Abstract
Sediments overlying a brine pool methane seep in the Gulf of Mexico (Green Canyon 205) were analyzed using molecular and geochemical approaches to identify geochemical controls on microbial community composition and stratification. 16S rRNA gene and rRNA clone libraries, as well as mcrA gene clone libraries, showed that the archaeal community consists predominantly of ANME-1b methane oxidizers; no archaea of other ANME subgroups were found with general and group-specific PCR primers. The ANME-1b community was found in the sulfate-methane interface, where undersaturated methane concentrations of ca. 100 to 250 μM coexist with sulfate concentrations around 10 mM. Clone libraries of dsrAB genes and bacterial 16S rRNA genes show diversified sulfate-reducing communities within and above the sulfate-methane interface. Their phylogenetic profiles and occurrence patterns are not linked to ANME-1b populations, indicating that electron donors other than methane, perhaps petroleum-derived hydrocarbons, drive sulfate reduction. The archaeal component of anaerobic oxidation of methane is comprised of an active population of mainly ANME-1b in this hypersaline sediment.
The anaerobic oxidation of methane (AOM) is responsible for oxidizing nearly all of the 70.3 to 85.3 Tg of methane produced in anoxic marine sediments per year globally (50). In addition to its importance in controlling greenhouse gases, AOM has implications for investigations of early life on Earth, since anaerobic methane oxidizers and methanogens comprise a methane cycle suitable for oxidant-poor, early-Earth-like environments (19). The presence of isotopically light organic carbon in the Archean Eon suggests that AOM may have been an important microbial pathway on early Earth (19).
Although the organisms responsible for AOM in marine sediments are not yet isolated in pure culture, stable carbon isotopic studies and environmental genomic studies have shown that the process relies on archaea capable of reverse methanogenesis that often form close associations with sulfate-reducing bacteria (SRB) (5, 14, 18, 21). The archaeal anaerobic methane oxidizers (ANMEs) fall into three phylogenetic groups called ANME-1 (with subgroups a and b), ANME-2 (with subgroups a, b, and c), and ANME-3 (14, 20, 29, 43-45, 62). The sulfate-reducing bacterial partners usually fall within the Desulfosarcina/Desulfococcus (DSS) cluster of the Desulfobacteraceae or the Desulfobulbaceae. Most of the previous microbiological investigations of AOM have focused on heterogeneous systems with high methane flux, which contain high biomass of both ANME-1 and ANME-2. The presence of a mosaic of ANME phylotypes at these sites makes it difficult to determine the factors causing their ecological zonation.
Here, we analyzed the microbial community associated with AOM in Gulf of Mexico (GOM) sediments, where methane is completely oxidized at the sulfate-methane interface (SMI) and subsurface brines intrude into the upper tens of centimeters. The microbial census of the archaeal and bacterial communities both at the SMI and in surface sediments, was based on 16S rRNA gene, 16S rRNA, mcrA, and dsrAB clone libraries.
Although 16S rRNA genes provide only indirect physiological information through phylogenetic assumptions, genes that encode specific functional enzymes provide a direct assay of potential physiologies of an environmental community. All known orders of methanogens use the enzyme methyl coenzyme M reductase (MCR) to catalyze the last step of methanogenesis (33, 55). In this step, a methyl group bound to coenzyme M is reduced, releasing methane. MCR is composed of subunits α, β, and γ, which are encoded by the genes mcrA, mcrB, and mcrG, respectively (51). So far, no homologues of these genes appear to be present in cultured species that are not capable of methanogenesis (55). The phylogenetic groupings of mcrA mirror the phylogeny of the 16S rRNA genes for all known orders of methanogens (33, 55). Therefore, these functional genes can be linked to organismal phylogeny. Recently, mcrA has been identified in enrichments of the anaerobic methane oxidizers ANME-1 and ANME-2 (17, 18), and an MCR protein has been isolated from microbial mats where AOM occurs (30). Thus, mcrA is a useful and specific marker gene for detecting and identifying anaerobic methane oxidizers, as well as methanogens, in the environment.
A key enzyme for sulfite and sulfate reduction, dissimilatory (bi)sulfite reductase (DSR), catalyzes the six-electron reduction of sulfite to sulfide, the energy-conserving step in the dissimilatory sulfate reduction pathway (40). DSR is present in all major known groups of sulfate reducers and is encoded by the α and β subunits of the dsr gene, comprising dsrAB. In most cases, the phylogeny of dsrAB follows that of 16S rRNA genes (65). However, lateral gene transfer has occurred in the cases of Thermodesulfobacteriales, Archaeoglobales, and low-G+C gram-positive sulfate reducers (27, 68). With the exception of these lateral gene transfer events, dsrAB gene phylogeny generally represents the organismal phylogeny of all known sulfate reducers.
Due to the persistence of DNA sequences from inactive microorganisms in anoxic sediments, DNA clone libraries often do not represent the diversity of the active population (38). rRNA is more rapidly degraded and more accurately represents the diversity of active microbial populations in marine sediments (7). Therefore, we also determined sequences of 16S rRNA at the SMI depth of these sediments.
This study focused on shallow sediments near a mud volcano in Green Canyon (GC) lease block 205, Genesis Field, Gulf of Mexico. Methane in this area is 33 to 55% archaeal in origin, deriving from methanogenic degradation of organic compounds in deep petroleum deposits (53). Sediments of Green Canyon overlie a salt diapir whose movement creates fractures in sediments that act as conduits for the upward migration of fluids rich in petroleum, methane, and other degraded hydrocarbons. The seafloor expressions of this fluid migration are mud volcanoes, cold methane seeps, and gas hydrates (26, 53). Lipid and nucleic acid biomarker analyses have shown that AOM is a key contributor to archaeal biomass in and around gas hydrates in nearby sediments from GC234 and Atwater lease block 425 (31, 37, 38, 67). Our sedimentary samples from GC205 lack hydrates and methane ebullition, thereby allowing the establishment of a zone of methane and sulfate overlap driven by steady-state diagenesis.
This work identifies geochemical controls that shape the composition of the archaeal and bacterial communities in these hypersaline and relatively quiescent sediments. By linking molecular (16S rRNA gene/rRNA and functional-gene sequences) and geochemical approaches, we identified environmentally relevant information about the diversity, distributions, and activities of bacterial and archaeal communities involved in methanogenesis, AOM, and sulfate reduction.
MATERIALS AND METHODS
Site description and sample collection.
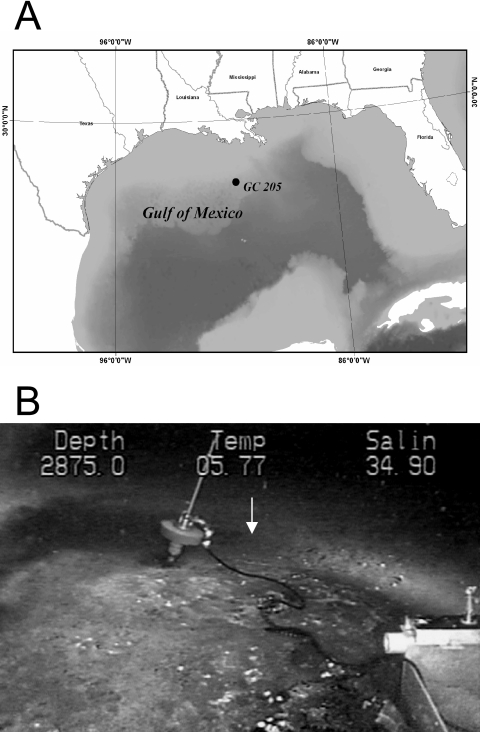
Core 5 from dive 4566 (4566-5) was retrieved from Green Canyon 205, Gulf of Mexico (27.71672778°N, −90.5334627°W), using the Johnson-Sea-Link submarine in the summer of 2003 (Fig. 1A). Visual observation of a mud volcano was made at GC205 during previous submersible reconnaissance of the site. Sediments were sampled at the site of a subsurface salt diapir in ∼876-m water depth, with a seawater temperature of ∼6°C. Although orange and white microbial mats were present near the sampling site, the core was taken from bare sediment adjacent to the mats (Fig. 1B). A push core (30 cm) was deployed from the Johnson-Sea-Link submersible to collect sediment and pore water. The core contained no gas hydrates, and the bottom contained visible carbonate crusts and petroleum. Onboard the ship, the core was subsectioned into 3-cm intervals. From each interval, a sediment plug was collected with a 3-ml-cutoff plastic syringe. Plugs were placed in 30-ml glass serum vials, sealed with rubber stoppers and aluminum caps, and frozen for future analysis of methane concentrations and stable carbon isotopes. The remaining sediment from each interval was then collected in sterile 15-ml centrifuge tubes and frozen at −20°C.
