Abstract
A total of 636 vancomycin-resistant Enterococcus faecium (VRE) isolates obtained between 1994 and 1999 from the Medical School Hospital of the University of Michigan were tested for bacteriocin production. Of the 277 (44%) bacteriocinogenic strains, 21 were active against E. faecalis, E. faecium, E. hirae, E. durans, and Listeria monocytogenes. Of those 21 strains, a representative bacteriocin of strain VRE82, designated bacteriocin 43, was found to be encoded on mobilizable plasmid pDT1 (6.2 kbp). Nine open reading frames (ORFs), ORF1 to ORF9, were presented on pDT1 and were oriented in the same direction. The bacteriocin 43 locus (bac43) consists of the bacteriocin gene bacA (ORF1) and the immunity gene bacB (ORF2). The deduced bacA product is 74 amino acids in length with a putative signal peptide of 30 amino acids at the N terminus. The bacB gene encodes a deduced 95-amino-acid protein without a signal sequence. The predicted mature BacA protein (44 amino acids) showed sequence homology with the membrane-active class IIa bacteriocins of lactic acid bacteria and showed 86% homology with bacteriocin 31 from E. faecalis YI717 and 98% homology with bacteriocin RC714. Southern analysis with a bac43 probe of each plasmid DNA from the 21 strains showed hybridization to a specific fragment corresponding to the 6.2-kbp EcoRI fragment, suggesting that the strains harbored the pDT1-like plasmid (6.2 kb) which encoded the bacteriocin 43-type bacteriocin. The bac43 determinant was not identified among non-VRE clinical isolates.
Bacteriocins are produced by a wide variety of gram-positive and gram-negative bacteria. They are bacterial proteins which inhibit the growth of other bacteria that are closely related to the producer strains, and they usually exhibit a relatively narrow spectrum of activity. Bacteriocins are thought to provide the producer strain with an ecological or selective advantage over other strains. Bacteriocin production has been described for several genera of lactic acid bacteria (LAB) (12, 33). LAB bacteriocins can be classified into two main classes (34), i.e., class I, modified bacteriocins (the lantibiotics) and class II, the small, heat-stable nonlantibiotics, which are divided into subgroups IIa, pediocin-like bacteriocins with a strong antilisterial effect, and IIb, non-pediocin-like bacteriocins and those with two peptides that require the complementary action of both peptides for full antimicrobial activity. In the genus Enterococcus, Enterococcus faecalis and E. faecium bacteriocins have been genetically and biochemically well characterized. E. faecalis bacteriocins include the β-hemolysin/bacteriocin (cytolysin) (9, 17, 18, 21, 23, 24), the peptide antibiotic AS-48 (28), bacteriocin 21 (Bac 21) (43), and Bac 31 (42). These bacteriocins have been identified from E. faecalis clinical isolates (29, 42, 43, 47). The well-characterized E. faecium bacteriocins have been isolated from food grade organisms (6, 19) and include enterocins A (1), B (3), P (4), I (15), L50A, and L50B (5). These bacteriocins belong to the LAB class II bacteriocins and are active against Listeria monocytogenes (34). Enterocins A and P are pediocin-like bacteriocins (12).
In contrast to the bacteriocins obtained from E. faecalis clinical isolates, there have been few reports describing either the bacteriocins present in E. faecium clinical isolates, including vancomycin-resistant E. faecium (VRE), or the relationship between the bacteriocin determinant and a plasmid (10, 11, 25).
In our previous study (25), a total of 636 VRE strains were tested for bacteriocin production against various indicator strains. Two hundred seventy-seven (44%) of the 636 strains tested were bacteriocinogenic. The bacteriocinogenic strains were classified into four groups on the basis of their bacteriocin activity. Of the 277 bacteriocin producers tested for activity against enterococci, 21 strains (3.3%) showed bacteriocin activity against E. faecalis, E. faecium, E. hirae, E. durans, L. monocytogenes, and L. denitrificans; 193 strains (69.7%) showed activity against E. faecium, E. hirae, and E. durans; and 4 strains (0.6%) showed activity against E. faecalis. The remaining 59 bacteriocinogenic strains produced a small zone of bacteriolysis against E. hirae. In this study, we present an analysis of Bac 43, which was active against E. faecalis, E. faecium, E. hirae, E. durans, L. monocytogenes, and L. denitrificans.
MATERIALS AND METHODS
Bacteria, media, and reagents.
