about 130 bp of the 23S rRNA gene. The forward primer was labeled with Beckman Coulter fluorescent dye D4 (Proligo). PCRs were carried out according to a method described previously by Kennedy et al. (27). Aliquots (1 μl) of PCR products were mixed with 38.4 μl deionized formamide, 0.2 μl of Beckman Coulter size standard 600 (dye D1), and 0.4 μl of custom-made marker (containing ribotypes ranging from 600 to 1,200 bp in intervals of 20 bp and 1,000 to 1,200 bp in intervals of 50 bp, all labeled with Beckman Coulter dye D1) (Bioventures, Murfreesboro, TN).
Analysis of intergenic spacer profiles was performed using a Beckman Coulter automated sequencer (CEQ8000) and Beckman Coulter CEQ8000 fragment analysis software, algorithm v 2.1.3, according to a method described previously by Kennedy et al. (27). Ribotypes that differed by less than 0.5 bp in different profiles were considered identical.
Statistical analysis.
Data sets for pH, microbial biomass, microbial activity, and most abundant ARISA bacterial profiles were analyzed by one-way factorial analysis of variance (ANOVA) using Genstat version 6. The significance level was set at a P value of <0.05. Multivariate statistical analysis was performed on ARISA profiles using Genstat version 6 and Canoco for Windows version 4.5. Initial analysis by detrended correspondence analysis revealed that the data exhibited a unimodal, rather than linear, response to the environmental variables (plant species, SSU addition, and time), so canonical correspondence analysis (CCA) (Canoco for Windows version 4.5) was used. The resulting ordination biplot approximated the weighted average of each species (in this case, relative abundance of ribotypes) with respect to each of the environmental variables, which are represented as arrows. The lengths of these arrows indicates the relative importance of that environmental factor in explaining the variation in bacterial profiles, while the angle between the arrows indicates the degree to which they are correlated. A Monte Carlo permutation test based on 199 random permutations was used to test the null hypothesis that bacterial profiles were unrelated to environmental variables.
The similarities of ribotype profiles for each plant treatment were examined using the Bray-Curtis index as a measure of similarity. Bacterial community matrices were analyzed using Primer software (Primer-E Ltd., United Kingdom) and were used to create nonmetric multidimensional scaling (MDS) plots for each of the data sets. MDS is an ordination technique that represents the relationships inherent in multivariate data sets and attempts to preserve the ranked order of the similarity of any two communities as an inverse function of the distance between the points representing those communities on the plot (28). Therefore, the communities with the highest similarity in the data set are represented on the plot by points that are plotted closest together, and the two communities with the lowest similarity are represented on the plot by points located the furthest apart. The degree to which the plot matches the similarity matrix can be judged by examining stress (Kruskal's stress), with values of less than 0.1 representing good ordinations with little risk of misinterpretation of the data (8). The MDS plots described here were used to visualize the relationships between bacterial communities (as determined by their ribotype profiles) under different treatment conditions. MDS was performed using 30 random starting configurations of sample points, and in all cases, two-dimensional solutions are presented.
RESULTS
Soil pHs from microcosms harvested after application of SSU are presented in Table 1. ANOVA revealed that the application of SSU significantly increased soil pH (P < 0.001), typically increasing depending on the concentration of SSU applied, with higher pH values at the highest SSU concentration. pH declined with time after application, with soil pH significantly lowering by 50 days in comparison to that at 10 days after SSU application. pH also differed significantly depending upon plant species, with L. perenne displaying a significantly lower soil pH than either unplanted or A. capillaris soils, although L. perenne and A. capillaris soils differed by less than 0.2 pH units.
TABLE 1.