FIG. 1.
(A) Location of block GC205, the site of dive 4566 on the slope of the Gulf of Mexico; water depth, 876 m. (B) Johnson-Sea-Link image of core location (white arrow) just outside white microbial mats.
Sample preparation.
Three days after the cruise, the sediment tubes were thawed, homogenized, and centrifuged at 3,000 × g for 5 min. The resultant supernatant was transferred to 2-ml O-ring-sealed plastic microcentrifuge tubes and frozen for sulfate and chloride analysis. The remaining sediment pellet was returned to −20°C for total organic carbon (TOC) concentration and carbon-to-nitrogen (C/N) ratio analyses. After 1 month, the sediment pellets were transferred to a deep freezer (−80°C) for molecular analyses. The cores were originally frozen for geochemical analyses only; no subsamples were taken and fixed for cell-imaging capabilities.
Geochemical analyses.
Pore water sulfate and chloride concentrations were determined by ion chromatography of 1:100-diluted pore fluid samples using a 2010i Dionex Ion chromatograph (Sunnyvale, CA) as described by Martens et al. (35). The percent TOC (%TOC), stable carbon isotopic composition, and C/N ratios were measured on ∼30 mg of freeze-dried sediment pellets that were vapor acidified to remove inorganic carbon prior to combustion to CO2 and N2 on a Carlo-Erba NA 1500 (CE Elantech, Inc., Milan, Italy) elemental analyzer. The effluent gas stream was introduced to a Finnigan Mat 252 isotope ratio mass spectrometer (Thermo Finnigan, Bremen, Germany) via a modified Finnigan ConFlo interface (6). Methane concentrations and carbon stable-isotope ratios were analyzed for each depth interval using the same serum vial, as previously described. For methane concentrations, a 5-ml headspace aliquot was analyzed on a Shimadzu Mini II Gas Chromatograph (Kyoto, Japan) equipped with a flame ionization detector. Carbon stable-isotope ratios for dissolved methane were obtained using a preconcentrating system on line with a continuous-flow 5890 Hewlett-Packard gas chromatograph (Palo Alto, CA), capillary combustion, and isotope ratio mass spectrometry, as described by Rice et al. (52). The results are reported using the standard “del” notation: δ13C (‰) = (Rsample/Rstandard − 1) × 1,000, where R is the ratio of the heavy isotope to the light isotope. The precision for replicate measurements of single samples was ±3% for sulfate, chloride, and methane concentrations; 1% for TOC and methane isotopes; and 5 to 10% for %TOC and C/N.
Nucleic acid extraction, amplification, cloning, and sequencing.
Genomic DNA was extracted from three depth sections: 0 to 3 cm (SURF), 15 to 18 cm (SMI-1), and 18 to 21 cm (SMI-2). DNA was extracted from 0.5- to 1.3-g sediments using the MOBIO UltraClean Soil DNA Kit (MOBIO Labs, Inc., Solana Beach, CA), following the manufacturer's instructions, with the following modification: the sediments were subjected to four cycles of freezing at −80°C and then thawing at 65°C after addition of the inhibitor removal solution. Bacterial 16S rRNA genes and dsrAB genes were PCR amplified with the following primers: B8f-B1492r as general bacterial primers (62); DSR1f-DSR4r (65) with a subsequent nested reamplification with 1f1r (10) as dsrAB-specific primers. For amplification of mcrA from SMI-1, the primer set ME1-ME2 (16) was used; for SMI-2, mcrA was amplified from an MCRf-ME2 nested reamplification within ME1-MCRr (16, 33). For amplification of archaeal 16S rRNA genes from the SURF and SMI-2 sections, the first amplification with primers A8f and A1492r (62) was followed by nested reamplification with primer combination A21f-A915r (1, 8). This nested amplification was necessary in these sections in order to obtain sufficient PCR product for cloning and sequencing. For nonnested rRNA gene amplification in the SMI-1 section, general primers A8f-A1492r (62) and A21f-A915r (1, 8) were used, as well as the ANME-2-specific primers 8f-EelMS932r (5). A listing of all the primers used and their annealing temperatures can be found in Table 1. Primer combinations (nested and nonnested) and the resulting clone library compositions are listed in Tables 2, 3, and 4. Since the primer A21f proved to have 3′-terminal mismatches to multiple archaeal groups, the unnested clone library obtained with primers A21f-A915r may have been biased against these groups. Clone libraries based on the shorter, more general primer A8f (62), used as the first nested primer or as the sole forward primer, show a higher diversity of archaeal lineages (Table 2).
TABLE 1.
Primers used in this study
| Primer | Target gene | Predicted target group | Sequence (5′ to 3′)a | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| A8f | 16S | Archaea | TCC GGT TGA TCC TGC C | 55 or 59 | 62 |
| A1492r | 16S | Archaea | GGC TAC CTT GTT ACG ACT T | 55 | 62 |
| A21f | 16S | Archaea | TTC CGG TTG ATC CYG CCG GA | 64 | 8 |
| A915r | 16S | Archaea | GTG CTC CCC CGC CAA TTC CT | 64 | 8 |
| Ee1MS932r | 16S | ANME-2 | AGC TCC ACC CGT TGT AGT | 59 | 5 |
| B8f | 16S | Bacteria | AGR GTT TGA TCC TGG CTC AG | 50 or 55 | 62 |
| B1492r | 16S | Bacteria | CGG CTA CCT TGT TAC GAC TT | 50 or 55 | 62 |
| dsr1f | dsrAB | Sulfate reducers | ACS CAY TGG AAG CAC G | 54 | 65 |
| dsr4f | dsrAB | Sulfate reducers | GTG TAG CAG TTA CCG CA | 54 | 65 |
| 1F1 | dsrAB | Sulfate reducers | CAG GAY GAR CTK CAC CG | 48 | 10 |
| 1RI | dsrAB | Sulfate reducers | CCC TGG GTR TGR AYR AT | 48 | 10 |
| MCRf | mcrA | Methanogens/ANMEs | TAY GAY CAR ATH TGG YT | 50 or 55 | 55 |
| MCRr | mcrA | Methanogens/ANMEs | ACR TTC ATN GCR TAR TT | 50 or 55 | 55 |
| ME1 | mcrA | Methanogens/ANMEs | GCM ATG CAR ATH GGW ATG TC | 50 or 55 | 16 |
| ME2 | mcrA | Methanogens/ANMEs | TCA TKG CRT AGT TDG GRT AGT | 50 or 55 | 16 |
| ANME-2a-647r | 16S | ANME-2a | TCT TCC GGT CCC AAG CCT | 45 to 64 | 29 |
| ANME-2c-622r | 16S | ANME-2c | CCC TTC GCA GTC TGA TTG | 46 to 64 | 29 |
| ANME-2c-760r | 16S | ANME-2c | CGC CCC CAG CTT TCG TCC | 47 to 64 | 29 |
Y, C/T; R, A/G; M, A/C; W, A/T; K, G/T; H, A/C/T; D, A/G/T.