The strains and plasmids used in this study are listed in Table 1. A total of 640 VRE clinical isolates were obtained from different patients who had been admitted to the University of Michigan Medical School Hospital, Ann Arbor, between 1994 and 1999. The bacteriocinogenic strains among these isolates were previously classified into three groups on the basis of their bacteriocin activity (25). Of the 636 VRE clinical isolates tested, 21 strains showing bacteriocin activity against E. faecalis, E. faecium, E. hirae, E. durans, L. monocytogenes, and L. denitrificans were used in this study. The indicator strains used for the bacteriocin assay were Staphylococcus aureus FDA209P (32), E. faecalis FA2-2 (8) and OG1S (7), E. faecium BM4105RF (44), E. hirae ATCC 9790 (38), E. durans ATCC 49135, E. raffinosus JCM8733, E. gallinarum BM4174 (27), S. agalactiae, S. pyogenes, and L. monocytogenes. Enterococcus strains were grown in Todd-Hewitt broth (THB; Difco, Detroit, Mich.) or antibiotic medium 3 (Difco). Escherichia coli strains were grown in Luria-Bertani medium. Solid and soft media were prepared by the addition of 1.5% or 0.75% (wt/vol) agar, respectively. All cultures were grown at 37°C. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml for Enterococcus and 50 μg/ml for E. coli; vancomycin, 12.5 μg/ml; rifampin, 25 μg/ml; fusidic acid, 25 μg/ml; streptomycin, 250 μg/ml; gentamicin, 250 μl/ml; spectinomycin, 250 μg/ml; kanamycin, 500 μg/ml for Enterococcus and 40 μg/ml for E. coli; tetracycline, 12.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmid | Genotype or phenotype | Description | Reference or source |
|---|---|---|---|
| Strains | |||
| Enterococcus faecalis | |||
| FA2-2 | Rifr Fusr | Derivative of JH2 | 8 |
| JH2SS | Strr Spcr | Derivative of JH2 | 41 |
| OG1-10 (OG1S) | Strr | Derivative of OG1 | 7 |
| Enterococcus faecium | |||
| BM4105RF | Rifr Fusr | Derivative of plasmid-free E. faecium BM4105 | 44 |
| BM4105SS | Strr Spcr | Derivative of plasmid-free E. faecium BM4105 | 44 |
| VRE82 | pDT1(Bac 43) Kamr Genr Teir Vanr Ampr | Bacteriocinogenic clinical isolate | 44, this study |
| Enterococcus hirae ATCC 9790 | Penicillin susceptible | Wild type | 38 |
| Enterococcus durans ATCC 49135 | Penicillin susceptible | Wild type | |
| Enterococcus raffinosus JCM8733 | Penicillin susceptible | Wild type | |
| Enterococcus gallinarum BM4174 | Penicillin susceptible | Wild type | 27 |
| Staphylococcus aureus FDA209P | Penicillin susceptible | Wild type | 32 |
| Escherichia coli | |||
| DH5α | recA1 endA1 gyrA96 thi-1 relA1 hsdR17 supE44 φ80 lacZΔ M15 | Bethesda Research Laboratories | |
| TH688 | CSH57b thr::Tn5 | 40 | |
| Plasmids | |||
| pDT1 | Bac 43 | Mobilizable plasmid (6.2 kb) | This study |
| pAM401 | Cmr Tcr | E. coli-E. faecalis shuttle vector | 46 |
| pHTβ | Vanr | pMG1-like highly conjugative plasmid (53.7 kb) | 45 |
| pUC18 | AmprlacZ | E. coli vector | Nippon Gene Co. |
Soft-agar assay for bacteriocin production and immunity.
The bacteriocin production assay was performed as described previously (24). Fifty microliters of an overnight culture of the indicator strains grown in antibiotic medium 3 was added to 5 ml of molten soft agar (0.75%), which was then poured onto a THB plate. After solidification, each strain to be tested was inoculated into the soft agar with a toothpick. The halos of inhibition around the inoculated test strains were monitored after overnight culture.
To test immunity to the bacteriocin, a modification of the bacteriocin production test was performed. The indicator strain was used to test immunity. Inhibition of halo formation means that the indicator strain has immunity against the bacteriocin produced by the inoculated strain.
Mating procedures.
Solid-surface matings were performed as previously described (44). Overnight cultures of the donor and the recipient were mixed at a donor/recipient ratio of 1:10, and 10 μl of the mixed culture was dripped onto THB agar without drug. The plates were then incubated overnight (18 h) at 37°C. After incubation, the bacteria grown on the agar plates were scraped off and transferred into 1 ml of fresh broth and then 0.1-ml samples of the suspension were inoculated onto the appropriate selective agar plates. Colonies were counted after 48 h of incubation at 37°C.
Isolation and manipulation of plasmid DNA.
Plasmid DNA was isolated by the alkaline lysis method (36). Plasmid DNA was treated with restriction enzymes and subjected to agarose gel electrophoresis for analysis of DNA fragments. Restriction enzymes were obtained from Nippon Gene (Toyama, Japan); New England Biolabs, Inc. (Massachusetts); and Takara (Tokyo, Japan) and used in accordance with the suppliers' specifications. Agarose was obtained from Wako Chemicals (Osaka, Japan) and used at a 0.8% agarose concentration in agarose gel electrophoresis. DNA fragments were eluted from agarose gels with the Wizard SV Gel and the PCR Clean-Up System (Promega Corporation, Madison, WI). The eluted fragments were ligated to dephosphorylated, restriction enzyme-digested vector DNA with the DNA Ligation Kit Ver.2 (Takara, Tokyo, Japan) and then introduced into E. coli by electrotransformation (16). Transformants were selected on Luria-Bertani medium agar containing suitable antibiotics.
PCR methodology.
The PCR program, with an Ex Taq DNA polymerase (Takara), comprised 2 min at 95°C followed by 30 cycles of 2 min at 95°C, 2 min at 56°C, and 2 min at 72°C and then a final incubation at 4°C with a GeneAmp PCR system 9600 thermal cycler (Perkin-Elmer).
Determination of pDT1 restriction map.
The restriction map of pDT1 was constructed by double digestion and analysis by agarose gel electrophoresis. The restriction enzymes EcoRI, HindIII, EcoRV, XbaI, and BamHI were used for digestion.
Generation of transposon insertional mutants.
Tn5 (Kmr) insertion into pDT1 was performed as described elsewhere (39, 42). pDT1 was introduced into E. coli K-12 TH688 (with Tn5 in the thr locus) (40) by electrotransformation. Ten of the transformants were selected and spread onto selective medium containing 40 μg of kanamycin and 12.5 μg of tetracycline per ml, and the plates were left at room temperature for 10 days. The bacteria which grew on the selective plates were scraped off, and the plasmid DNA was then isolated and used to transform E. coli DH5α. The transformants were selected on plates containing kanamycin (40 μg/ml) and tetracycline (12.5 μg/ml) for selection of Tn5 kanamycin resistance and pTD1-borne tetracycline resistance, respectively. The transformants were purified and examined to determine the specific location of Tn5 within the plasmid. The precise location of the Tn5 insertion was determined by direct nucleotide sequencing with a synthetic primer shown in Table 2, which hybridized to the end of Tn5 (43).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence and restriction sitesa | Description |
|---|---|---|
| Bac43-1 | GAATTCAAAACTACTTTTTATGACG | Analysis of bac determinant |
| Bac43-2 | GAATTCTAGGAACTTGTCTAGCTGG | |
| Bac43-5′ | GAATTCTATGATAATTTTTCGGCTC | |
| Bac43-4′ | GAATTCGATAGTCATCTATAGTTGC | |
| Bac43-6′ | GAATTCAAGCCCATCCTCTATATAC | |
| Tn5 | CAGATTTAGCCCAGTCGG | Analysis of Tn5 insetion mutant |
| J1 | GAGTATTGCAACTTGCTCGC | Analysis of EcoRI junction of pTD1 |
| J2 | GCTACAAGAAGTGGTTCGGC | |
| C | TTGGTACAGGCGTTACTTGG | Analysis of bacA gene |
| E2 | ATCCGAATTCATAACCTCCCTACCACTACC | |
| H1 | CGAAAAGGAAAAACAATCATG | Analysis of bac43 determinants |
| H2 | TCCCATTTTCATTTTATTCC | |
| M1 | AAGGGTGGGACTTATGAGCG | Analysis of mob genes |
| M2 | TTGTTGGTAGTCTGCTCCTC |
Underlined letters indicate restriction sites (GAATTC; EcoRI).