Soil and microbial biomass measurements as affected by interactions between plant species, SSU additions, and timea
| Parameter | pH | Microbial activity (mg TPF g−1 dry soil) | Microbial biomass (mg biomass C g−1 dry soil) | Ribotype no. (means of ribotypes) |
|---|---|---|---|---|
| SSU treatments | 5.379 | 25.65 | 2.395 | 53.5 |
| Zero SSU | 6.021 | 22.05 | 2.485 | 61.2 |
| Low SSU | 6.275 | 41.68 | 2.532 | 53.5 |
| Medium SSU | 6.404 | 41.88 | 2.57 | 51.8 |
| High SSU | ||||
| P value | *** | *** | NS | NS |
| SED | 0.0292 | 2.445 | 0.118 | 5.66 |
| Plant species | ||||
| Bare soil | 6.031 | 31.44 | 2.508 | 50.8 |
| A. capillaris | 6.103 | 32.73 | 2.442 | 58.2 |
| L. perenne | 5.925 | 32.77 | 2.536 | 56.1 |
| P value | *** | NS | NS | NS |
| SED | 0.0253 | 2.117 | 0.1022 | 4.9 |
| Time | ||||
| Day 35 | 6.462 | 35.46 | 3.767 | 54.4 |
| Day 75 | 5.577 | 29.18 | 1.224 | 55.7 |
| P value | *** | *** | *** | NS |
| SED | 0.0207 | 1.729 | 0.0834 | 4 |
P values shown as *** indicate a P value of <0.001. NS, not significant; SED, standard error of difference; TPF, triphenylformozan.
Microcosm soils were assessed for microbial activity and biomass C using measurements of dehydrogenase and substrate-induced respiration, respectively (Table 1). Soils treated with zero and low concentrations of SSU had significantly (P < 0.001) lower microbial activity than soils treated with medium and high SSU concentrations. Time also had a significant (P < 0.001) effect on microbial activity, with activity decreasing between 10 and 50 days after the application of SSU. Plant species had no significant effect on microbial activity. Microbial biomass was not significantly affected either by the amount of SSU applied or by the presence of particular plant species. Nevertheless, there was a significant reduction (P < 0.001) in microbial biomass with time (Table 1). Ribotype number, however, was not significantly affected by plant species, SSU, or time.
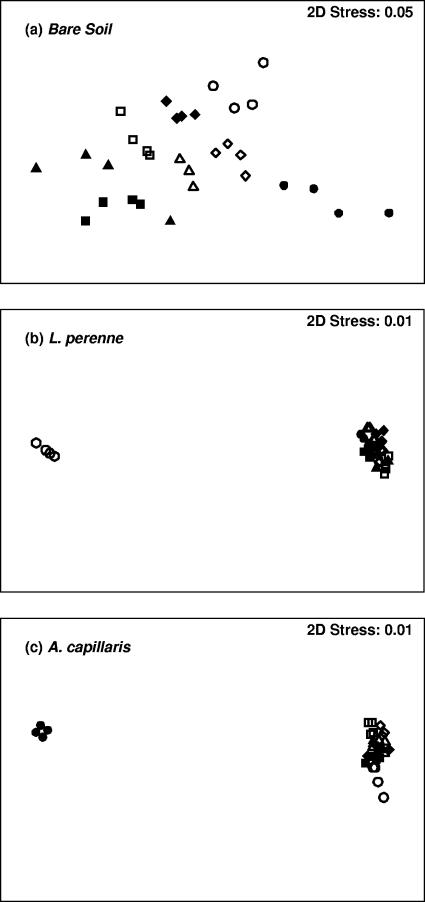
ARISA was used to generate bacterial community profiles consisting of the individual ARISA amplicons present and their relative abundances. Each amplicon indicated a sequence polymorphism, or ribotype. A total of 390 unique ribotypes were detected across all samples, and multivariate statistical analysis was performed to determine the main factors contributing to variation within the microcosm. MDS ordination was used to determine if overall bacterial community compositions varied across soils of different treatments based upon the presence or absence of ribotypes. Figure 1a to c shows MDS ordinations for individual replicates of all SSU treatments (both 10 days and 50 days after application) in unplanted soil (Fig. 1a) and where L. perenne (Fig. 1b) and A. capillaris (Fig. 1c) were planted.
FIG. 1.
MDS plots showing effects of synthetic sheep's urine (zero, low, medium, and high concentrations) and time after application (10 days and 50 days) on bacterial community compositions across different plant treatments. (a) Bare soil; (b) L. perenne; (c) A. capillaris. ▵, zero SSU at 10 days; □, low concentration of SSU at 10 days; ⋄, medium concentration of SSU at 10 days; ○, high concentration of SSU at 10 days; ▴, zero SSU at 50 days; ▪, low concentration of SSU at 50 days; ⧫, medium concentration of SSU at 50 days; •, high concentration of SSU at 50 days.