TABLE 2.
Phylogenetic affiliations of archaeal 16S rRNA genes and rRNA clone libraries at each depth below the seafloor and primer combination
| Phylogenetic affiliation | No. of clones from 16S rRNA genes
|
No. of clones from rRNA at 15-18 cm | ||||
|---|---|---|---|---|---|---|
| 0-3 cm | 15-18 cm
|
18-21 cm
|
||||
| A8f-A1492r/A21f-915r | A8f-Eel932r | A21f-A915r | A8f-A1492r | A8f-A1492r/ A21f-915r | A8f-A1492r | |
| Euryarchaeota | ||||||
| ANME-1b | 37 | 80 | 66 | 20 | ||
| Methanosarcinales (GoM Arc I) | 23 | 4 | 1 | 1 | ||
| Outside Methanosarcinales/ Methanomicrobiales | 1 | 10 | ||||
| Thermoplasmatales | ||||||
| MBG-D | 22 | 2 | 34 | 2 | ||
| Other Thermoplasmatales | 2 | |||||
| Crenarchaeota | ||||||
| MG-I | 2 | |||||
| MBG-B | 22 | 2 | ||||
| Total no. of clones | 46 | 63 | 36 | 86 | 79 | 21 |
TABLE 3.
Phylogenetic affiliations of dsrAB clone libraries at each depth below the seafloor
| Phylogenetic affiliation | No. of clones
|
||
|---|---|---|---|
| 0-3 cm | 15-18 cm | 18-21 cm | |
| GOM dsr I | 24 | 11 | 16 |
| Group IV | 7 | 4 | |
| Desulfobacterium anilini | 7 | 42 | |
| Syntrophobacteraceae | 6 | ||
| Desulfosarcina/Desulfococcus | 3 | 1 | |
| Total no. of clones | 47 | 58 | 16 |
TABLE 4.
Phylogenetic affiliations of bacterial 16S rRNA gene clone libraries at each depth below the seafloor
| Phylogenetic affiliation | No. of clones
|
|
|---|---|---|
| 0-3 cm | 15-18 cm | |
| Deltaproteobacteria | ||
| Desulfosarcina/Desulfococcus | 4 | 3 |
| Desulfobacteria (Eel-1) | 4 | |
| Syntrophus | 1 | |
| Desulfobulbus/Desulfocapsa | 4 | |
| Eel-2 | 13 | |
| Other deltaproteobacteria | 9 | |
| Epsilonproteobacteria | 1 | 1 |
| Gammaproteobacteria | 1 | |
| Acidobacteria | 7 | |
| Hypersaline group I | 3 | |
| JS1 | 14 | |
| Hypersaline group II | 6 | |
| Planctomycetales | 1 | 2 |
| OP8 | 1 | 2 |
| Actinobacteria | 4 | |
| Chloroflexi | 4 | |
| Hydrocarbon-associated group | 7 | |
| KB-1 | 5 | |
| Total no. of clones | 23 | 74 |
Total RNA was extracted following previously described methods (34, 56). Briefly, ∼2 ml sediment was mixed with 5 ml phenol (pH 5), 5 ml of extraction buffer (50 mM sodium acetate and 10 mM EDTA, pH 5), and 0.5 ml 20% sodium dodecyl sulfate. This mixture was subjected to four cycles of freezing at −80°C and then thawing at 65°C and then extracted with phenol, phenol-chloroform (1:1), and chloroform; precipitated in 7.5 mM NH4CH3COOH and isopropanol; and washed with ethanol. PCR amplification of reverse-transcribed cDNA (RT-PCR) was performed using the general archaeal primers A8f-A1492R (62).
Each 25-μl PCR and RT-PCR mixture contained 1 μl DNA template, 2.5 μl primer solution (10 pmol/μl), 1 μl bovine serum albumin (10 mg/ml), and 1 μl deoxynucleotide triphosphate (10 mM each dATP, dCTP, dGTP, and dTTP). PCR mixtures included an additional 2.5 μl 10× PCR buffer (Promega) and 0.15 μl Taq polymerase in storage buffer B (Promega); RT-PCR mixtures included an additional 5 μl 5× RT-PCR buffer (QIAGEN) and 1 μl One-Step RT-PCR enzymes (QIAGEN). PCR and RT-PCR amplifications were performed in a Bio-Rad iCycler. The conditions for PCR were as follows: an initial denaturation at 94°C for 2 min, followed by 30 cycles, each consisting of 30 s of denaturation at 94°C, 1 min at primer-annealing temperature (Table 1), and 1 to 3 min of elongation at 72°C. RT-PCR required the following protocol: 50°C incubation for reverse transcription for 30 min, 95°C polymerase activation for 2 min, followed by 40 cycles of 94°C denaturation for 30 s, 55°C annealing for 1 min, and 72°C extension for 1.5 min, plus a final cycle of 94°C for 30 s, 55°C for 3 min, and 72°C for 10 min. Extracted total RNA subjected to forward PCRs did not yield visible product on a 1.5% agarose gel, indicating the absence of significant coextracted DNA. Parallel blank extractions, serving as controls for contamination during DNA or RNA extraction, also yielded no visible products. All PCR and RT-PCR products were purified using a MoBio PCR cleanup kit, A tailed, and cloned using the TOPOXL PCR cloning kit before being transformed into Escherichia coli following the manufacturer's protocols (Invitrogen, San Diego, Calif.). Sequences were obtained at the Josephine Bay Paul Center at the Marine Biological Laboratory (Woods Hole, MA), using an ABI 3730 sequencer and M13 universal primers SP010 and SP030 (Operon). The sequences were converted to text files with Chromas 1.45 and assembled.
PCR check for ANME-2.
To test rigorously for the presence of the two most common phylotypes of ANME-2, forward primers ANME-2a-622, ANME-2c-647, and ANME-2c-760 (29) were each paired with reverse primer EelMS932r (5) for a low-stringency PCR amplification of SMI-1 DNA extract. Low annealing temperatures allow primers to bind more easily to templates at the expense of specific binding (24). In addition, increasing the cycle number increases the probability of producing amplicons from a smaller quantity of starting template at the expense of template bias minimization (57). Therefore, a high PCR cycle number and a range of annealing temperatures were used to increase the probability of amplifying ANME-2 sequences, even at low template numbers. The PCR mixtures were identical to those used in clone libraries, except that HotStarTaq (QIAGEN) was used in order to increase the sensitivity to low gene copy numbers. The following PCR protocol was used: enzyme activation at 95°C for 15 min, then 40 cycles of 94°C for 30 s; 45°C, 47°C, 50°C, 51°C, 53°C, 55.5°C, 59.3°C, 62°C, or 64°C for 1 min; and 72°C for 1.5 min, plus a final 5 min at 72°C. The PCR products were then visualized on a 1.5% agarose gel using an ethidium bromide stain.
Phylogenetic analysis.
Archaeal and bacterial 16S rRNA gene sequences were aligned with BLAST (basic local alignment search tool) hits from GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) using the ARB software (http://www.arb-home.de) fast aligner utility, followed by manual adjustments. The sequences were screened for chimeras by comparing neighbor-joining trees made of the first and second halves of all sequences. Any clones that had different groupings in the first and second halves were then checked using the Pintail program (2) and excluded from the clone libraries. Of a total of 623 sequences (including those from 16S rRNA genes, 16S rRNA, and functional genes), 25 sequences were found to be chimeras. Unalignable regions (helices 6, 11, and 17 for archaeal 16S and helices 6, 10, 11, and 18 for bacterial 16S) and chimeras were excluded from analysis. A hierarchical likelihood ratio test from Modeltest v3.7 (48) used within PAUP* version 4.b10 (58) was used to determine the distance correction that best described the sequence data. For archaeal 16S rRNA genes, a Tamura-Nei distance model was used with unequal base frequencies, a gamma shape of 0.6757, and a percent invariable sites value of 0.1773. For bacterial 16S rRNA genes, a general time-reversible model was used with gamma shape 0.7539 and a percent invariable sites value of 0.2141. Nonchimeric mcrA and dsrAB sequences were translated into amino acids using SeqPup 0.9 (http://iubio.bio.indiana.edu/soft/molbio/seqpup/java/seqpup-doc.html) and then aligned using ClustalW (63). Manual adjustments of mcrA and dsrAB sequence alignments were performed using SeqPup 0.9. Unalignable regions were not included in phylogenetic inferences. PAUP* version 4.b10 was used for all tree construction and bootstrap calculations. Trees were visualized with Treeview 1.6.6 (46).