DNA sequence analysis.
Nucleotide sequence analysis was carried out as previously described (37). To determine the entire sequence of pDT1, shotgun sequencing was performed. Fragmented DNA libraries were constructed by sonication of EcoRI-digested pDT1, followed by ligation into the SmaI-digested pUC18 vector plasmid. pUC18 plasmids containing 0.5- to 1.0-kb inserts were used to transform E. coli DH5α. The resulting constructs were sequenced in both orientations with an ABI Prism 377 sequencer (Applied Biosystems). The BigDye Terminator Ver.1.1 cycle sequencing kit (Applied Biosystems) and primers 21M13 and M13Rev (Perkin-Elmer) were used for the sequencing reaction. Open reading frame (ORF) analysis was performed with Genetyx, version 6.1 (Genetyx Corp., Tokyo, Japan). The DNA Data Bank of Japan (DDBJ; National Institute of Genetics, Mishima, Japan) was used for homology analysis of nucleotide and amino acid sequences.
Direct sequencing was performed to confirm the sequence near the EcoRI junction of pDT1 and the structures of the insertion and deletion mutants. The PCR products were eluted from agarose gels as described above and sequenced in both orientations with an ABI Prism 310 sequencer (Applied Biosystems). The BigDye Terminator Ver.1.1 cycle sequencing kit (Applied Biosystems) was used for the sequencing reaction with PCR primers (Table 2).
Deletion mutant analysis.
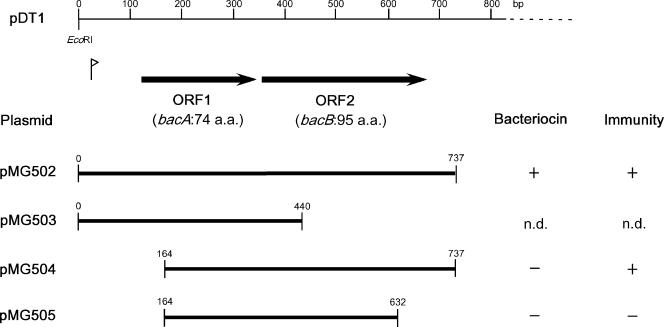
The deletion mutants shown in Fig. 3 were constructed by PCR with pDT1 as the template. The primers used to construct each subclone are listed in Table 2. Subclones of pMG502, pMG503, pMG504, and pMG505 were constructed with primer pairs Bac43-1 and -5′, Bac43-1 and -4′, Bac43-2 and -5′, and Bac43-2 and -6′, respectively. PCR products were digested with EcoRI and cloned into shuttle vector pAM401. Each subclone was introduced into E. faecalis FA2-2 and tested in the soft-agar assay. The sequences of all subclones were confirmed by direct DNA sequencing.
FIG. 3.
Cloning of PCR products from the region of the bacteriocin determinant of pDT1. Thick lines represent the cloned PCR product. The numbers at the ends of the thick lines represent the 5′ and 3′ ends of the segment on the map (base pairs). The vertical bar with an arrowhead is the potential promoter. a.a., amino acids; n.d., the pasmid did not transform E. faecalis FA2-2.
Southern hybridization.
Southern hybridization was performed with the digoxigenin (DIG)-based nonradioisotope system of Boehringer GmbH (Mannheim, Germany), and all procedures were based on the manufacturer's manual and standard protocols (36). Hybridization was performed overnight at 42°C in the presence of 50% formamide. The PCR product generated between primers C and E2 with the PCR DIG synthesis kit (Roche Diagnostics, Mannheim, Germany) was used as the probe for bacA. The nucleotide sequences of the primer pair are shown in Table 2. Signals were detected with the DIG chemiluminescence detection kit (Boehringer GmbH). CSPD (Boehringer GmbH) was used as a substrate for alkali phosphatase conjugated to the anti-DIG antibody.
Pulsed-field gel electrophoresis.
Genomic DNA was prepared as previously described (31). A gel block containing genomic DNA was incubated overnight with 10 U of SmaI. Electrophoresis was then carried out with a 1% agarose gel with 0.5% Tris-borate-EDTA, and the settings applied were 1 to 21 s, 6 V/cm2, and 20 h. The gel was stained with ethidium bromide for UV observation. The results were classified as closely related, possibly related, or different types (31).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this article are available from the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB178871.
RESULTS
Identification of a plasmid-coded bacteriocin active against E. faecalis, E. faecium, E. hirae, E. durans, and L. monocytogenes.
