Abstract
Previous studies failed to detect c-type cytochromes in Pelobacter species despite the fact that other close relatives in the Geobacteraceae, such as Geobacter and Desulfuromonas species, have abundant c-type cytochromes. Analysis of the recently completed genome sequence of Pelobacter carbinolicus revealed 14 open reading frames that could encode c-type cytochromes. Transcripts for all but one of these open reading frames were detected in acetoin-fermenting and/or Fe(III)-reducing cells. Three putative c-type cytochrome genes were expressed specifically during Fe(III) reduction, suggesting that the encoded proteins may participate in electron transfer to Fe(III). One of these proteins was a periplasmic triheme cytochrome with a high level of similarity to PpcA, which has a role in Fe(III) reduction in Geobacter sulfurreducens. Genes for heme biosynthesis and system II cytochrome c biogenesis were identified in the genome and shown to be expressed. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels of protein extracted from acetoin-fermenting P. carbinolicus cells contained three heme-staining bands which were confirmed by mass spectrometry to be among the 14 predicted c-type cytochromes. The number of cytochrome genes, the predicted amount of heme c per protein, and the ratio of heme-stained protein to total protein were much smaller in P. carbinolicus than in G. sulfurreducens. Furthermore, many of the c-type cytochromes that genetic studies have indicated are required for optimal Fe(III) reduction in G. sulfurreducens were not present in the P. carbinolicus genome. These results suggest that further evaluation of the functions of c-type cytochromes in the Geobacteraceae is warranted.
Pelobacter species seem to be an anomaly within the family Geobacteraceae. They are phylogenetically intertwined with Geobacter and Desulfuromonas species and have the capacity to use Fe(III) as an electron acceptor (29, 34), yet they were previously found to lack c-type cytochromes (34, 43-47), which are abundant in Geobacter and Desulfuromonas species and are thought to be important in electron transfer to Fe(III) in these organisms (8, 25, 28, 30, 35, 41). This has led to questions about the evolution of the different genera within the Geobacteraceae and about the true role of c-type cytochromes in Fe(III) reduction in this family. In fact, the apparent lack of c-type cytochromes in Pelobacter but conservation of the structural gene for electrically conductive pilin is one line of evidence suggesting that pili serve as the electrical conduit between the outer surface of Geobacteraceae cells and Fe(III) oxides (40).
Pelobacter species are common in anaerobic subsurface environments (16, 22, 37, 50, 52). Pelobacter carbinolicus, which grows by fermentation of butanediol, acetoin, and ethylene glycol to ethanol and acetate, was isolated from marine mud (43). P. carbinolicus can also grow by oxidizing ethanol and other alcohols (i) in coculture with H2-oxidizing methanogens or acetogens (43) or (ii) with Fe(III) or So as an electron acceptor (34). However, these organic electron donors are only incompletely oxidized to acetate, in contrast to the ability of Geobacter and Desulfuromonas species to completely oxidize acetate and other organic electron donors to carbon dioxide (32). Yet P. carbinolicus appears to be capable of conserving energy from electron transfer to Fe(III) because it can grow via Fe(III) reduction with H2 as the electron donor (34), and the cell yields per unit of Fe(III) reduced with both organic electron donors and H2 are equivalent to those for Geobacter species (E. S. Shelobolina, unpublished data).
Analysis of the recently completed genome sequence of P. carbinolicus DSM2380 (www.jgi.doe.gov) led to the surprising finding that this organism contains genes predicted to encode c-type cytochromes, as well as genes for heme biosynthesis and cytochrome c biogenesis. Here we report that most of these c-type cytochrome genes are expressed under one or more growth conditions and that low levels of c-type cytochromes can be detected biochemically.
MATERIALS AND METHODS
Bioinformatics.
The predicted protein-encoding sequences in the P. carbinolicus genome sequence (www.jgi.doe.gov) were searched for CXXCH heme c binding motifs using the FindPatterns algorithm of the Genetics Computer Group Wisconsin Package, version 10.3 (Accelrys Inc., San Diego, CA). The subcellular location of the CXXCH-containing putative proteins was predicted using several programs, including PSORTb (12), SubLoc (19), TMPred (15), and SignalP (5). Conserved LXXC lipoprotein binding motifs in signal sequences were used to predict outer membrane cytochromes (14). Proteins that were predicted to be cytoplasmic membrane associated, periplasmic, or outer membrane associated were further analyzed by BLASTP (2) to predict the function and location based on similarity to other proteins, with a cutoff E value of <10−5. The predictions were based solely on sequence analysis and were not confirmed experimentally. Proteins predicted to catalyze heme biosynthesis and cytochrome c biogenesis were identified on the basis of similarity to such proteins in other bacteria (4, 23, 26).
Media and culture conditions.