Nucleotide sequence accession numbers.
The sequences are available from GenBank under the following numbers: 16S rRNA gene and rRNA sequences, DQ521754 to DQ521825; dsrAB genes, DQ521826 to DQ521856; mcrA genes, DQ521857 to DQ521864.
RESULTS
Geochemical conditions.
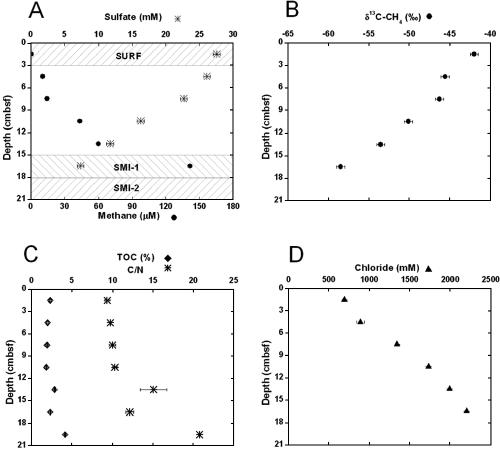
The deep subsurface of GC205 encompasses a salt diapir that allows petroleum to migrate upward through fractures in the sediments (26, 53). Accordingly, petroleum deposits and small carbonate crusts could be seen at the bottom of core 4566-5. Geochemical evidence for microbial activity was present in pore water chemical constituents. Sulfate concentrations decreased from 28 mM at the sediment-water interface to 7 mM at the SMI (Fig. 2A). This decrease was not caused by upwardly advecting sulfate-free brine and therefore indicated the activity of sulfate-reducing bacteria (32). Conversely, methane concentrations increased with depth in the sediment column from 0 mM at the sediment-water interface to ∼220 μM in the SMI (Fig. 2A). Upwardly diffusing methane is therefore removed in the presence of sulfate within the SMI. The δ13C of methane increased as it diffused toward the sediment-water interface (Fig. 2B), indicating that anaerobic methane oxidizers preferentially removed the lighter isotope.
FIG. 2.
Geochemical pore water and sediment measurements with depth in core 5, dive 4566, GC205, Gulf of Mexico. (A) Concentrations of sulfate (*) and methane (•); the shaded regions labeled SURF, SMI-1, and SMI-2 indicate sediment sections used for clone libraries. (B) δ13C of methane (•). (C) %TOC (⧫) and C/N ratio (*). (D) Chloride concentration (▴). The error bars in panels A and B represent the 95% confidence intervals of instrumental error. The error bars in panels C and D represent the 95% confidence intervals for triplicate measurements of each sample.
The %TOC and C/N ratios both increased with depth, indicating a sizable N-depleted source of subsurface organic carbon that could be petroleum derived (Fig. 2C). The δ13C of sedimentary TOC in the SMI at 18-cm depth (−26‰) was 13C depleted relative to the background signal of −20.2‰ to −22‰, measured at non-methane seep Gulf of Mexico sediments at similar water depths and locations (15, 32). Therefore, other 13C-depleted carbon sources, such as petroleum at −26.7‰ (26) or methane at −60‰ (this study), contributed to the TOC in the SMI. Evidence for the influence of subsurface salt deposits comes from chloride profiles, which showed an increase to 2,200 mM at 18 cm (Fig. 2D), roughly four times that of local seawater, which is 550 mM (32). These trends in %TOC, C/N ratio, and chloride show that these sediments are directly affected by a subsurface intrusion of a salt diapir and petroleum deposits.
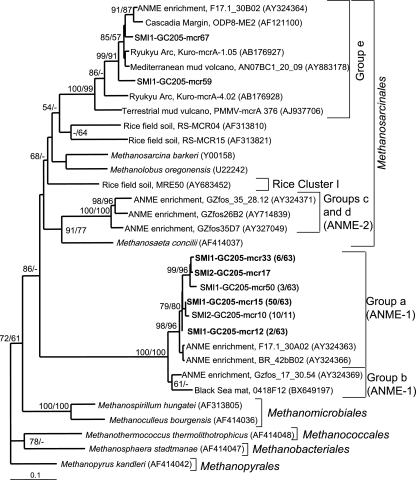
Diversity of mcrA.
The nomenclature used to describe the diversity of mcrA lineages found in this study follows that of Hallam et al. (17). Methanogen and ANME community compositions, as indicated by the distribution of mcrA clones, were similar between sediments in the SMI-1 and SMI-2 sections (Fig. 3). Both sections were dominated by clones that grouped closely with the mcrA genes from fosmids called group a (97% of 63 clones for SMI-1 and 100% of 11 clones from SMI-2), which are contiguous with 16S rRNA gene sequences from ANME-1 (17, 18). The remaining two clones (from SMI-1) fell into group e within the Methanosarcinales, along with sequences from Cascadia Margin hydrates, Kuroshima Knoll methane seeps, a terrestrial mud volcano, and methane-oxidizing enrichments containing ANME-1 and ANME-2 archaea. While an association with AOM cannot be ruled out for group e sequences (17), they could equally likely represent typical methanogen members of the Methanosarcinales, which can disproportionate acetate and methylated compounds. Utilization of noncompetitive substrates might allow these members of the Methanosarcinales to be present in the AOM zone, where sulfate reducers compete for organic carbon sources. No mcrA genes from ANME-2 archaea were amplified from either SMI layer, even though the primer sets used in this study have been shown to amplify mcrA from members of the ANME-2 group in other marine sediments (17, 23). No mcrA genes were amplified from the surface sediments, even though two primer sets and nested-PCR reamplifications were employed.
FIG. 3.
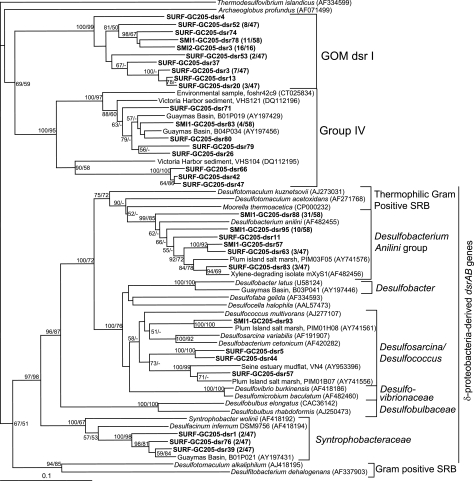
Least-squares distance tree based on translated partial amino aid sequences of PCR-amplified mcrA genes from GC205 sediment depths SMI-1 (15 to 18 cm) and SMI-2 (18 to 21 cm) (boldface). Group names are based on those of Hallam et al. (17). Bootstrap values (percent) for 1,000 repetitions each of distance (first) and parsimony (second) are listed for values of >50%. Groups that have >99% similar nucleic acid sequences within each sediment depth are represented by a single sequence, with the number of clones out of the total in parentheses. Environmental clones are included for comparison and are designated by their location or environmental type, clone name, and GenBank accession number. The scale bar represents 10% estimated distance.