MDS ordinations are typically interpreted based on the distance between ordinate points; where treatments (and replicates) appear close together, these can be regarded as having similar bacterial community compositions. Figure 1a showed that for unplanted soil, time did not appear to greatly alter the bacterial community composition. On the other hand, SSU treatment did affect how replicates clustered, with a gradual shift in clustering as the SSU concentration increased (a shift rightwards). The effects of the addition of plants (as well as SSU) to microcosms were more pronounced. Figure 1b showed that L. perenne soils treated with high concentrations of SSU had a significantly different bacterial community composition only 10 days after SSU application but not after 50 days. In the case of A. capillaris, communities from an area treated with a high concentration of SSU 50 days after the SSU application clustered separately from all other treatments, indicating a markedly different bacterial community composition (Fig. 1c). The stress values calculated for the three ordinations indicated that the plots were very reliable representations of the data.
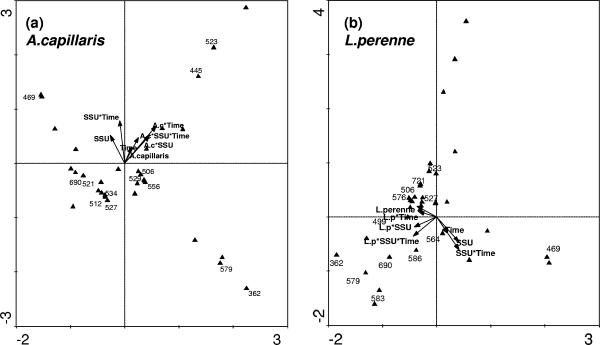
As MDS analysis gives a broad view of treatment effects on bacterial community composition, another multivariate approach was also used to elucidate the effects of individual environmental factors on bacterial community structure. CCA was used to explain data from L. perenne and A. capillaris microcosms separately. For both analyses, the species-environment correlations were all in excess of 0.96, indicating that the sample data were strongly correlated with the environmental parameters. The cumulative species-environment correlations for axes 1 and 2 (45.1 for L. perenne and 46.3 for A. capillaris) indicated that these axes accounted for almost 50% of the variance in the data that could be attributed to environmental factors (plant species, SSU, and time). Monte Carlo significance tests revealed that for both analyses, both the first axis and all axes combined explained a significant amount of variation (P value of <0.002).
The CCA ordinations are presented in Fig. 2a and b (A. capillaris and L. perenne, respectively). On ordination plots, environmental factors and their interactions are represented by arrows, and the most abundant ribotypes (representing 35% of the total abundance for both plant species) are represented as triangles. The relative length of arrows compares the relative importance of an environmental factor or interaction, and the position of ribotypes in relation to that arrow indicates how a particular ribotype/population is influenced by a given environmental factor/interaction. Both ordinations include data from unplanted soil. The ordination plot for A. capillaris microcosms is given in Fig. 2a and shows that two interactions (A. capillaris-SSU-time and SSU-time) were the most influential factors/interactions influencing bacterial ribotypes. On the ordination plots, ribotypes found to be significantly affected by one of the environmental factors or their interactions according to ANOVA (see the supplemental material) are numbered on the basis of their base pair lengths (ribotypes not significantly affected are not numbered). In general, from the positioning of individual ribotypes on the A. capillaris ordination plot, many ribotypes were negatively correlated with environmental factors/interactions.
FIG. 2.
CCA ordination diagrams, with environmental variables and the interactions represented as arrows and ribotypes represented as triangles. (a) A. capillaris; (b) L. perenne.
Figure 2b gives the ordination plot for the L. perenne microcosm. In this case, the most influential factors were SSU and SSU-time. Again, those ribotypes that were significantly affected by factors/interactions according to ANOVA are numbered (base pairs). In this case, ribotypes were positioned differently on the ordination plot, with a much higher proportion being orientated to suggest positive correlation with many of the factors/interactions (i.e., positioned in the same direction as arrow directions). Correlations between the top 50 most abundant ribotypes and environmental variables were explored further by ANOVA. Broadly, few ribotypes were significantly correlated with specific variables (plant, time, or SSU), but the majority of ribotypes were significantly correlated to some extent with interactions between variables at some level.
DISCUSSION
Application of synthetic sheep urine to grassland soil microcosms resulted in substantial changes in bacterial community structure, depending upon the grass species present, the amount of SSU applied, and the time after application. In general, changes in bacterial populations were rarely driven by single environmental parameters and were most often linked to interactions between one or more of the environmental parameters considered.