P. carbinolicus DSM2380 was cultured at 30°C under strictly anaerobic conditions in media containing (per liter) 9.0 g of NaCl, 2.7 g of MgCl2 · 6H2O, 2.5 g of NaHCO3, 0.25 g of NH4Cl, 0.6 g of NaH2PO4 · H2O, 0.1g of KCl, and 0.14 g of CaCl2 · 2H2O. Vitamins and minerals (10 ml liter−1 each) were added from stock solutions (31). Media were dispensed into anaerobic pressure tubes or bottles, and the tubes or bottles were gassed with 80% N2-20% CO2, sealed with butyl rubber stoppers, and autoclaved. Media were reduced with sterile Na2S at a final concentration of 0.02 mM. Electron donors were added from sterile, anaerobic stock solutions at a final concentration of 10 mM (acetoin [3-hydroxy-2-butanone]) or 2 mM (ethanol). Fe(III) was provided in ethanol-containing cultures in the form of Fe(III) nitrilotriacetic acid (NTA) at a final concentration of 5 mM (41). Due to the salinity of the medium, much of the Fe(III) was insoluble. Cell growth on acetoin was monitored by measuring the optical density at 600 nm with a Genesys 2 spectrophotometer (Spectronic Instruments, Rochester, NY). Fe(II) production in Fe(III) NTA cultures was monitored with ferrozine (33).
Nucleic acid extraction.
Cultures used for DNA or RNA extraction were grown to mid-log phase, transferred to 50-ml conical tubes, and centrifuged at 3,150 × g for 20 min at 4°C. Cell pellets used for RNA extraction were frozen in liquid nitrogen and stored at −80°C. Chromosomal DNA was extracted from acetoin-grown P. carbinolicus cells using a FastDNA Spin kit for soil (QBiogene, Irvine, CA) or a MasterPure DNA purification kit (EPICENTRE Biotechnologies, Madison, WI). Total RNA was isolated using protocols described previously (17, 18). For acetoin-grown cells, cell pellets from 50 ml of culture were resuspended in 4 ml of Tris-EDTA-sucrose buffer (18), and RNA was extracted by the protocol used for RNA extraction from the surfaces of current-harvesting electrodes (18). For Fe(III) NTA-ethanol-grown cells, cell pellets from 50 ml of culture were resuspended in 8 ml TPE buffer (17), and RNA was extracted by using the protocol for RNA extraction from sediments (17), except that yeast tRNA was omitted. To confirm that RNA was not contaminated with DNA, PCRs were performed using RNA as the template.
PCR.
Specific primers were designed for the genes for the 14 putative c-type cytochromes, as well as for cytochrome c biogenesis genes and selected heme biosynthesis genes (Table 1). Each 50-μl PCR mixture contained 10 μl of 5× Q solution (QIAGEN), 1.5 mM MgCl2, 5 μl of 10× PCR buffer (QIAGEN), each deoxynucleoside triphosphate (Sigma) at a concentration of 0.02 mM, 50 pmol of each primer, 0.2 mg/ml bovine serum albumin (New England BioLabs), 1 U of Taq DNA polymerase (QIAGEN), and 50 ng of chromosomal DNA. PCR amplification was carried out with a PTC-200 thermal cycler (MJ Research, Waltham, MA) as follows: denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 45 s, 55 or 60°C for 1 min, and 72°C for 1 min and then a final extension step at 72°C for 10 min. The annealing temperature used for each primer set is shown in Table 1. PCR products were purified from agarose gels with a QIAquick gel extraction kit (QIAGEN) and were cloned with a TOPO TA cloning kit, version R (Invitrogen). Plasmids were extracted from 8 to 12 colonies per PCR and sequenced with the M13F primer to confirm that the correct gene was amplified.
TABLE 1.
Primers used for RT-PCR analysis of P. carbinolicus cytochromes and cytochrome cbiosynthesis genes
| Gene | Description | Forward primer (5′-3′) | Reverse primer (5′-3′) | Annealing temp for PCR (oC) |
|---|---|---|---|---|
| Cytochromes | ||||
| PCAR2944 | Glutamate synthase, large subunit | CAGCATGCCATCAAGTTCGT | ATAATGCGTACATCGGCCG | 60 |
| PCAR2570 | Cytochrome c family protein | TTCGCTATCTCCCTCGTTCAA | TGGCCAAAGTACGGACAATG | 60 |
| PCAR2549 | Hypothetical protein | CGGCTTGCTGTTTGGTACTGT | TGAACGGCATCGAATGGTTAC | 60 |
| PCAR2529 | caa3-type cytochrome coxidase, subunit II | GAGGCGGTGGACAAGGTTTT | GGAAGGCCGGGATATAAAGG | 60 |
| PCAR0558 | Cytochrome c family protein | CTTGACGGTGATGGCATGC | CGCAAGGATATGTTCAGCCAC | 60 |
| PCAR0192 | Molybdopterin oxidoreductase/precorrin-4 methylase | CTGGCGAAAATCCTTGAACAA | AGGTAGGTGGCAAAGCGCT | 60 |