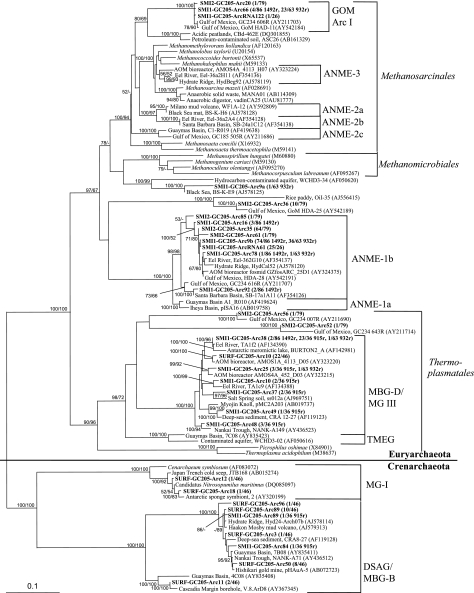
Diversity of archaeal 16S rRNA genes and rRNA.
Clones from the ANME-1b group dominated the archaeal 16S clone libraries from the SMI-1 and SMI-2 sediment horizons (Table 2 and Fig. 4). The ANME-1a and ANME-1b designations are based on the ANME phylogeny described by Knittel et al. (29). No ANME-1 sequences were present in general archaeal 16S clone libraries of surface sediments, indicating that the ANME-1b sequences were associated only with the SMI (Table 2). No ANME-2 sequences were amplified from any sediment layer with archaeal 16S rRNA gene primers. The SMI-1 sediment layer was reexamined for the presence or absence of ANME-2 groups, using primer EelMS932, a probe specific for ANME-2 (5), in combination with the general forward primer A8f. This primer combination yielded ANME-1b sequences and other members of the Methanosarcinales, but not ANME-2 (Table 2). In order to more carefully ascertain the absence of ANME-2, PCR of the SMI-1 section was performed using one forward primer specific for ANME-2a and two specific for ANME-2c (29), in combination with EelMS932r. Amplicons of the correct size were never detected, even using a temperature range of 45 to 65°C and 40 cycles per PCR run. However, bands about 100 base pairs larger than the size of the correct amplicon at 47°C using the ANME-2a forward primers were examined through band excision, cloning, and sequencing and were not found to contain rRNA genes.
FIG. 4.
Least-squares distance tree of archaeal 16S rRNA gene and rRNA clones from GC205 sediment depths SURF (0 to 3 cm), SMI-1 (15 to 18 cm), and SMI-2 (18 to 21 cm) (boldface). The bootstrap values, >99% similarity groupings, distance scale, and environmental-clone names follow the conventions in Fig. 3. For SMI-1 the >99% similar clone numbers are listed for each reverse primer used. Sequences including “RNA” in their names are cDNAs that were reverse transcribed from total RNA; all other sequences were PCR amplified from DNA. TMEG, terrestrial miscellaneous euryarchaeotal group (61); DSAG, deep-sea archaeal group. See the text for explanations of other phylogenetic group acronyms.
The next largest group of clones from SMI-1 fell within GOM arc I in the Methanosarcinales, along with other sequences from methane hydrate-rich sediments in the Gulf of Mexico (Table 2). This group was previously called ANME-2d (37, 38); however, it is not monophyletic with ANME-2a, 2b, 2c, or 3, nor has it been shown to assimilate methane and/or to form consortia with sulfate reducers. In the absence of these tests, 16S rRNA gene similarities alone do not support conferring an AOM metabolism on GOM arc I. The closest sister lineages to this group are uncultured clones from methane-producing, nonmarine, hydrocarbon-rich soils (Fig. 4), making it likely that GOM Arc I is a group of uncultured methanogenic Methanosarcinales. Eleven more sequences from the SMI fall just outside of the Methanosarcinales and Methanomicrobiales and group with sequences from other hydrocarbon-rich sediments. They are represented by clones SMI1-GC205-Arc9 and SMI2-GC205-Arc36 (Fig. 4).
The only archaeal group found in all three sediment depths was marine benthic group D (MBG-D) (64), synonymous with marine group III (MG-III) (9), a lineage within the Thermoplasmatales that generally occurs in marine sediments and hydrothermal environments (14, 38, 59). In our study, clones of the MBG-D archaea were mostly obtained from surface sediments (48% of 46 clones) and from the A21f-915r amplification of the SMI-1 section (94% of 36 clones) (Table 2). Two sequences from the SMI-2 section also formed sister lineages to MBG-D (Fig. 4).
Most of the crenarchaeotal clones fell within MBG-B (64), which is synonymous with the deep-sea archaeal group (22). This group is mostly found in surficial and deep marine sediments and hydrothermal environments (22, 29). MBG-B archaea dominated the surface clone library (48% of 46 clones) and were represented by two sequences in the SMI. All other crenarchaeotal sequences from surface sediments fell within MG-I, a pelagic group that includes the sponge symbiont Cenarchaeum symbiosum and the ammonia oxidizer Nitrosopumilus maritimus (Table 2; Fig. 4).
To elucidate the phylogenetic composition of the population that was most likely active, total rRNA was extracted from the SMI-1 section, followed by RT-PCR to amplify 16S rRNA sequences. Out of 21 clones, 20 grouped within the ANME-1b, and 1 clone fell within the GOM arc I group in the Methanosarcinales (Table 2). The general agreement between rRNA and rRNA gene sequences in A8f-A1492r clone libraries indicated that the ANME-1b and the Methanosarcinales-related rRNA gene clones represented active organisms at this depth.
Diversity of dsrAB.
dsrAB sequences were more diverse than mcrA sequences at all three depths, with a total of five distinct groups represented and no large-scale changes in sulfate reducer community composition with depth (Fig. 5). The largest group of dsrAB sequences retrieved from SMI-1 (72% of 58 clones) grouped with dsrAB sequences from the aromatic-hydrocarbon degraders Desulfobacterium anilini and isolate mXyS1 (Table 3). All cultured members of this group obligately degrade hydrocarbons commonly found in natural petroleum deposits.
FIG. 5.
Least-squares distance tree based on translated partial amino aid sequences of PCR-amplified dsrAB genes from GC205 sediment depths SURF (0 to 3 cm), SMI-1 (15 to 18 cm), and SMI-2 (18 to 21 cm) (boldface). The bootstrap values, >99% similarity groupings, distance scale, and environmental-clone names follow the conventions in Fig. 3.
A large proportion of sequences from all three depths fell into two deeply branching clades, group IV (10) and GOM dsr I (Table 3 and Fig. 5). Group IV also contains sequences retrieved from Guaymas Basin, a sedimented, petroleum-rich hydrothermal-vent site; Plum Island, a New England salt marsh; Kysing Fjord, a shallow brackish fjord in Denmark; and other marine sediments. At present, group GOM dsr 1 does not have cultured relatives or closely related dsrAB phylotypes from other environments; it may represent a sulfate-reducing lineage with specific adaptations to the biogeochemical regime in GC205 sediments. This cluster is well supported (100% bootstrap) and appears to form a sister group to group IV, with moderate bootstrap support (around 70%) (Fig. 5).
Only a few clones from both surface sediments and SMI-1 were within the DSS group, although with poor bootstrap support. In the 16S rRNA gene phylogeny, the DSS group contains the putative SRB syntrophs for AOM (5, 43). Cultured members of this group can oxidize a wide range of substrates completely to CO2 and can utilize organic and inorganic carbon sources.
The only group of dsrAB genes found at a single depth was the Syntrophobacteraceae in the surface sediments (Table 3). Cultured members of the family Syntrophobacteraceae include syntrophic propionate oxidizers, which can also reduce sulfate (genus Syntrophobacter), as well as thermophilic sulfate reducers that access a wide variety of organic acids (genus Desulfacinum).
Diversity of bacterial 16S rRNA genes.