The application of SSU affected soil physicochemical properties, with pH increasing as higher concentrations of SSU were applied. Additionally, as more SSU was applied, microbial activity increased, although the microbial biomass remained relatively static. Upon application to soils, urea is hydrolyzed by increasing soil ammonium pools and raising soil pH, particularly where soil water is not limiting (10). It is not clear whether changes in microbial activity or biomass were mediated through increased nitrogen availability, pH change, or a combination of both. Microbial activity and biomass measurements are not able to discriminate between bacterial and fungal components. In these acidic upland soils, fungal biomass is known to be higher than that for bacteria (3, 7) and is much less responsive to environmental changes in comparison to bacterial communities (27). This may suggest that the observed increase in microbial activity might reflect an increase in bacterial activity, with the stability of the microbial biomass reflecting little change in the biomass-dominant fungal community. Neale et al. (31) noted that the addition of lime to soils (leading to an increase in soil pH) increased soil respiration and microbial biomass, probably reflecting an increase in acid-intolerant bacteria. Kennedy et al. (26) suggested that in acidic upland grassland soils, an increase in soil pH promoted the biomass and dominance of neutrophilic species, but they also showed that the application of N alone was much less effective in promoting microbial biomass.
ARISA is a PCR-based approach, and community fingerprints from this method need to be interpreted with caution because of species differences in rRNA gene copy numbers, biases generated during DNA extraction and PCR amplification, and the difficulty in standardizing the amount of DNA analyzed per replicate (11, 12). However, recent studies on bacterial community profiling found that terminal restriction fragment length polymorphism analysis, a method similar to ARISA, gave a quantitative view of microbial communities not affected by PCR bias, particularly when nondegenerate primers (such as the ones used in this study) were used (29, 38). In this study, all samples were subject to the same biases, and comparisons were made on a relative basis after standardization of ARISA fragment peak heights on a proportion basis.
MDS revealed that the overall bacterial community composition was affected by the application of SSU. In the absence of plants (bare soil), there was some evidence that bacterial composition changed with SSU concentration, with a left-to-right (low SSU to high SSU) gradation across the MDS plot. This would suggest that SSU application alone may cause some change in the bacterial community structure. This might reflect the pH and N increase in this soil selecting for neutrophilic bacterial species as discussed above. In the presence of plants (A. capillaris and L. perenne), MDS revealed that only high concentrations of applied SSU modified the overall bacterial community composition (Fig. 1). Interestingly, each rhizosphere responded differently, with the bacterial community associated with A. capillaris being different 50 days after application only, whereas L. perenne-associated communities were different 10 days after application but not after 50 days.
The difference in the responses of the two plant species to high SSU concentrations may partially lie in how the individual plant species responded to SSU application. A. capillaris plants were severely scorched at medium and high SSU concentrations, which occurred much less severely for L. perenne. Scorching of A. capillaris by sheep urine has been widely reported previously (46) and may contribute to the delayed changes in bacterial community structure in the A. capillaris rhizosphere. Nevertheless, this response was different from the unplanted response and may indicate the effect of an interaction between plant species and some other environmental parameter. There have been several studies that examined the fate of urine in soils, and most studies found that soil urea concentrations decline rapidly after urine deposition (10). Typically, urea is hydrolyzed to ammonium within days, which then is mineralized by microbial biomass or undergoes nitrification, with little of the original N being directly taken up by plant roots (13, 22, 42). The observed differences in plant growth response and bacterial community structure may indicate that L. perenne responds differently to sheep urination than A. capillaris, giving L. perenne a competitive advantage in grazing systems where improvement is occurring in acidic upland grasslands. In a previous study by Personeni and Loiseau (34), the importance of plant root morphology in competing for nutrients was highlighted. Those authors suggested that the thick root system of L. perenne plants may have a competitive advantage over other plant species when high concentrations of soil N are present. Scorching of plants is attributed to root death due to exposure to NH3 (46), and the low decay rate exhibited by L. perenne roots may be a reason why scorching was not so apparent on these plants. Sheep urine may therefore affect the competitiveness of plant species in pastures, perhaps favoring mesotrophic species such as L. perenne over indigenous acidic grassland species such as A. capillaris. The deposition of highly concentrated SSU and the subsequent increase in soil pH may leave A. capillaris at a disadvantage, reflecting its adaptation to and distribution in nutrient-poor, acidic soils.