| PCAR2867 | Cytochrome c nitrite reductase, nrfH | ACTTCAGTACGGACCCGACG | CCCGTTGTCATGAACTCTTCC | 60 |
| PCAR2866 | Cytochrome c nitrite reductase nrfA | TCGAGAAGACCTGGGATGAGA | ACTTCAGTGCGGCAATTTCC | 60 |
| PCAR2550 | Cytochrome c | CACGGTTCCCAAAAATTCCA | CCATCCCATTTTCAACGAGC | 60 |
| PCAR1628 | Cytochrome c7, ppcA | CTGTTCCGACGCCATATCAA | GCAACCACCGCAGTCTGTC | 60 |
| PCAR0152 | Hypothetical protein | GGTACCGGCATCGCTTTTC | CATCAAACAACCGGGCATC | 60 |
| PCAR2984 | Cytochrome c family protein | ATGAAAAAATGCCTTTGGATGCT | GCAACGGAAGCAGGGTTCT | 60 |
| PCAR2745 | Hypothetical protein | TGGTGGCGGATTTTCTTCA | TGCAGAACATCTTCCCGGATA | 60 |
| PCAR2069 | Protein-disulfide isomerase | AACAGGCCGACAAGGTTTTG | GCGGTTACAGACGATGCTCTT | 60 |
| Heme biosynthesis | ||||
| PCAR3065 | ccsA homolog | GCGCTGGTGCTGATGATTTT | TCCCGAAAGCAGCAGATTG | 55 |
| PCAR3064 | Glutamyl tRNA synthetase, hemA | GTCCTATGCGGCGGTGG | AAACGCACGATTTCCTGCTC | 55 |
| PCAR0266 | Glutamate 1-semialdehyde aminotransferase, hemL | CAGCCCTGTTCGTGCATTTA | ACCCTGCCCAGATCGTTGT | 55 |
| PCAR0770 | Ferrochetalase, hemH | GGCCGTGATTATGCGCTACT | ACGCCCTCATCCACCAAG | 55 |
| Cytochrome c biogenesis | ||||
| PCAR2229 | Heme transport and ligation, ccsB | GCCTGCGTTCCCTGAAACT | CGCCGAGCAATATGATCAAA | 55 |
| PCAR2228 | Heme transport and ligation, ccsA | GATTGCGTCGCGAAGGG | AATTGACATCATCCAGCGTGG | 55 |
| PCAR1954 | Thioredoxin, resA | TTTCTGGTGCTGTCTGTCGG | GCGCAATGCTCCGGC | 55 |
| PCAR1953 | Thioredoxin, ccdA | GCCCTGTGTATTGCCGCT | GAGAGCAAAAACGGCAATCC | 55 |
RT-PCR.
Reverse transcription (RT) was performed with an Enhanced Avian First Strand synthesis kit (Sigma) as described previously (17). Two negative controls lacking reverse transcriptase or RNA were included for each gene. PCRs were performed as described above, using 5 μl of the RT reaction mixture as the template for PCR. As described above for PCR products, RT-PCR products were gel purified, TOPO cloned, and sequenced.
Proteomics.
Late-log-phase acetoin-grown P. carbinolicus cells or fumarate-acetate-grown Geobacter sulfurreducens cells were harvested by centrifugation at 3,150 × g for 20 min at 4°C, and the cell pellets were stored at −20°C. Cells were washed in 50 mM Tris-HCl buffer (pH 8.0) containing 1 mM MgCl2 and Complete protease inhibitor cocktail (Roche). Washed cells were suspended in the same buffer at a concentration of 0.2 g (wet weight) of cells per ml of buffer. The cells were lysed by sonication at 4°C. The cell debris was removed by centrifugation at 9,000 × g for 15 min, and the supernatant contained whole-cell protein. Ten micrograms of protein from each organism was separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and c-type cytochromes were heme stained as described previously (11, 51).
Heme-stained protein bands were excised from the gel and washed with 500 μl of MilliQ water for 30 min at 20°C to remove the chemical residue. The gel pieces were dehydrated in 200 μl of 50 mM ammonium bicarbonate in 50% acetonitrile for 1 h, followed by acetonitrile for 30 min. Each gel piece was digested in 20 to 40 μl digestion buffer (20 mM ammonium bicarbonate containing 75 ng trypsin) at 37°C overnight. The supernatant was recovered, and the remaining peptides were extracted from the gel piece by washing it with 80% acetonitrile-1% formic acid in water. The extracts were pooled, and the volume was reduced to 5 to 10 μl with a Speedvac (Vacufuge, Germany). The eluted tryptic peptides were desalted and concentrated with a commercial ZipTip C18 pipette tip (Millipore). Peptides were detected by matrix-assisted laser desorption ionization-time of flight mass spectrometry as previously described (20). Peptide mass fingerprints were analyzed by using the MS-FIT Protein Prospector program (UCSF Mass Spectrometry Facility).
RESULTS AND DISCUSSION
c-Type cytochrome genes.
The P. carbinolicus genome sequence contains 58 CXXCH motifs in 42 predicted proteins. Individual predicted proteins contain between one and five CXXCH motifs. Twenty-eight of the putative c-type cytochromes are predicted to be cytoplasmic and therefore not expected to bind heme c. Of the remaining 14 protein sequences, 4 are predicted to be associated with the cytoplasmic membrane, 6 are predicted to be periplasmic, and the other 4 may be outer membrane associated (Table 2).
TABLE 2.