Of the 277 (44%) bacteriocinogenic strains identified among a total of 636 VRE strains, 21 bacteriocinogenic strains that are active against E. faecalis, E. faecium, E. hirae, E. durans, and L. monocytogenes (3.3%) were used in this study. The drug resistance patterns of the 21 strains are shown in Table 3. The enterococcal bacteriocins are usually carried on plasmids, some of which are self-transferable and some of which can be mobilized by coresident conjugative plasmids. To examine whether bacteriocin production was cotransferred with drug resistance, mating experiments were performed between each of the 21 bacteriocinogenic strains and recipient strain E. faecium BM4105RF on a solid surface (filter mating). Vancomycin or gentamicin was used as a selective marker for transconjugants (Table 3), and rifampin and fusidic acid were used for counterselection against the donor strain. Vancomycin- or gentamicin-resistant transconjugants were obtained at frequencies of 10−5 to 10−8 per donor cell with 12 of the 21 strains (Table 3). Bacteriocin activities were examined in the transconjugants from each of these strains. The drug resistance transconjugants exhibited bacteriocin activities at a relatively high frequency (Table 3). The bacteriocin activities were identical to that of the donor strain. Of these bacteriocinogenic strains, VRE82 was chosen as a representative for further analysis. With the VRE and bacteriocinogenic E. faecium BM4105RF transconjugant of VRE82 as the initial donor, repeated experiments to transfer bacteriocin production were performed between E. faecium BM4105RF and E. faecium BM4105SS. Vancomycin resistance was used as a selective marker for the transconjugants. Vancomycin-resistant transconjugants were tested for bacteriocin activity against E. faecalis, E. faecium, E. hirae, E. durans, and L. monocytogenes. About 80 to 95% of the transconjugants were bacteriocinogenic and showed bacteriocin activity identical to that of donor strain VRE82, and the remaining transconjugants showed no bacteriocin activity. Plasmid DNA was isolated from each of the transconjugants, treated with the EcoRI restriction enzyme, and analyzed by agarose gel electrophoresis. All of the nonbacteriocinogenic vancomycin-resistant transconjugants exhibited two major bands, and all of the bacteriocinogenic and vancomycin-resistant transconjugants exhibited an additional DNA band with a molecular size of 6.2 kbp in their agarose gel electrophoresis profiles (Fig. 1).
TABLE 3.
Drug resistance patterns of bacteriocinogenic strains and transferabilities of bacteriocin activity with drug resistance
| Straina | Drug resistance patternb | Transfer frequencyc (% of bacteriocinogenic transconjugants)
|
|
|---|---|---|---|
| Vamr | Genr | ||
| VRE74 | Apc Gen Kan Tei Van | 2 × 10−8 (50) | <1 × 10−8 |
| VRE78 | Apc Gen Kan Tei Van | 2 × 10−6 (100) | <1 × 10−8 |
| VRE82 | Apc Gen Kan Tei Van | 2 × 10−8 (90) | <1 × 10−8 |
| VRE83 | Apc Gen Kan Tei Van | 2 × 10−7 (80) | <1 × 10−8 |
| VRE94 | Apc Gen Kan Tei Van | <1 × 10−8 | <1 × 10−8 |
| VRE252 | Apc Gen Kan Tei Van | <1 × 10−8 | 4 × 10−8 (75) |
| VRE272 | Apc Gen Kan Str Tei Van | <1 × 10−8 | 4 × 10−8 (100) |
| VRE278 | Apc Gen Kan Str Tei Van | 2 × 10−7 (75) | 4 × 10−8 (5) |
| VRE319 | Apc Gen Kan Str Tei Van | 6 × 10−8 (100) | 4 × 10−8 (35) |
| VRE330 | Apc Gen Kan Str Tei Van | <1 × 10−8 | <1 × 10−8 |
| VRE351 | Apc Gen Kan Str Tei Van | 2 × 10−7 (75) | 9 × 10−5 (8) |
| VRE367 | Apc Kan Str Tei Van | 1 × 10−7 (100) | NTd |
| VRE418 | Apc Kan Str Tei Van | <1 × 10−8 | NT |
| VRE419 | Apc Gen Kan Str Tei Van | <1 × 10−8 | <1 × 10−8 |
| VRE424 | Apc Gen Kan Str Tei Van | <1 × 10−8 | 3 × 10−8 (50) |
| VRE437 | Apc Gen Kan Str Tei Van | <1 × 10−8 | <1 × 10−8 |
| VRE455 | Apc Gen Kan Str Tei Van | <1 × 10−8 | <1 × 10−8 |
| VRE477 | Apc Gen Kan Str Tei Van | <1 × 10−8 | 2 × 10−8 (75) |
| VRE506 | Apc Kan Mino Tet Tei Van | <1 × 10−8 | NT |
| VRE576 | Apc Gen Kan Tet Tei Van | <1 × 10−8 | <1 × 10−8 |
| VRE595 | Apc Gen Kan Tet Tei Van | <1 × 10−8 | <1 × 10−8 |
The strains exhibited bacteriocin activity against E. faecalis, E. faecium, E. durans, L. monocytogenes, and L. denitrificans.
Abbreviations: Apc, ampicillin; Gen, gentamicin; Kan, kanamycin; Mino, minocycline; Str, streptomycin; Tet, tetracycline; Tei, teicoplanin; Van, vancomycin.
The frequency was calculated as the number of selected transconjugants per donor cell.
NT, not tested.
FIG. 1.