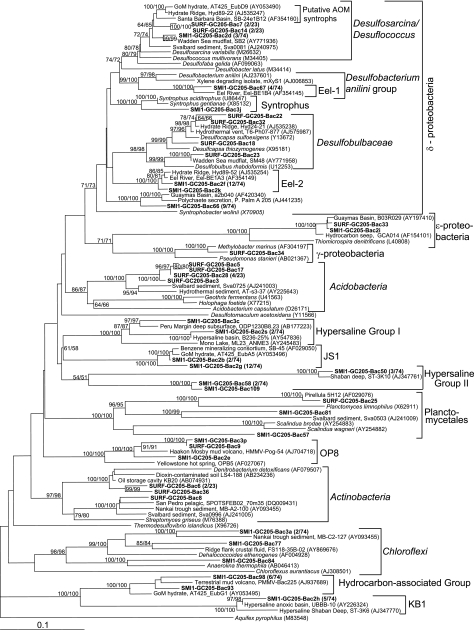
Bacterial 16S rRNA genes are more diverse than archaeal 16S rRNA genes in both surface and SMI-1 clone libraries (Fig. 6 and Table 4). Clone libraries of bacterial 16S rRNA genes were not made for SMI-2, since archaeal libraries looked very similar to those of SMI-1. Bacterial clones in surface sediments (34% of 23 clones) and the SMI-1 layer (40% of 74 clones) fell predominantly within the deltaproteobacteria, a group containing many SRB (Table 4). From both sediment depths, a few clones formed branches parallel to the putative AOM syntrophic sulfate reducers within the DSS group (43). The remaining deltaproteobacterial clones from the surface sediments fell within the propionate-oxidizing Desulfobulbaceae next to other uncultured sequences from other anoxic marine sediments (Fig. 6). A few clones from SMI-1 were members of the Eel-1 group, a sister lineage to Desulfobacterium anilini and the xylene-degrading isolate mXyS1. Clones from SMI-1 also fell within the Eel-2 group, which has no close cultured relatives. Both the Eel-1 and Eel-2 groups are characteristic of marine methane-rich sediments and often occur in sediments where ANME-1 phylotypes dominate over ANME-2 (43, 62), as was the case in the current study. A single clone from SMI-1 formed a sister taxon to Syntrophus gentianae and Syntrophus aciditrophus; cultured members of this genus ferment benzoate and often live syntrophically with H2-consuming methanogens. Nine near-identical SMI-1 clones (represented by SMI1-GC205-Bac66) (Fig. 6) formed a distinct, new deltaproteobacterial lineage, together with sequences obtained from Guaymas Basin sediments and tubeworm secretions.
FIG. 6.
Least-squares distance tree of bacterial 16S rRNA gene clones from GC205 sediment depths SURF (0 to 3 cm) and SMI-1 (15 to 18 cm) (boldface). The bootstrap values, >99% similarity groupings, distance scale, and environmental-clone names follow the conventions in Fig. 3.
Other proteobacterial clones include a gammaproteobacterial clone near the methyl-oxidizing genus Methylobacter and two clones within the epsilonproteobacteria; cultured species of this group are generally chemolithotrophic O2- or NO3-respiring H2 or S oxidizers (Fig. 6) (60).
Among nonproteobacterial clones, most clones in the surface sediments (31%) were members of the Holophaga/Acidobacteria, a cosmopolitan lineage in marine sediments (49). Cultured representatives of this group share little physiological similarity with each other and thus do not allow physiological inferences for uncultured relatives; Holophaga foetida is an acetogen, and Geothrix fermentens is a nonacetogenic Fe(III) reducer. The surface sediments also harbored members of the Firmicutes/Actinobacteria, which are abundant in marine, freshwater, and terrestrial habitats and have a range of physiologies.
SMI-1 harbored several uncultured bacterial lineages that are characteristic of marine sediments and hypersaline habitats: chloroflexi occur frequently in hydrocarbon-rich sediments and the deep subsurface (62); the JS1 group is found in marine sediments and the subsurface (66); the Kebrit Deep I (KB-I), hypersaline group I (HS-I), and HS-II from hypersaline marine basins and seeps (11), and hydrocarbon group I from hydrocarbon-rich sediments (31). The HS-I, HS-II, and hydrocarbon group I designations are proposed in this study to simplify discussion of these groups. Two clones from SMI-1 and one clone from surface sediments belong to the candidate division OP8, an uncultured phylum-level lineage found in hydrothermal vents, hot springs, mud volcanoes, oil spills, and marine sediments.
Three clones from the sediment surface and SMI-1 grouped within the Planctomycetes, a phylum that includes the genera Pirellula and Planctomyces, aerobic or nitrogen-respiring marine heterotrophs, and the anaerobic ammonia-oxidizing genus Scalindua.
DISCUSSION
The ANME-1b archaeal community.
Studies of the microbial populations responsible for AOM almost always find cooccurring populations of ANME-1a, ANME-1b, various subgroups of ANME-2, and sometimes ANME-3 (14, 20, 29, 31, 36, 38, 39, 41, 43-45, 62). So far, communities known to be dominated by ANME-1 are limited to benthic microbial mats and carbonate reefs in the Black Sea (4, 29, 36). To our knowledge, our study is the first to describe a natural sedimentary ANME population consisting only of ANME-1b archaea.
In the GC205 sediments, ANME-2 sequences could not be detected with two sets of general archaeal 16S and mcrA primers that are known to amplify these groups. Furthermore, the ANME-2-targeted probe EelMS932r (5, 44) amplified only clones from ANME-1b and other euryarchaeota (Table 2). EelMS932r, in combination with forward primers targeting the most common ANME-2 subgroups, ANME-2a and ANME-2c, amplified no ANME-2 sequences, even though low-stringency PCR techniques were used. Therefore, it appears that these SMI sediment layers harbor an AOM community whose archaeal component comprises only ANME-1b archaea. Since ANME-1b dominates the rRNA library, as well as that of rRNA genes, ANME-1b is not only present but may be active in these sediments. This result also has implications for the identification of mcrA phylotypes. Due to the absence of the ANME-1a subgroup in 16S rRNA gene and rRNA clone libraries, the tightly clustered ANME-1 mcrA sequences reported here should represent the ANME-1b subgroup (Fig. 3).
Geochemical controls at site GC205 may be responsible for the existence of only ANME-1b archaea in this population of anaerobic methane oxidizers. In previous studies, methane concentrations and the presence of oxygen have been postulated as selective factors for ANME-1 and ANME-2 (4, 29). Knittel et al. (29) proposed that ANME-1 is more sensitive to oxygen than is ANME-2. According to this hypothesis, Black Sea carbonate structures contain predominantly ANME-1 because, unlike cold methane seeps, hydrothermal vents, and methane hydrates, the Black Sea mats are exposed only to anoxic bottom waters. Similarly, the sediments at site GC205 are permanently anoxic, and no resuspension by gas ebullition or hydrate outcropping was observed at the time of sampling. However, oxygen concentration profiles and time series for oxygen dynamics in ANME-rich hydrate sites and methane seeps are needed to substantiate this hypothesis.
Blumenberg et al. (4) suggested that ANME-1 outcompetes ANME-2 in the interior sections of Black Sea carbonate chimneys, where methane is presumed to be limiting. However, ANME-1 and ANME-2 archaea usually coexist near methane hydrates or seeps with high concentrations of methane (23, 31, 37, 43). In fact, laboratory enrichments of AOM show that ANME-1 archaea outcompete ANME-2 under a high methane flow rate (13). In addition, ANME-1 was slightly more numerous than ANME-2 at site GC232 in the Gulf of Mexico, even under high millimolar concentrations of methane (41). At Gulf of Mexico sites GC233 and GC232, ANME-2 and ANME-1 coexist at methane concentrations both higher and lower than those in the current study at site GC205 (41). Therefore, both ANME-1 and ANME-2 archaea appear to tolerate a wide range of methane concentrations and fluxes. At GC205, the methane concentration at SMI-1 is relatively low for a methane seep site (∼220 μM), but the upward trend of methane with depth indicates a substantial methane flux into the SMI. Therefore, the dominance of ANME-1b at GC205 is not sufficiently explained by the specific methane concentrations and fluxes at our study site.