Predicted c-type cytochromes in P. carbinolicus and mRNA expression determined by RT-PCR
| Gene | Annotation | No. of CXXCH motifs | Associated gene(s) | Function | mRNA expression
|
|
|---|---|---|---|---|---|---|
| Acetoin | Fe(III) NTA-ethanol | |||||
| Cytoplasmic membrane associated | ||||||
| PCAR2944 | Glutamate synthase, large subunit | 1 | None | Unknown | − | + |
| PCAR2570 | Cytochrome c family protein | 1 | PCAR2571 | In operon with another membrane-bound protein | − | − |
| PCAR2549 | Hypothetical protein | 1 | PCAR2550 to PCAR2553 | In operon with quinol:cytochrome coxidoreductase | + | + |
| PCAR2529 | caa3-type cytochrome c oxidase, subunit II | 1 | PCAR2526 to PCAR2528 | Terminal oxidase | + | + |
| Soluble periplasmic | ||||||
| PCAR0558 | Cytochrome c family protein | 1 | None | Unknown | + | + |
| PCAR0192 | Molybdopterin oxidoreductase/precorrin-4 methylase | 1 | PCAR0190 to PCAR0195 | Unknown | + | + |
| PCAR2867 | Cytochrome c nitrite reductase, nrfH | 4 | PCAR2866 | Nitrite reduction, electron transfer | + | + |
| PCAR2866 | Cytochrome c nitrite reductase, nrfA | 5 | PCAR2867 | Nitrite reduction, catalytic subunit | + | + |
| PCAR2550 | Cytochrome c | 5 | PCAR2549 to PCAR2553 | Electron transfer from quinol:cytochrome c oxidoreductase | − | + |
| PCAR1628 | Cytochrome c7, ppcA | 3 | None | Periplasmic electron carrier | − | + |
| Outer membrane associated | ||||||
| PCAR0152 | Hypothetical protein | 1 | PCAR0153 to PCAR0159 | Associated with Fe3+-siderophore ABC transporter | + | − |
| PCAR2984 | Cytochrome c family protein | 2 | PCAR2983 | Associated with pyruvate kinase | + | + |
| PCAR2745 | Hypothetical protein | 3 | None | Unknown | + | + |
| PCAR2069 | Protein disulfide isomerase | 1 | None | Unknown | + | + |
One gene (PCAR1628) is predicted to encode a cytochrome c7 that belongs to a family of well-conserved cytochromes in the Geobacteraceae, which includes cytochrome c7 in Desulfuromonas acetoxidans (3, 6) and Geobacter metallireducens (1) and PpcA in G. sulfurreducens (28). In G. sulfurreducens this cytochrome appears to function as a periplasmic intermediary electron carrier between cytoplasmic electron donors and outer membrane-associated Fe(III) reductase (28).
PCAR2984 encodes a putative outer membrane diheme cytochrome c and is conserved in Geobacteraceae genomes upstream of a gene for pyruvate kinase. A mutant lacking the homolog in G. sulfurreducens (GSU3332) was incapable of reducing poorly crystalline Fe(III) oxides and was deficient in reduction of U(VI), suggesting that this cytochrome may play a role in electron transfer to extracellular electron acceptors (E. S. Shelobolina, unpublished data).
PCAR2867 and PCAR2866 encode proteins that are homologous to the two subunits of cytochrome c nitrite reductase, which catalyze nitrite reduction to ammonia in many bacteria (48) and are conserved in the Geobacteraceae. In P. carbinolicus PCAR2866 the unique CXXCK heme binding motif (48) is replaced by a CXXCH motif. The role of cytochrome c nitrite reductase in P. carbinolicus, which is not known to use nitrite as an electron acceptor, could be similar to that in Desulfovibrio vulgaris, in which nitrite reduction is not coupled to growth and cytochrome c nitrite reductase is used for nitrite detoxification (13, 39).
The periplasmic pentaheme cytochrome c encoded by PCAR2550 is part of a conserved operon (PCAR2550 to PCAR2553) that is predicted to encode a cytoplasmic membrane-bound quinol:cytochrome c oxidoreductase. A homologous complex was purified from Chloroflexus aurantiacus, and homologous operons are found in individual members of seven bacterial phyla, suggesting that the complex has been laterally transferred (53). A similar but distinct complex has been found in the genome sequences of G. metallireducens, D. vulgaris, and Desulfovibrio desulfuricans but not in the G. sulfurreducens sequence (53) or in the draft sequence of Desulfuromonas acetoxidans (www.jgi.doe.gov). The gene encoding cytoplasmic membrane-bound monoheme cytochrome c, PCAR2549, is immediately upstream of PCAR2550, but this cytochrome has no significant similarity to other proteins.
PCAR2529 encodes subunit II of a caa3-type cytochrome c oxidase (encoded by PCAR2526 to PCAR2529), which is homologous to the oxidases in other members of the Geobacteraceae, as well as the characterized oxidase in Rhodothermus marinus (42). G. sulfurreducens has a cytochrome c oxidase (36) and has been shown to grow with low levels of oxygen as a terminal electron acceptor (27).
PCAR0152 encodes a putative outer membrane cytochrome with no significant similarity to other proteins. This gene is located between the genes for an ABC-type transporter with homology to the Escherichia coli Fep ferric enterobactin transporter (7) and therefore may play a role in the uptake of chelated Fe(III).