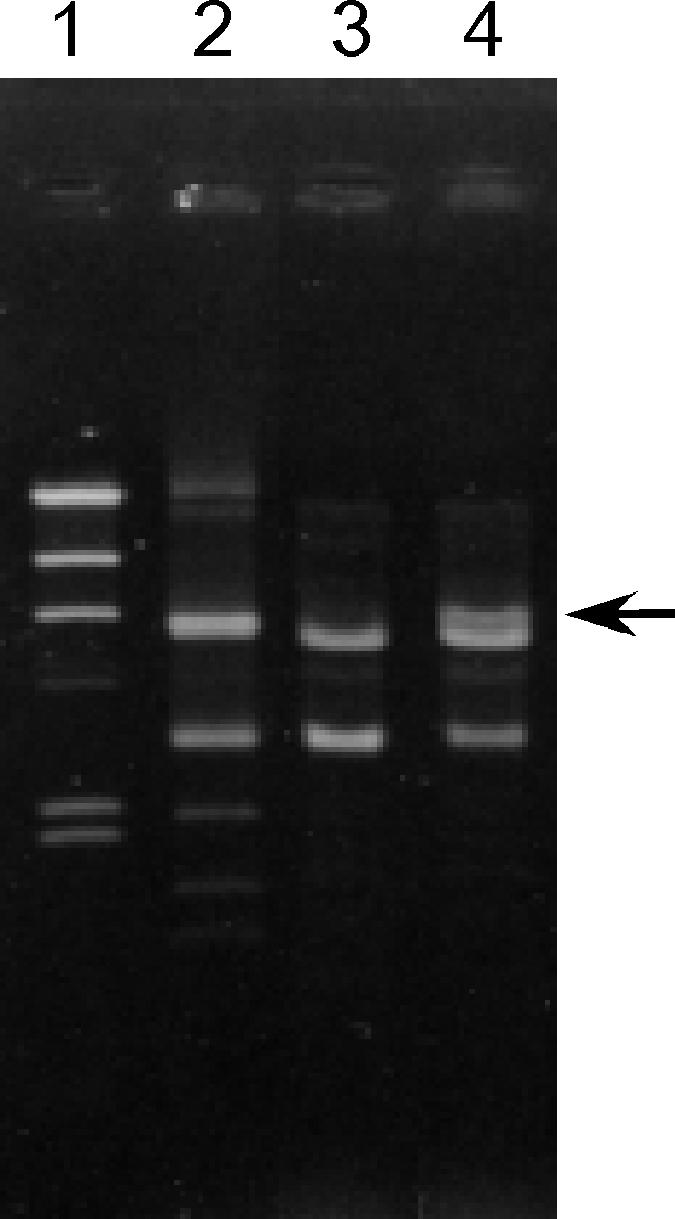
Agarose gel electrophoresis of EcoRI-digested plasmid DNAs of bacteriocinogenic strain VRE82 and transconjugants. Lanes: 1, HindIII-digested lambda DNA; 2, E. faecium VRE82 (wild-type VRE strain); 3, nonbacteriocinogenic VRE BM4105RF transconjugant; 4, bacteriocinogenic VRE BM4105RF transconjugant. Arrow, 6.2-kb band.
The 6.2-kb EcoRI fragment obtained by agarose gel electrophoresis was eluted from the agarose gel and ligated with shuttle vector pAM401. The cloned 6.2-kbp EcoRI fragment was introduced into E. coli DH5α, and the clone pAM401::6.2-kbp EcoRI fragment was designated pMG501. E. faecalis FA2-2 and E. hirae ATCC 9790 were transformed with pMG501. The transformants expressed bacteriocin activity identical to that of wild-type strain VRE82. These results implied that bacteriocinogenic VRE82 harbored a 6.2-kbp plasmid that conferred bacteriocin activity and had one EcoRI site. The 6.2-kbp plasmid was designated pDT1, and the bacteriocin encoded by pDT1 was designated Bac 43.
DNA sequence of pDT1.
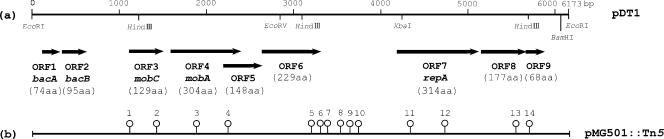
The DNA sequence of pDT1 was determined with plasmid pMG501. There was a possibility that another small EcoRI fragment lay in the gap formed by the single EcoRI site of pDT1, but this was too small to detect by agarose gel electrophoresis. PCR was also performed with the plasmid DNAs of VRE82 and the J1 and J2 primers (Table 2), which lie on either side of the single EcoRI site of pDT1 (Fig. 2). Sequence analysis of the PCR products confirmed that there was no other fragment lying in the gap formed by the EcoRI site of the 6.2-kbp plasmid. pDT1 was found to be 6,173 bp in length. Computer analysis revealed the presence of eight ORFs (ORF1 to ORF8) in pDT1, and all were oriented in the same direction. Figure 2a shows the ORFs that had a good ribosome binding site within a 20-base region upstream of the predicted start codon. Homology analysis of each ORF was performed with the DDBJ data bank. Each of the predicted proteins encoded by ORF5, ORF6, ORF8, and ORF9 showed no significant homology to the reported proteins. The ORF7 protein showed significant homology to the replication proteins of the plasmid found in gram-positive bacteria and designated the repA gene of pTD1. The analyses of the remaining four ORFs (ORF1 to ORF4) are described later.
FIG. 2.
Physical map of pDT1 showing deduced ORFs and transposon insertions into pDT1 of pMG501 (pAM401::pDT1). (a) Physical map of pDT1 (6.2 kbp) and deduced ORFs. Thick horizontal arrows indicate ORFs on pDT1 and the direction of transcription. (b) Map of Tn5 insertions into pDT1 of pAM401::pDT1. Open circles indicate Tn5 insertion mutants. Numbers beside symbols are mutant identification numbers. aa, amino acids.
Generation of Tn5 insertion mutants.
Tn5 insertion mutant forms of the pMG501 clone containing pDT1 were generated. Fourteen insertions in pDT1 were obtained (Fig. 2b). Inserts were obtained in each of the ORFs, except ORF1 and ORF2. All of the insertion mutants expressed bacteriocin activity and immunity at the same level as wild-type pMG501 in an E. faecium BM4105RF background with respect to the bacteriocin activity obtained by soft-agar assay. The result implied that seven ORFs (ORF3 to ORF9) were not related to the expression of Bac 43. Although we could not exclude any potential polar effects on the adjacent gene(s) by transposon insertion, it was probable that ORF1 and ORF2 were the bacteriocin determinant.
Cloning of PCR products that confer bacteriocin production.
The PCR products that corresponded to the 0- to 700-bp region of the map position and contained ORF1 and ORF2 were cloned into pAM401. Transformation of E. faecalis FA2-2 was performed with pAM401 carrying the PCR products. The transformants were selected on a selective agar plate containing chloramphenicol for selection of pAM401 and examined for bacteriocin activity. The results are shown in Fig. 3. pMG502 carried a 737-bp fragment and contained both ORF1 and ORF2. E. faecalis FA2-2 containing pMG502 expressed bacteriocin activity and immunity. pMG503 contains ORF1 and the N-terminal region of ORF2. pMG503 could not transform E. faecalis FA2-2. pMG504 had a deletion in the N-terminal region of ORF1 and contained the C-terminal region of ORF1 and all of ORF2. E. faecalis FA2-2 containing pMG504 did not express bacteriocin activity but expressed immunity. pMG505 had a deletion in the N-terminal region of ORF1 and the C-terminal region of ORF2 and contained the C-terminal region of ORF1 and the N-terminal region of ORF2. E. faecalis FA2-2 containing pMG505 expressed neither bacteriocin activity nor immunity. These results indicated that the fragment containing both ORF1 and ORF2 conferred bacteriocin activity and immunity on the E. faecalis strain. ORF1 encoded the bacteriocin, and ORF2 encoded immunity against this bacteriocin. ORF1 and ORF2 were designated bacA and bacB, respectively.