CCA revealed that no environmental factor individually influenced the community; rather, a number of environmental parameters and their interactions determined community structure. ANOVA was performed to detail effects of environmental parameters and their interactions on specific ribotypes; the data from this analysis supported the finding by CCA analysis (Fig. 2) that interactions between factors are more influential on community structure (interpreted as effects on individual ribotypes) than individual parameters (plant, SSU, or time). Overall, it is not possible to come to a general conclusion as to which factor predominates, with each ribotype appearing to have a quite individual response to the microcosm treatments.
There have been very few studies combining sophisticated multivariate analysis and molecular estimates of microbially diverse populations to examine the N relations of upland grassland soils. Kennedy et al. (26) used a similar approach to assess the effects of nitrate additions on the microbial community structure. In that case, nitrate additions were found to be very influential in the microbial community structure, which was different from the findings of this study. This could indicate that the effect of pH, with increases in soil ammonium increasing pH (rather than the soil pH lowering the effect of nitrate), may also have a range of effects within the microcosm. It is likely that the effect of SSU in this case may have been much more far reaching than that of nitrate addition reported by Kennedy et al. (26), for example, affecting soil nutrient availability to both plants and microbial communities as pH increases. Increased soil pH is known to affect the availability of a wide range of soil nutrients, including phosphorus (15).
These data give some insight into the effects of urine deposition on upland grasslands. Ritz et al. (35) found that community structure had a high degree of spatial variability, which they linked to sheep urine patches, while Nunan et al. (33) also found that in a field trial, there were changes in microbial community structure but not in composition in response to the application of sheep urine, as shown by denaturing gradient gel electrophoresis profiles. From our microcosm study, it is suggested that the response of bacterial communities within a urine patch is dependent upon the grass species present and the concentration of sheep urine added. MDS analysis suggested that community composition was heavily influenced at high SSU concentrations, with differential time responses between the two plant species. Sheep urine typically adds nitrogen to pastures at a rate of around 500 kg N ha−1, corresponding to treatment with a medium concentration of SSU in this study, but can approach 1,200 kg N ha−1 (14), mostly as urea. It is at the upper end of this range that there appear to be effects on microbial community structure; higher amounts of sheep urine being deposited on pastures as a consequence of intensification may contribute to the switch from species-rich acidic grasslands to L. perenne-dominated pastures. Nevertheless, this study also demonstrates that a complex of other environmental factors may also have a significant effect on the development of grassland soil microbial community structures.
Supplementary Material
Acknowledgments
This work was supported by Environmental Protection Agency grant 2002-PHD2-16 under the Environmental Research Technological Development and Innovation (ERTDI) program.
We acknowledge valuable discussions with Lorraine Muckian and Mary Murphy for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alef, K., and P. Nannipieiri. 1995. Methods in applied soil microbiology and biochemistry. Academic Press Ltd., London, United Kingdom.