The functions of the remaining six putative cytochromes cannot be predicted by sequence homology, either because they have no significant BLASTP hits or because they are similar to proteins that are not known to bind heme c. These cytochromes include two that are predicted to be cytoplasmic membrane bound, the cytochrome encoded by PCAR2570 and the glutamate synthase homolog encoded by PCAR2944; two periplasmic cytochromes, one encoded by PCAR0558 and one encoded by PCAR0192, the latter of which is homologous to molybdopterin oxidoreductase in the N terminus and precorrin-4 methylase in the C terminus; and two that are predicted to be outer membrane bound, the triheme cytochrome encoded by PCAR2745 and the protein disulfide isomerase encoded by PCAR2069.
Heme biosynthesis and cytochrome c biogenesis genes.
In order for the putative cytochromes to be functional, heme c must be covalently bound to the proteins in the periplasm. Formation of c-type cytochromes requires heme biosynthesis, transport of heme and apoprotein to the periplasm, and covalent attachment of heme to CXXCH motifs. P. carbinolicus possesses all of the genes required for heme biosynthesis in four regions of the genome (Table 3). Consistent with its anaerobic physiology, P. carbinolicus contains a homolog of the oxygen-independent protoporphyrinogen oxidase HemG (encoded by PCAR0772), which catalyzes the penultimate step in heme biosynthesis, but not HemY, which catalyzes the same reaction in an oxygen-dependent manner (10). Likewise, it contains a homolog of the oxygen-independent coproporphyrinogen III oxidase HemN (encoded by PCAR0110), but it lacks the oxygen-dependent form, HemF (10). hemACD are located in an operon with genes encoding phosphoheptose isomerase, siroheme synthase, and a cytochrome biogenesis protein homolog (PCAR3062 to PCAR3067). hemB is located downstream of this operon in a dicistronic operon containing a gene encoding a conserved hypothetical protein (PCAR3060 and PCAR3061).
TABLE 3.
Predicted P. carbinolicus heme biosynthesis and cytochrome c biogenesis genes and mRNA expression determined by RT-PCR
| Gene | Designation | Description | mRNA expression (acetoin) |
|---|---|---|---|
| Heme biosynthesis | |||
| PCAR3065 | CcsA homolog | + | |
| PCAR3064 | hemA | Glutamyl tRNA synthase | + |
| PCAR3063 | hemC | Porphobilinogen deaminase | NDa |
| PCAR3062 | hemD | Uroporphyrinogen-III synthase/methyltransferase | ND |
| PCAR3061 | hemB | ALA dehydratase | ND |
| PCAR0266 | hemL | Glutamate 1-semialdehyde aminotransferase | + |
| PCAR0769 | hemE | Uroporphyrinogen-III decarboxylase | ND |
| PCAR0770 | hemH | Ferrochetalase | + |
| PCAR0110 | hemN | Coporphyrinogen-III oxidase | ND |
| PCAR0772 | hemG | Protoporphyrinogen-IX oxidase | ND |
| Cytochrome c biogenesis | |||
| PCAR2229 | ccsB | Heme transport and ligation | + |
| PCAR2228 | ccsA | Heme transport and ligation | + |
| PCAR1954 | resA | Thioredoxin for reduction of CXXCH cysteines | + |
| PCAR1953 | ccdA | Thioredoxin for reduction of ResA | + |
ND, not determined.
The steps after heme biosynthesis are collectively referred to as the cytochrome c biogenesis pathway. There are three different systems for cytochrome c biogenesis, called systems I, II, and III (24, 38). System II is found in gram-positive bacteria, β- and ɛ-Proteobacteria, and chloroplasts (23, 24). P. carbinolicus, along with other members of the Geobacteraceae, has genes for the system II cytochrome c biogenesis pathway (49). The four genes fall into two operons (Table 3) encoding four integral membrane proteins. PCAR2228 and PCAR2229 encode CcsA and CcsB homologs, which transport heme to the periplasm and may catalyze the covalent linkage of heme to apocytochrome c (24). ResA (encoded by PCAR1954) is a thioredoxin that reduces the cysteines of the apocytochrome c so that heme can be attached, and CcdA (encoded by PCAR1953) rereduces ResA (24).
mRNA expression.
Most of the putative c-type cytochrome genes were expressed during fermentation and/or during Fe(III) reduction; the only exception was PCAR2570 (Table 2). PCAR0152, encoding the cytochrome associated with genes for the Fe3+-siderophore ABC transporter, was expressed during fermentation but not during Fe(III) reduction, supporting the hypothesis that this cytochrome has a role in iron uptake during iron limitation. Three cytochrome genes were expressed during Fe(III) reduction but not during fermentation, including PCAR1628 encoding the periplasmic cytochrome c7 and PCAR2550 encoding the periplasmic pentaheme cytochrome in the quinol:cytochrome c oxidoreductase. Together, these two cytochromes could transport electrons from the quinone pool to the outer membrane. The gene encoding a cytoplasmic membrane-bound cytochrome related to glutamate synthase was also expressed only during Fe(III) reduction, but the role of this cytochrome is not clear.