DNA sequence analysis of ORF1 (bacA) and ORF2 (bacB).
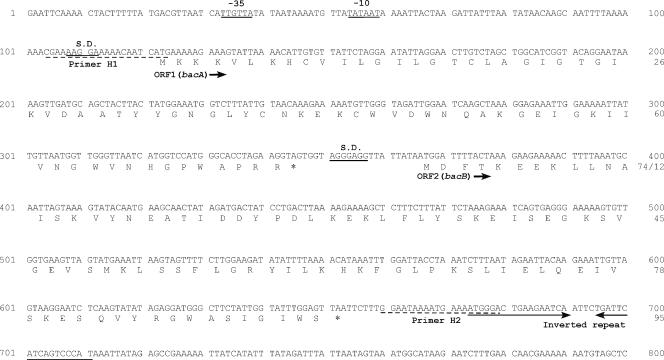
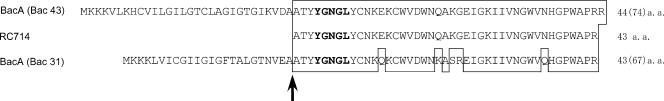
A homology search of bacA and bacB was performed against the DDBJ protein database. bacA encoded a 74-amino-acid protein. The ATG start codon was preceded by a potential Shine-Dalgarno (S.D.) ribosome binding site (AAGGAA) at a location 9 bp upstream (Fig. 4). The deduced BacA protein had a span of hydrophobic residues typical of a signal sequence in its N-terminal region, and a potential signal peptidase processing site corresponding to the V-D-A sequence was located at positions 28 to 30 (Fig. 5). Comparison of the primary structure of the deduced amino acid sequence of the mature BacA protein, which was composed of 44 amino acid residues, showed 50 to 86% homology with the mature Bac 31 (42), enterocin SE-K4 (13), enterocin P (4), divercin V41 (30), and listerocin 743A (26) proteins, which belong to the class IIa bacteriocins produced by LAB (14, 22), and 98% homology with Bac RC714 (43 amino acids), which corresponds to a protein lacking the last amino acid residue (44th Arg) of Bac 43 (10). As with the class IIa bacteriocins, the BacA protein had a hydrophobic N-terminal region, contained the consensus sequence Tyr-Gly-Asn-Gly-Lys(Val) (YGNGL[V]), and had a relatively hydrophilic C-terminal region (Fig. 5). The putative signal sequences (30-amino-acid sequences) did not show any significant homology with any other reported proteins or leader peptides.
FIG. 4.
Nucleotide sequence of bacA and bacB of bacteriocin 43 and deduced amino acid sequence. Potential promoters (−10 and −35) and S.D. ribosome binding sequences are underlined. The inverted repeat sequence is indicated by horizontal arrows. Primers H1 and H2, which were used for PCR analysis of the bac43 determinant in clinical isolates, are indicated by dashed lines. The accession number is AB178871.
FIG. 5.
Comparison of the amino acid sequence of the predicted BacA protein of bacteriocin 43 with the amino acid sequences of homologous bacteriocins. The sequences of the predicted BacA protein and other class IIa bacteriocins are shown. The consensus sequence Tyr-Gly-Asn-Gly-Lys (Val) (YGNGL[V]) of class IIa bacteriocins is indicated in boldfaced letters. The vertical arrow and dashed line indicate the cleavage site in the prebacteriocins. Identical amino acids (a.a.) are boxed.
bacB encoded a 95-amino-acid deduced protein without a putative signal sequence. The ATG start codon was preceded by a potential S.D. ribosome binding site (AGGGAG) at a location 9 bp upstream (Fig. 4). Comparison of the primary structure of the deduced amino acid sequence of the BacB protein showed 50% and 25% homology with the immunity proteins of bacteriocin 31 and enterocin SE-K4, respectively.
Identification of a Bac 43 determinant in VRE strains producing the same bacteriocin spectrum as Bac 43.
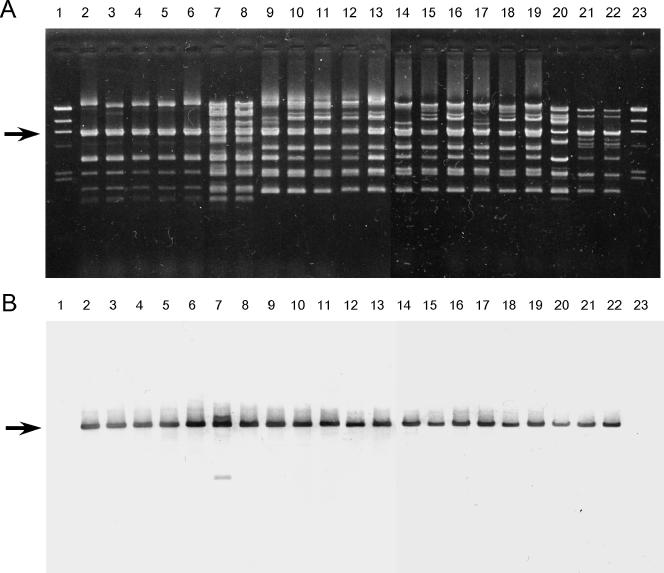
Plasmid DNAs isolated from each of the 21 VRE strains that showed the same bacteriocin activity as that of Bac 43 were examined for the presence of the bac43 determinant by PCR analysis with specific primers H1 and H2 for the bacA and bacB genes of bac43, respectively. The PCR primers are shown in Table 2 and Fig. 4. The 21 strains gave rise to the expected 576-bp product of bacA and bacB by PCR analyses (data not shown). Each of the PCR products specific for the bacA and bacB genes was sequenced. The nucleotide sequences of the genes from the strains were identical to those of the genes carried by pDT1 (data not shown). These indicated that the Bac 43-type bacteriocinogenic 21 strains carried bacA and bacB of bacteriocin 43 on the plasmid. Plasmid DNAs isolated from each of the 21 VRE strains were also examined for the presence of the bac43 determinant by Southern analysis with a specific probe (Table 2). EcoRI fragments of plasmid DNAs from each of the 21 strains were separated by agarose gel electrophoresis (Fig. 6A). The gel was Southern blotted and hybridized with the bac43 determinant (Fig. 6B). The probe hybridized to a specific EcoRI fragment with a molecular size of 6.2 kbp in each of the 21 strains that had been confirmed to carry the bac43 determinant by PCR analysis and DNA sequencing.