The dominance of ANME-1b at GC205 may be explained by the site's uniquely high salinity compared to previous studies. The chloride concentration in the SMI is 2,200 mM, which is roughly four times that of seawater and sediments near the brine pool at GC233 (both around 35 ppt, or 550 mM Cl−) (41). However, natural samples of predominantly ANME-1 or ANME-2 archaea (from Black Sea carbonates and Hydrate Ridge sediments at seawater salinities) share peak sulfate reduction rates at salinities around 470 mM, near seawater salinity, and show a sharp decline around 785 to 945 mM (39). The complete absence of ANME-2 and ANME-1a at salinities of 2,200 mM in our samples suggests that a high-salt-adapted subpopulation of ANME-1b may persist where other ANME groups cannot. Salinity is a geochemical control factor on the bacterial community as well; 16S rRNA gene sequences fell within the uncultured groups KB-1, HS-I, and HS-II, which contain sequences only from hypersaline environments (11).
Relationship between SRB and AOM.
Unlike the archaeal 16S rRNA gene/rRNA and mcrA clone libraries, the bacterial 16S rRNA gene and dsrAB clone libraries are not dominated by sequences of organisms previously associated with AOM. According to the dsrAB clone libraries, the SRB community composition does not change appreciably between surface sediments and those at the sulfate-methane interface, indicating that the community composition is not altered by increasing methane concentrations downcore. The majority of dsrAB sequences from both the upper and the lower sections group with GOM dsr I, the Desulfobacterium anilini group, and group IV (Fig. 5 and Table 3). This diversity of dsrAB genes far exceeds the known range of sulfate-reducing bacterial syntrophs driving AOM, and no groups are specifically associated with the SMI, indicating that these sulfate-reducing communities grow independently of anaerobic methane oxidation.
This decoupling of sulfate reducers from AOM has been observed in similar Gulf of Mexico sediments, where rates of sulfate reduction are much higher than rates of AOM (25, 42) and petroleum and other hydrocarbons are the likely carbon sources for sulfate reduction (12, 25). Petroleum degradation is probably occurring at site GC205, since the organic matter became N depleted with depth and the δ13C values for TOC at site GC205 reflected a petroleum source. The bacterial community composition is consistent with this process; many bacterial 16S rRNA gene sequences from site GC205 grouped closely with sequences from other hydrocarbon-rich areas and hydrocarbon-degrading enrichments. Group IV sulfate reducers, for instance, are often found in organic-rich marine sediments, rich in decaying plant biomass and/or petroleum hydrocarbons (3, 10). A similar bacterial utilization of ancient hydrocarbons was observed in Guaymas Basin, where the majority of living biomass is petroleum derived, as determined by radioactive and stable carbon isotopic signatures (47). An alternative scenario, methane oxidation by sulfate-reducing bacteria alone without syntrophic ANME groups, is unlikely, given that we did not find any mcrA genes that were not accounted for by archaeal 16S rRNA gene sequences. So far, no sulfate reducer has been reported to oxidize methane directly, whereas petroleum-derived hydrocarbons are a viable electron and carbon source for sulfate reduction (47).
A few 16S rRNA gene and dsrAB clones fell within the DSS group, which contains members that form consortia with ANME archaea (5, 44). However, free-living members of the DSS group are also widespread in marine sediments and often constitute a dominant SRB population (3, 28, 49). To our knowledge, no studies have positively differentiated dsrAB genes of DSS syntrophs associated with AOM from dsrAB genes of free-living DSS members that are abundant and ubiquitous in marine sediments. Likewise, the dsrAB sequences in this study, although related to the DSS group, cannot be assigned to AOM syntrophs with certainty. The 16S rRNA gene and dsrAB clone libraries yielded DSS clones, not only from the SMI, but also from surface sediments, where low methane concentrations make AOM improbable, no ANME groups were found, and a background population of free-living DSS group bacteria is very likely. In brief, free-living and syntrophic dsrAB phylotypes cannot be identified with certainty without more specific techniques, such as mRNA-targeted fluorescence in situ hybridization.
Implications.
Using 16S rRNA genes and functional genes, we have identified how uncultured groups of bacteria and archaea interact with the specific geochemical gradients that characterize their habitat. Our work shows that AOM in hypersaline conditions is mediated by a previously recognized clade of ANMEs (ANME-1b) and does not require the presence of a novel group with evolutionarily divergent mcrA genes. This does not appear to be the case for hypersaline/alkaline Mono Lake, where AOM is possibly mediated by organisms that are unrelated to the known ANME clades (54). Future studies should investigate (i) whether ANME-1b dominance applies to larger areas of the seafloor and (ii) which specific adaptations enable the ANME-1b group to meet the energy challenges of maintaining osmoregularity using the low energy-yielding process of AOM.
In contrast to the highly specific ANME-1b community supported by AOM at the sulfate-methane interface, sulfate reduction supports a phylogenetically diverse group of bacteria that probably degrade hydrocarbons and/or petroleum throughout the sediment column. The presence of deeply branching dsrAB sequences at GC205 raises the possibility that some of the uncultured 16S rRNA gene bacterial lineages are sulfate reducers. The sulfate reducers identified by dsrAB gene sequencing may represent novel forms of halophilic or halotolerant petroleum-degrading organisms, although isolations and isotope determinations are required to establish the link to petroleum.
Acknowledgments
We thank Ian MacDonald and Miriam Kastner (cruise co-chief scientists), the captain and crews of R/V Seward Johnson II and the Johnson-Sea-Link submersible, and Katrin Knittel, who provided advice on experiments. We also thank the Josephine Bay Paul Sequencing Center and their technicians Jillian Ward and Leslie Graham for their rapid, high-quality sequencing work.
This study was supported by the NASA Astrobiology Institutes “Environmental Genomes” (NCC 2-1054) and “Subsurface Biospheres” (NCC 2-1275) (A.T. and K.G.L.) and the EPA-STAR fellowship (L.L.). This portion of the Gulf of Mexico Gas Hydrate Research Consortium cruise was funded by US Minerals Management Services.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Amann, R., L. R. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahr, M., B. C. Crump, V. Klepac-Ceraj, A. Teske, M. L. Sogin, and J. E. Hobbie. 2005. Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ. Microbiol. 7:1175-1185. [DOI] [PubMed] [Google Scholar]
- 4.Blumenberg, M., R. Seifert, J. Reitner, T. Pape, and W. Michaelis. 2004. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. USA 101:11111-11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 6.Brenna, J. T., T. N. Corso, H. J. Tobias, and R. J. Caimi. 1997. High-precision continuous-flow isotope ratio mass spectrometry. Mass Spectrom. Rev. 16:227-258. [DOI] [PubMed] [Google Scholar]
- 7.Danovaro, R., A. Dell'Anno, A. Pusceddu, and M. Fabiano. 1999. Nucleic acid concentrations (DNA, RNA) in the continental and deep-sea sediments of the eastern Mediterranean: relationships with seasonally varying organic inputs and bacterial dynamics. Deep Sea Res. I 46:1077-1094. [Google Scholar]
- 8.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E. F. 1998. Everything in moderation: archaea as ‘non-extremophiles’. Curr. Opin. Genet. Dev. 8:649-654. [DOI] [PubMed] [Google Scholar]
- 10.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eder, W., M. Schmidt, M. Koch, D. Garbe-Shonberg, and R. Huber. 2002. Prokaryotic phylogenetic diversity and corresponding geochemical data of the brine-seawater interface of the Shaban Deep, Red Sea. Environ. Microbiol. 4:748-763. [DOI] [PubMed] [Google Scholar]
- 12.Formolo, M. J., T. W. Lyons, C. Zhang, C. A. Kelley, R. Sassen, J. Horita, and D. R. Cole. 2004. Quantifying carbon sources in the formation of authigenic carbonates at gas hydrate sites in the Gulf of Mexico. Chem. Geol. 205:253-264. [Google Scholar]
- 13.Girguis, P. R., A. E. Cozen, and E. F. DeLong. 2005. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl. Environ. Microbiol. 71:3725-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. DeLong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goñi, M. A., K. C. Ruttenberg, and T. I. Eglinton. 1997. Sources and contribution of terrigenous organic carbon to surface sediments in the Gulf of Mexico. Nature 389:275-278. [Google Scholar]
- 16.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 19.Hinrichs, K.-U. 2002. Microbial fixation of methane carbon at 2.7 Ga: was an anaerobic mechanism possible? Geochem. Geophys. Geosyst. 3:10.1029. [Google Scholar]
- 20.Hinrichs, K.-U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 21.Hoehler, T. M., and M. J. Alperin. 1996. Anaerobic methane oxidation by a methanogen-sulfate reducer consortium: geochemical evidence and biochemical considerations, p. 326-333. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Boston, Mass.