Expression of selected genes in the heme biosynthesis pathway was determined by RT-PCR. These genes included the genes encoding the enzymes for first two steps in the pathway (hemA and hemL) and the final step (hemH), all of which were expressed during fermentation (Table 3). A ccsA homolog in the same operon as hemA was also expressed. The four cytochrome c biogenesis genes were expressed as well (Table 3). Therefore, all of the genes necessary for heme biosynthesis and cytochrome c biogenesis are present and expressed in P. carbinolicus, which allows functional c-type cytochromes to be produced.
Protein expression.
Previous attempts to detect cytochromes in P. carbinolicus by difference spectroscopy of crude cell extracts and membrane fractions of fermentatively grown cells (43) or of intact washed cells (34) were unsuccessful. Likewise, no cytochromes were detected by difference spectroscopy of fermentatively grown cells, but when whole-cell protein from these cells was electrophoresed on SDS-PAGE gels and heme stained, three bands were detected (Fig. 1). The same amount of protein from G. sulfurreducens produced many more bands and contained much more heme-containing protein (Fig. 1). Proteins in the three heme-stained P. carbinolicus bands were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry, and the results were in agreement with the RT-PCR results. The following three identified cytochromes were all predicted to be soluble: cytochrome c nitrite reductase encoded by PCAR2866, molybdopterin oxidoreductase/precorrin-4 methylase encoded by PCAR0192, and the cytochrome c family protein encoded by PCAR0558.
FIG. 1.
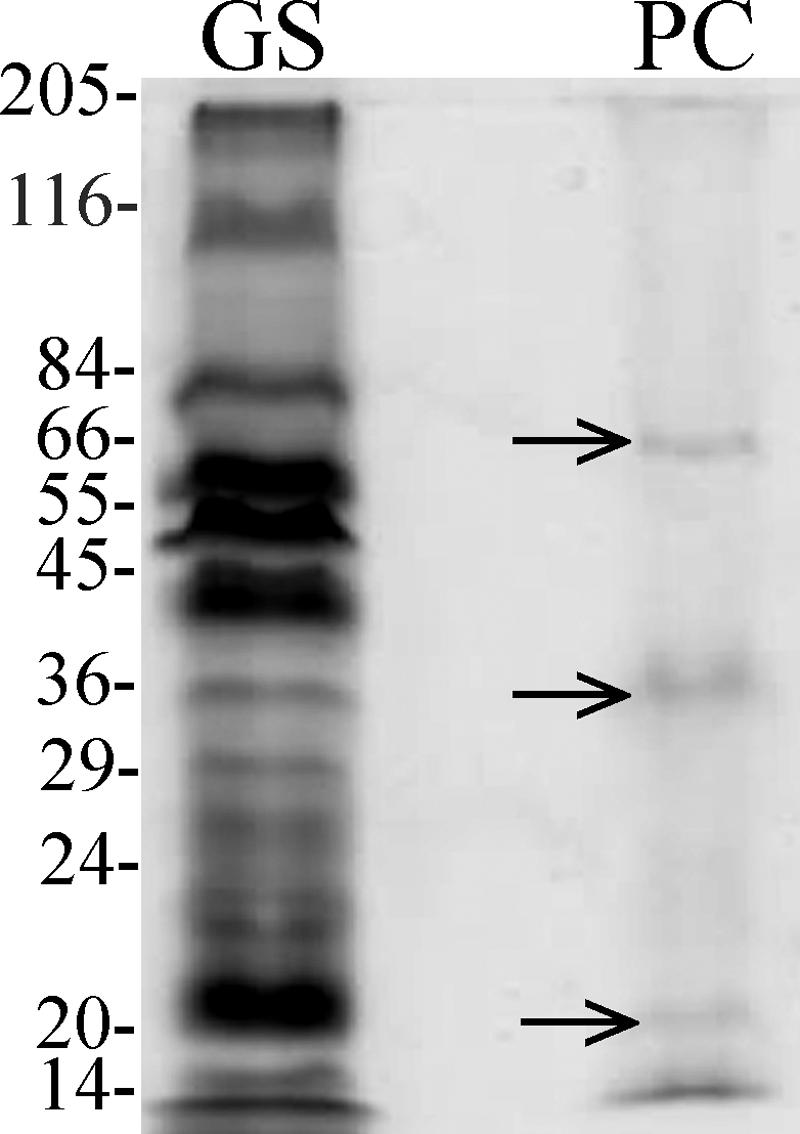
Heme-stained SDS-PAGE of whole-cell protein extracted from P. carbinolicus (lane PC) and G. sulfurreducens (lane GS). Each lane contained 10 μg of protein. The bands indicated by arrows were excised for identification by mass spectrometry.
Implications for Pelobacter physiology.
Our results demonstrate for the first time that P. carbinolicus contains c-type cytochromes. As noted above, the functions of some of these cytochromes can be inferred from homology with cytochrome genes encoding known functions in other organisms. However, definitive elucidation of the functions of the genes in P. carbinolicus with genetic approaches has not been possible yet because techniques for generating specific mutations via homologous recombination that have been successful in G. sulfurreducens (8, 9, 20, 25, 28, 35) have not worked well in P. carbinolicus.