FIG. 6.
EcoRI-digested plasmid DNAs isolated from 21 VRE strains that showed bacteriocin activity against E. faecalis, E. faecium, E. hirae, E. durans, and L. monocytogenes. (A) Agarose gel electrophoresis of EcoRI-digested plasmid DNAs. (B) The gel was Southern blotted and hybridized with the bacA probe. Lanes: 1 and 23, HindIII-digested lambda DNA; 2 to 22, strains 74, 78, 82, 83, 94, 252, 272, 278, 319, 330, 351, 367, 418, 419, 424, 437, 455, 477, 506, 576, and 595, respectively. Arrows, 6.2-kb bands.
The banding patterns obtained by pulsed-field gel electrophoresis of SmaI-digested genomic DNA(s) were used to compare the 21 strains, which showed 12 different patterns. These data suggested that the Bac 43-type bacteriocin of each of the 21 strains was encoded on a pDT1-type plasmid and that the pDT1-type plasmid had been disseminated among different E. faecium VRE strains in the clinical environment.
Identification of the mobilization determinant.
To examine the determinant for the mobilization of pDT1, each of the Tn5 insertion mutant forms of pMG501 in E. faecalis FA2-2 shown in Fig. 2b was tested for the ability to be mobilized by the coresident vancomycin resistance-encoding conjugative plasmid pHTβ (63.7 kb) (45) (Table 4). E. faecalis JH2SS was used as the recipient strain. Each insertion mutant ORF, with the exceptions of ORF3 and ORF4, was mobilized by the pHTβ plasmid (Table 4). These results implied that ORF3 and ORF4 conferred the ability to mobilize the pDT1 plasmid.
TABLE 4.
Mobilization of Tn5 insertion mutant forms of pDT1 (pMG501::Tn5) by pHTβa
| # or Strain no.b | Plasmid | Location of Tn5 insertion | Position of insertion (bp) on pDT1 map | Transfer frequencyc
|
|
|---|---|---|---|---|---|
| Cmr | Vanr | ||||
| Vector | pAM401 | <1.0 × 10−6 | 6 × 10−2 | ||
| Wild type | pMG501 (pAM401::pDT1) | 9.5 × 10−4 | 5 × 10−2 | ||
| 1 | pMG501-01 | mobC (ORF3) | 1106 | <1.0 × 10−6 | 3 × 10−2 |
| 2 | pMG501-02 | mobC (ORF3) | 1420 | <1.0 × 10−6 | 2 × 10−2 |
| 3 | pMG501-03 | mobA (ORF4) | 1884 | <1.0 × 10−6 | 4 × 10−2 |
| 4 | pMG501-04 | mobA (ORF4) and ORF5 | 2228 | <1.0 × 10−6 | 6 × 10−2 |
| 5 | pMG501-05 | ORF6 | 3197 | 1.2 × 10−4 | 5 × 10−2 |
| 6 | pMG501-06 | ORF6 | 3296 | 4.1 × 10−5 | 6 × 10−2 |
| 7 | pMG501-07 | Downstream of ORF6 | 3338 | 2.2 × 10−4 | 8 × 10−2 |
| 8 | pMG501-08 | Between ORF6 and ORF7 | 3535 | 1.1 × 10−3 | 5 × 10−2 |
| 9 | pMG501-09 | Between ORF6 and ORF7 | 3641 | 3.4 × 10−4 | 3 × 10−2 |
| 10 | pMG501-10 | Between ORF6 and ORF7 | 3725 | 2.1 × 10−4 | 4 × 10−2 |
| 11 | pMG501-11 | ORF7 | 4329 | 1.0 × 10−4 | 4 × 10−2 |
| 12 | pMG501-12 | ORF7 | 4730 | 1.2 × 10−4 | 6 × 10−2 |
| 13 | pMG501-13 | ORF8 | 5546 | 2.0 × 10−4 | 5 × 10−2 |
| 14 | pMG501-14 | ORF9 | 5703 | 4.0 × 10−4 | 5 × 10−2 |
Mating experiments were performed with E. faecalis FA2-2 carrying plasmids pMG501::Tn5 and pHTβ as the donor strain and E. faecalis JH2SS as the recipient strain. The donor strain harbored both pHTβ (Vanr) as a mobilizer plasmid and each of the pAM401 derivatives (Cmr) containing a Tn5 insertion mutant form of pMG501 as the tester plasmid.
The pMG501 derivative numbers correspond to the insertion mutant numbers in Fig. 2b.
The frequency was calculated as the number of selected transconjugants per donor cell.
DNA sequence analysis of ORF3 and ORF4 was performed by DDBJ against the protein database. ORF3 encoded a 129-amino-acid protein. The GTG start codon was preceded by a potential S.D. ribosome binding site (AGGA) at a location 13 bp upstream. ORF4 encoded a 304-amino-acid protein. The ATG start codon was preceded by an S.D. ribosome binding site (AAGGAG) at a location 12 bp upstream. Comparison of the primary structures of the deduced amino acid sequences of the ORF3 and ORF4 proteins showed 55% homology with the MobC protein encoded by S. aureus plasmid pRJ9 (35) and 45% homology with the MobA protein encoded by E. faecalis plasmid pEF1071 (2), respectively. The reported MobC and MobA proteins were the relaxosome and nickase for plasmid DNA, respectively (2, 35). ORF3 and ORF4 were designated mobC and mobA, respectively.