- 2.Anderson, J. P. E., and K. H. Domsch. 1978. A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol. Biochem. 10:215-221. [Google Scholar]
- 3.Bardgett, R. D., P. J. Hobbs, and A. Frostegard. 1996. Changes in soil fungal:bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol. Fertil. Soils 22:261-264. [Google Scholar]
- 4.Bardgett, R. D., D. K. Leemans, R. Cook, and P. J. Hobbs. 1997. Seasonality of the soil biota of grazed and ungrazed hill grasslands. Soil Biol. Biochem. 29:1285-1294. [Google Scholar]
- 5.Blackstock, T. H. C. A. Rimes, D. P. Stevens, R. G. Jefferson, H. J. Robertson, J. Mackintosh, and J. J. Hopkins. 1999. The extent of semi-natural grassland communities in lowland England and Wales: a review of conservation surveys 1978-96. Grass Forage Sci. 54:1-18. [Google Scholar]
- 6.Brodie, E., S. Edwards, and N. Clipson. 2002. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 44:260-270. [DOI] [PubMed] [Google Scholar]
- 7.Brodie, E., S. Edwards, and N. Clipson. 2003. Soil fungal community structure in a temperate upland grassland soil. FEMS Microbiol. Ecol. 45:105-114. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, K. R. 1993. Non-parametric multivariate analysis of changes in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 9.Clegg, C. D., R. D. L. Lovell, and P. J. Hobbs. 2003. The impact of grassland management regime on the community structure of selected bacterial groups in soils. FEMS Microbiol. Ecol. 43:263-270. [DOI] [PubMed] [Google Scholar]
- 10.Clough, T. J. 1996. Fate of urine nitrogen on mineral and peat soils in New Zealand. Plant Soil 178:141-152. [Google Scholar]
- 11.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-802. [DOI] [PubMed] [Google Scholar]
- 12.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frame, J. 1991. Herbage production and quality of a range of secondary grass species at 5 rates of fertiliser application. Grass Forage Sci. 46:139-151. [Google Scholar]
- 14.Fraser, P. M., C. K. Cameron, and R. R. Sherlock. 1994. Lysimeter study of the fate of nitrogen in animal urine returns to irrigated pasture. Eur. J. Soil Sci. 45:439-447. [Google Scholar]
- 15.Gahoonia, T. S., N. Claasen, and A. Jungk. 1992. Mobilisation of phosphate in different soils by ryegrass supplied with ammonium or nitrate. Plant Soil 140:241-248. [Google Scholar]
- 16.Grayston, S. J., C. D. Campbell, R. D. Bardgett, J. L. Mawdsley, C. D. Clegg, K. Ritz, B. S. Griffiths, J. S. Rodwell, S. J. Edwards, and W. J. Davies. 2004. Assessing shifts in microbial community structure across a range of grasslands of differing management intensity using CLPP, PLFA and community DNA techniques. Appl. Soil Ecol. 25:63-84. [Google Scholar]
- 17.Grayston, S. J., G. S. Griffith, J. L. Mawdsley, C. D. Campbell, and R. D. Bardgett. 2001. Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol. Biochem. 33:533-551. [Google Scholar]
- 18.Grayston, S. J., S. Wang, C. Campbell, and A. Edwards. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30:369-378. [Google Scholar]
- 19.Green, B. H. 1990. Agricultural intensification and the loss of habitat, species and amenity in British grasslands: a review of historical change and assessment of future prospects. Grass Forage Sci. 45:365-372. [Google Scholar]
- 20.Grime, J. P. 1998. Benefits of plant diversity to ecosystems: immediate, filter and founder effects. J. Ecol. 86:902-910. [Google Scholar]
- 21.Haynes, R. J., and P. H. Williams. 1993. Nutrient cycling and soil fertility in the grazed pasture ecosystem. Adv. Agron. 49:119-199. [Google Scholar]
- 22.Holland, P. T., and C. During. 1977. Movement of nitrate-N and transformations of urea-N under field conditions. N. Z. J. Agric. Res. 20:479-488. [Google Scholar]
- 23.Hooper, D. U., and P. M. Vitousek. 1997. The effects of plant composition and diversity on ecosystem processes. Science 277:1302-1305. [Google Scholar]
- 24.Ibekwe, A. M., A. M. Kennedy, P. S. Frohne, S. K. Papiernik, C. H. Yang, and D. E. Crowley. 2002. Microbial diversity along a transect of agronomic zones. FEMS Microbiol. Ecol. 26:151-163. [DOI] [PubMed] [Google Scholar]
- 25.Jarvis, S. C., and B. F. Pain. 1990. Ammonia volatilisation from agricultural land. Proceedings no. 298. The Fertilizer Society, London, United Kingdom.