The detection of transcripts for three c-type cytochrome genes during growth on Fe(III) but not under fermentative conditions suggests that the cytochromes may be specifically involved in Fe(III) reduction. It is notable that one of these differentially expressed cytochrome genes encodes a triheme, periplasmic cytochrome that is highly conserved in the Geobacteraceae and is essential for optimal Fe(III) reduction in G. sulfurreducens (28). However, the triheme cytochrome is much less abundant in P. carbinolicus than in G. sulfurreducens, and G. sulfurreducens contains five homologs of this protein (36) compared to just one homolog in P. carbinolicus. Furthermore, P. carbinolicus lacks genes for many c-type cytochromes that have been found to be required for optimal Fe(III) reduction in G. sulfurreducens. These cytochromes include the inner membrane cytochrome MacA (8) and the outer membrane cytochromes OmcB (25), OmcS (35), and OmcE (35) that are thought to be involved in electron transfer to Fe(III). Also missing are OmcF, OmcG, and OmcH, which are outer membrane cytochromes that may play a regulatory role during Fe(III) reduction (20, 21). Not only is the overall number of c-type cytochrome genes in P. carbinolicus much lower than that in G. sulfurreducens, 14 versus 111 (36), but also the number of hemes in P. carbinolicus cytochromes, 5 or less, is generally lower than the number found in many G. sulfurreducens cytochromes, which can have as many as 27 hemes.
The differences between the c-type cytochrome contents of P. carbinolicus and G. sulfurreducens could conceivably be linked to factors related to metabolism of acetate, which G. sulfurreducens can use as a sole electron donor for Fe(III) reduction but P. carbinolicus cannot use (32). Alternatively, they could be related to differences in the environmental conditions in the preferred habitats of the organisms. However, further studies to definitively determine the functions of the c-type cytochromes in both organisms are necessary before substantive conclusions can be made.
Acknowledgments
This research was supported by grant DE-FC02-02ER63446 from the Genomics: GTL Program of the Office of Science (BER), U.S. Department of Energy.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Afkar, E., and Y. Fukumori. 1999. Purification and characterization of triheme cytochrome c7 from the metal-reducing bacterium, Geobacter metallireducens. FEMS Microbiol. Lett. 175:205-210. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Aubert, C., E. Lojou, P. Bianco, M. Rousset, M.-C. Durand, M. Bruschi, and A. Dolla. 1998. The Desulfuromonas acetoxidans triheme cytochrome c7 produced in Desulfovibrio desulfuricans retains its metal reductase activity. Appl. Environ. Microbiol. 64:1308-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckett, C. S., J. A. Loughman, K. A. Karberg, G. M. Donato, W. E. Goldman, and R. G. Kranz. 2000. Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol. Microbiol. 38:465-481. [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Bruschi, M., M. Woudstra, B. Guigliarelli, M. Asso, E. Lojou, Y. Petillot, and C. Abergel. 1997. Biochemical and spectroscopic characterization of two new cytochromes isolated from Desulfuromonas acetoxidans. Biochemistry 36:10601-10608. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 8.Butler, J. E., F. Kaufmann, M. V. Coppi, C. Nunez, and D. R. Lovley. 2004. MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J. Bacteriol. 186:4042-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dailey, H. A. 2002. Terminal steps of haem biosynthesis. Biochem. Soc. Trans. 30:590-595. [DOI] [PubMed] [Google Scholar]
- 11.Francis, R. T., Jr., and R. R. Becker. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136:509-514. [DOI] [PubMed] [Google Scholar]
- 12.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. L. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 13.Haveman, S. A., E. A. Greene, C. P. Stilwell, J. K. Voordouw, and G. Voordouw. 2004. Physiological and gene expression analysis of inhibition of Desulfovibrio vulgaris Hildenborough by nitrite. J. Bacteriol. 186:7944-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22:451-471. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 16.Holmes, D. E., D. R. Bond, R. A. O'Neil, C. E. Reimers, L. R. Tender, and D. R. Lovley. 2004. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb. Ecol. 48:178-190. [DOI] [PubMed] [Google Scholar]
- 17.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 70:7251-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes, D. E., K. P. Nevin, R. A. O'Neil, J. E. Ward, L. A. Adams, T. L. Woodard, H. A. Vrionis, and D. R. Lovley. 2005. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl. Environ. Microbiol. 71:6870-6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua, S., and Z. Sun. 2001. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721-728. [DOI] [PubMed] [Google Scholar]
- 20.Kim, B. C., C. Leang, Y. H. Ding, R. H. Glaven, M. V. Coppi, and D. R. Lovley. 2005. OmcF, a putative c-type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J. Bacteriol. 187:4505-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, B. C., X. Qian, C. Leang, M. V. Coppi, and D. R. Lovley. 2006. Two putative c-type multiheme cytochromes required for the expression of OmcB, an outer membrane protein essential for optimal Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 188:3138-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovacik, W. P., Jr., K. Takai, M. R. Mormile, J. P. McKinley, F. J. Brockman, J. K. Fredrickson, and W. E. Holben. 2006. Molecular analysis of deep subsurface Cretaceous rock indicates abundant Fe(III)- and So-reducing bacteria in a sulfate-rich environment. Environ. Microbiol. 8:141-155. [DOI] [PubMed] [Google Scholar]