- 22.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inagaki, F., U. Tsunogai, M. Suzuki, A. Kosaka, H. Machiyama, K. Takai, T. Nunoura, K. H. Nealson, and K. Horikoshi. 2004. Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, Southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes. Appl. Environ. Microbiol. 70:7445-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii, K., and M. Fukui. 2001. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 67:3753-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joye, S. B., A. Boetius, B. N. Orcutt, J. P. Montoya, H. N. Schulz, M. J. Erickson, and S. K. Lugo. 2004. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem. Geol. 205:219-238. [Google Scholar]
- 26.Kennicutt, M. C. I., J. M. Brooks, and G. J. Denoux. 1988. Leakage of deep, reservoired petroleum to the near surface on the Gulf of Mexico continental slope. Mar. Chem. 24:39-59. [Google Scholar]
- 27.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klepac-Ceraj, M. Bahr, B. C. Crump, A. Teske, J. E. Hobbie, and M. F. Polz. 2004. High overall diversity and dominance of microdiverse relationships in salt marsh sulphate-reducing bacteria. Environ. Microbiol. 6:686-698. [DOI] [PubMed] [Google Scholar]
- 29.Knittel, K., T. Lösekann, A. Boetius, R. Kort, and R. Amann. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krüger, M., A. Meyerdierks, F.-O. Glöckner, R. Amann, F. Widdel, M. Kube, R. Reinhardt, J. Kahnt, R. Böcher, R. K. Thauer, and S. Shima. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878-881. [DOI] [PubMed] [Google Scholar]
- 31.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lapham, L. L. 2006. Ph.D. thesis. Biogeochemical and physical controls on methane and sulfate in cold seep environments. University of North Carolina, Chapel Hill.
- 33.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 34.MacGregor, B. J., D. P. Moser, E. W. Alm, K. H. Nealson, and D. A. Stahl. 1997. Crenarchaeota in Lake Michigan sediment. Appl. Environ. Microbiol. 63:1178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens, C. S., D. B. Albert, and M. J. Alperin. 1999. Stable isotope tracing of anaerobic methane oxidation in the gassy sediments of Eckernforde Bay, German Baltic Sea. Am. J. Sci. 299:589-610. [Google Scholar]
- 36.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, R. Amann, B. B. Jørgensen, F. Widdel, J. Peckmann, N. V. Pimenov, and M. B. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 37.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 38.Mills, H. J., R. J. Martinez, S. Story, and P. A. Sobecky. 2005. Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl. Environ. Microbiol. 71:3235-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nauhaus, K., T. Treude, A. Boetius, and M. Krüger. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-1 and ANME-II communities. Environ. Microbiol. 7:98-106. [DOI] [PubMed] [Google Scholar]
- 40.Odom, J. M., and H. D. Peck. 1984. Hydrogenase, electron-transfer proteins, and energy coupling in the sulfate-reducing bacteria Desulfovibrio. Annu. Rev. Microbiol. 38:551-592. [DOI] [PubMed] [Google Scholar]
- 41.Orcutt, B., A. Boetius, M. Elvert, V. Samarkin, and S. B. Joye. 2005. Molecular biogeochemistry of sulfate reduction, methanogenesis and the anaerobic oxidation of methane at Gulf of Mexico cold seeps. Geochim. Cosmochim. Acta 69:4267-4281. [Google Scholar]
- 42.Orcutt, B. N., A. Boetius, S. K. Lugo, I. R. MacDonald, V. A. Samarkin, and S. B. Joye. 2004. Life at the edge of methane ice: microbial cycling of carbon and sulfur in Gulf of Mexico gas hydrates. Chem. Geol. 205:239-251. [Google Scholar]
- 43.Orphan, V. J., K.-U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 45.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 47.Pearson, A., J. S. Seewald, and T. I. Eglinton. 2005. Bacterial incorporation of relict carbon in the hydrothermal environment of Guaymas Basin. Geochim. Cosmochim. Acta 69:5477-5486. [Google Scholar]
- 48.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 53:793-808. [DOI] [PubMed] [Google Scholar]
- 49.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reeburgh, W. S. 1996. “Soft spots” in the global methane budget, p. 334-342. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Boston, Mass.
- 51.Reeve, J. N., J. Nolling, R. M. Morgan, and D. R. Smith. 1997. Methanogenesis: genes, genomes and who's on first? J. Bacteriol. 179:5975-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice, A. L., A. A. Gotoh, H. O. Ajie, and S. C. Tyler. 2001. High-precision continuous-flow measurement of δ13C and δD of atmospheric CH4. Anal. Chem. 73:4104-4110. [DOI] [PubMed] [Google Scholar]
- 53.Sassen, R., A. V. Milkov, E. Ozgul, H. H. Roberts, J. L. Hunt, M. A. Beeunas, J. P. Chanton, D. A. DeFreitas, and S. T. Sweet. 2003. Gas venting and subsurface charge in the Green Canyon area, Gulf of Mexico continental slope: evidence of a deep bacterial methane source? Org. Geochem. 34:1455-1464. [Google Scholar]
- 54.Scholten, J. C. M., S. B. Joye, J. T. Hollibaugh, and J. C. Murrell. 2005. Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microb. Ecol. 50:29-39. [DOI] [PubMed] [Google Scholar]
- 55.Springer, E., M. S. Sachs, C. R. Woese, and D. R. Boone. 1995. Partial gene sequences for the A subunit of methyl-coenzyme M reductase (mcrA) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45:554-559. [DOI] [PubMed] [Google Scholar]
- 56.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (and other methods), version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 59.Takai, K., and K. Horikoshi. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated ε-proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 61.Takai, K., D. P. Moser, M. DeFlaun, T. C. Onstott, and J. K. Fredrickson. 2001. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 67:5750-5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. de Vera Gomezqq, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A.-L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster, G., R. J. Parkes, J. C. Fry, and A. J. Weightman. 2004. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl. Environ. Microbiol. 70:5708-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, C. L., R. D. Pancost, R. Sassen, Y. Qian, and S. A. Macko. 2003. Archaeal lipid biomarkers and isotopic evidence of anaerobic methane oxidation associated with gas hydrates in the Gulf of Mexico. Org. Geochem. 34:827-836. [Google Scholar]
- 68.Zverlov, V., M. Klein, L.-H. Lin, S. Lücker, M. W. Friedrich, J. Kellermann, D. A. Stahl, A. Loy, and M. Wagner. 2005. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol. 187:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]