- 26.Kennedy, N., J. Connolly, and N. Clipson. 2004. Impact of lime, nitrogen and plant species on fungal community structure in grassland microcosms. Environ. Microbiol. 7:780-788. [DOI] [PubMed] [Google Scholar]
- 27.Kennedy, N. M., D. E. Gleeson, J. Connolly, and N. J. W. Clipson. 2005. Seasonal and management influences on bacterial community structure in an upland grassland soil. FEMS Microbiol. Ecol. 53:329-337. [DOI] [PubMed] [Google Scholar]
- 28.Kruskal, J. B. 1964. Multidimensional scaling by optimizing goodness of fit to a non-metric hypothesis. Psychometrika 29:1-27. [Google Scholar]
- 29.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marriot, C. A., G. Hudson, D. Hamilton, R. Neilson, B. Boag, L. Handley, J. Wishart, C. M. Scrimgeour, and D. Robinson. 1997. Spatial variability of total soil C and N and their stable isotopes in an upland Scottish grassland. Plant Soil 196:151-162. [Google Scholar]
- 31.Neale, S. P., Z. Shah, and W. Adams. 1997. Changes in microbial biomass and nitrogen turnover in acidic organic soils following liming. Soil Biol. Biochem. 29:1463-1474. [Google Scholar]
- 32.Normand, P., C. Ponsonnet, X. Nesme, M. Neyra, and P. Simonet. 1996. ITS analysis of prokaryotes, p. 3.4.5, 1-12. In A. D. Akkermans, J. D. van Elsas, and E. I. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Press, Amsterdam, The Netherlands.
- 33.Nunan, N., B. Singh, E. Reid, B. Ord, A. Papert, J. Squires, J. I. Prosser, R. E. Wheatley, J. McNicol, and P. Millard. 2006. Sheep-urine-induced changes in soil microbial community structure. FEMS Microbiol. Ecol. 56:310-320. [DOI] [PubMed] [Google Scholar]
- 34.Personeni, E., and P. Loiseau. 2005. Species strategy and N fluxes in grassland soil. A question of root litter quality or rhizosphere activity? Eur. J. Agron. 22:217-229. [Google Scholar]
- 35.Ritz, K., J. W. McNicol, N. Nunan, S. Grayston, P. Millard, D. Atkinson, A. Gollotte, D. Habeshaw, B. Boag, and C. D. Clegg. 2004. Spatial structure in soil chemical and microbiological properties in an upland grassland. FEMS Microbiol. Ecol. 49:191-205. [DOI] [PubMed] [Google Scholar]
- 36.Rodwell, J. S. 1992. British plant communities: grasslands and montane communities. Cambridge University Press, Cambridge, United Kingdom.
- 37.Sessitch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2006. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, B., S. Munro, E. Reid, B. Ord, J. M. Potts, E. Paterson, and P. Millard. 2006. Investigating microbial community structure in soils by physiological, biochemical and molecular fingerprinting methods. Eur. J. Soil Sci. 57:72-82. [Google Scholar]
- 39.Sparks, D. L., P. A. Hlemke, R. H. Loeppert, P. N. Soltanpour, and M. A. Tabatabai (ed.). 1996. Methods of soil analysis: chemical methods, part 3. Soil Science Society of America, Madison, Wis.
- 40.Stoate, C., N. D. Boatman, R. J. Borralho, C. R. Carvalho, G. R. Snoo, and P. Eden. 2001. Ecological impacts of arable intensification in Europe. J. Environ. Manage. 63:337-365. [DOI] [PubMed] [Google Scholar]
- 41.Thalmann, A. 1968. Zur methodik ber Bestimmung der dehydrogenaseactivitat im Boden mittels triphenyltetrazoliumchlorid (TTC). Landwirtsch Forsch. 21:249-258. [Google Scholar]
- 42.Thomas, R. J. 1988. Transformations and fate of sheep urine-N applied to an upland U.K. pasture at different times during the growing season. Plant Soil 107:173-181. [Google Scholar]
- 43.Torsvik, V., F. L. Daae, R. A. Sandaa, and L. Vreas. 1998. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 44.West, A. W., and G. P. Sparling. 1986. Modifications to the substrate-induced respiration method to permit measurement of microbial biomass in soils of differing water contents. J. Microbiol. Methods 5:177-189. [Google Scholar]
- 45.Williams, B. L., L. A. Dawson, S. J. Grayston, and C. A. Shand. 2003. Impact of defoliation on the distribution 15</N-labelled synthetic sheep urine between shoots and roots of Agrostis capillaris and soil N pools. Plant Soil 251:269-278. [Google Scholar]
- 46.Williams, B. L., C. A. Shand, S. Sellers, and M. E. Young. 1999. Impact of synthetic sheep urine on N and P in two pastures in the Scottish uplands. Plant Soil 214:93-103. [Google Scholar]
- 47.Williams, B. L., S. J. Grayston, and E. J. Reid. 2000. Influence of synthetic sheep urine on the microbial biomass, activity and community structure in two pastures in the Scottish uplands. Plant Soil 225:175-185. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.