- 23.Kranz, R. G., C. S. Beckett, and B. S. Goldman. 2002. Genomic analyses of bacterial respiratory and cytochrome c assembly systems: Bordetella as a model for the system II cytochrome c biogenesis pathway. Res. Microbiol. 153:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Kranz, R. G., R. Lill, B. S. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29:383-396. [DOI] [PubMed] [Google Scholar]
- 25.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Brun, N. E., J. Bengtsson, and L. Hederstedt. 2000. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 36:638-650. [DOI] [PubMed] [Google Scholar]
- 27.Lin, W. C., M. V. Coppi, and D. R. Lovley. 2004. Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl. Environ. Microbiol. 70:2525-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, J. R., C. Leang, A. L. Hodges Myerson, M. V. Coppi, S. Cuifo, B. Methe, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. Phillips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 31.Lovley, D. R., R. C. Greening, and J. G. Ferry. 1984. Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl. Environ. Microbiol. 48:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 33.Lovley, D. R., and E. J. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lovley, D. R., E. J. Phillips, D. J. Lonergan, and P. K. Widman. 1995. Fe(III) and S0 reduction by Pelobacter carbinolicus. Appl. Environ. Microbiol. 61:2132-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta, T., M. V. Coppi, S. E. Childers, and D. R. Lovley. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:8634-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Methe, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 37.Mussmann, M., K. Ishii, R. Rabus, and R. Amann. 2005. Diversity and vertical distribution of cultured and uncultured Deltaproteobacteria in an intertidal mud flat of the Wadden Sea. Environ. Microbiol. 7:405-418. [DOI] [PubMed] [Google Scholar]
- 38.Page, M. D., Y. Sambongi, and S. J. Ferguson. 1998. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem. Sci. 23:103-108. [DOI] [PubMed] [Google Scholar]
- 39.Pereira, I. A., J. LeGall, A. V. Xavier, and M. Teixeira. 2000. Characterization of a heme c nitrite reductase from a non-ammonifying microorganism, Desulfovibrio vulgaris Hildenborough. Biochim. Biophys. Acta 1481:119-130. [DOI] [PubMed] [Google Scholar]
- 40.Reguera, G., K. D. McCarthy, T. Mehta, J. S. Nicoll, M. T. Tuominen, and D. R. Lovley. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098-1101. [DOI] [PubMed] [Google Scholar]
- 41.Roden, E. E., and D. R. Lovley. 1993. Dissimilatory Fe(III) reduction by the marine microorganism Desulfuromonas acetoxidans. Appl. Environ. Microbiol. 59:734-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santana, M., M. M. Pereira, N. P. Elias, C. M. Soares, and M. Teixeira. 2001. Gene cluster of Rhodothermus marinus high-potential iron-sulfur protein: oxygen oxidoreductase, a caa3-type oxidase belonging to the superfamily of heme-copper oxidases. J. Bacteriol. 183:687-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schink, B. 1984. Fermentation of 2,3-butanediol by Pelobacter carbinolicus sp. nov. and Pelobacter propionicus sp. nov., and evidence for propionate formation from C2 compounds. Arch. Microbiol. 137:33-41. [Google Scholar]
- 44.Schink, B. 1985. Fermentation of acetylene by an obligate anaerobe, Pelobacter acetylenicus sp. nov. Arch. Microbiol. 142:295-301. [Google Scholar]
- 45.Schink, B., and N. Pfennig. 1982. Fermentation of trihydroxybenzenes by Pelobacter acidigallici gen. nov. sp. nov., a new strictly anaerobic, non-sporeforming bacterium. Arch. Microbiol. 133:195-201. [Google Scholar]
- 46.Schink, B., and M. Stieb. 1983. Fermentative degradation of polyethylene glycol by a strictly anaerobic, gram-negative, nonsporeforming bacterium, Pelobacter venetianus sp. nov. Appl. Environ. Microbiol. 45:1905-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnell, S., A. Brune, and B. Schink. 1991. Degradation of hydroxyhydroquinone by the strictly anaerobic fermenting bacterium Pelobacter massiliensis sp. nov. Arch. Microbiol. 155:511-516. [Google Scholar]
- 48.Simon, J. 2002. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26:285-309. [DOI] [PubMed] [Google Scholar]
- 49.Stevens, J. M., O. Daltrop, J. W. A. Allen, and S. J. Ferguson. 2004. c-type cytochrome formation: chemical and biological enigmas. Acc. Chem. Res. 37:999-1007. [DOI] [PubMed] [Google Scholar]
- 50.Thamdrup, B., R. Rossello-Mora, and R. Amann. 2000. Microbial manganese and sulfate reduction in Black Sea shelf sediments. Appl. Environ. Microbiol. 66:2888-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 52.Vrionis, H. A., R. T. Anderson, I. Ortiz-Bernad, K. R. O'Neill, C. T. Resch, A. D. Peacock, R. Dayvault, D. C. White, P. E. Long, and D. R. Lovley. 2005. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl. Environ. Microbiol. 71:6308-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanyushin, M. F., M. C. del Rosario, D. C. Brune, and R. E. Blankenship. 2005. New class of bacterial membrane oxidoreductases. Biochemistry 44:10037-10045. [DOI] [PubMed] [Google Scholar]