Identification of the mobilization determinant in 21 VRE strains producing the same bacteriocin spectrum as Bac 43.
Plasmid DNAs isolated from each of the 21 VRE strains that showed the same bacteriocin activity as Bac 43 were examined for the presence of the mobilization determinant by PCR analysis with primers M1 and M2, which are specific for mobC and mobA, respectively. The PCR primers are shown in Table 2. The 21 strains gave rise to the expected 1,274-bp product by PCR analysis (not shown). This suggested that all of the 21 strains producing the same bacteriocin spectrum as Bac 43 possessed the mobC and mobA genes on a pDT1-type plasmid.
Analysis of the Bac 43 determinant in vancomycin-sensitive E. faecium and E. faecalis isolates.
The plasmid DNAs of 149 vancomycin-sensitive E. faecium and E. faecalis isolates were examined for the presence of the bac43 determinant by PCR analysis with primers specific for the bacA and bacB genes of bac43. Of the 149 isolates tested, 46 E. faecium isolates were isolated from healthy Japanese medical students between 2002 and 2003 and 56 E. faecium isolates and 47 E. faecalis isolates were isolated at Gunma University Hospital, Japan, between 1990 and 1993. One E. faecium strain from a student gave rise to the expected 576-bp product and produced a bacteriocin with the same spectrum as Bac 43. The bac43 determinant was not identified in other strains.
DISCUSSION
Bac 43 was identified in the VanA-type VRE strain designated VRE82. Bac 43 was active against E. faecalis, E. faecium, E. hirae, E. durans, and L. monocytogenes strains and was carried by plasmid pDT1 (6.2 kbp), which was efficiently mobilized to the recipient E. faecalis or E. faecium strain at a frequency of 10−5 to 10−7 per donor cell with the coresident conjugative vancomycin resistance plasmid. The Bac 43 determinant consisted of the bacteriocin structural gene bacA and the immunity gene bacB.
The deduced mature BacA protein showed 86% homology with the mature Bac 31 protein isolated from an E. faecalis strain (42) and 98% homology with the mature Bac RC714 protein isolated from VRE RC714 (10). RC714 is a 43-amino-acid protein and is identical to the mature BacA protein but lacks the last residue (44th Arg) at the C-terminal region. There was no homology between the deduced amino acid sequence of the leader peptides of BacA of Bac 43 and Bac 31 (Fig. 5). The deduced BacB protein of Bac 43 showed 50% homology with the BacB protein of Bac 31. Bac 31 is active against E. faecium, E. hirae, E. durans, and L. monocytogenes but is not active against E. faecalis (42). This implied that the six-amino-acid difference in the bacteriocin proteins of Bac 31 and Bac 43 resulted in the different bacteriocin activity spectra, as well as differences in the immunity proteins, as an adaptation in their bacteriocin activities.
Bacteriocinogenic E. faecium strain RC714 has been isolated from a VanA-type resistant E. faecium VRE clinical isolate (10). Mature Bac RC714 has been purified and characterized (10). As described above, the deduced BacA protein of Bac 43 showed 98% homology with Bac RC714 and was almost identical to RC714. Bac RC714 has been isolated only from one E. faecium VRE clinical isolate, and Bac 43 was also isolated only from VRE isolates, with the exception of one isolate from a healthy student. These data suggested that there would be a tendency for Bac RC714 or the Bac 43-type bacteriocin to be isolated in VRE clinical isolates than in vancomycin-sensitive isolates.
Two main types of bacteriocins were identified in the 277 (44%) bacteriocinogenic strains of the 636 VRE strains that were tested, and they were classified according to their bacteriocin activities (25). Bac 32 and Bac 32-type bacteriocins, which are active against E. faecium, E. hirae, and E. durans and are determined by bac32, were identified in 193 (70%) of the 277 bacteriocinogenic VRE strains (25). The other type of bacteriocin that was identified is active against E. faecalis, E. faecium, E. hirae, E. durans, and L. monocytogenes and was detected in 21 (3.3%) of the 277 bacteriocinogenic VRE strains (25). In this study, we showed that Bac 43 was representative of the Bac 43-type bacteriocins produced by the 21 bacteriocinogenic VRE isolates. The Bac 43 or Bac 43-type bacteriocinogenic VRE strains were the second most prevalent isolates after the Bac 32 or Bac32-type bacteriocinogenic VRE strains. However, the isolation frequency of Bac 43 or Bac 43-type bacteriocinogenic strains was far lower than that of the Bac 32 and Bac 32-type bacteriocinogenic strains. Both bacteriocins are carried by mobilizable plasmids and could be efficiently transferred to another strain by conjugative plasmids harbored by the VRE strains. The bacteriocinogenic VRE strains showed multiple-drug resistance. These characteristics indicated that Bac 32- and Bac 43-type bacteriocinogenic strains might have the same selective advantage in a clinical environment. The only difference in bacteriocin activity between Bac 43 and Bac 32 was that Bac 43 was active against E. faecalis and L. monocytogenes, whereas Bac 32 was not (25).
The well-characterized E. faecium bacteriocins (i.e., enterocins) are produced by food grade organisms that have been isolated from fermented foods (1, 3-6, 15, 19). Bacteriocinogenic food grade organisms are characteristically active against L. monocytogenes (34), which is a frequent cause of food-borne listeriosis (20). These food grade bacteriocinogenic E. faecium strains might have a selective advantage in their particular ecological niche. Bac 32 is not active against L. monocytogenes and is prevalent among the bacteriocins in E. faecium clinical isolates (25). The present study supports the previous hypothesis that the dominant type of bacteriocin in E. faecium clinical isolates might differ from the dominant type of bacteriocin found in food grade E. faecium isolates, which are active against L. monocytogenes (25).
Acknowledgments
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology [Tokuteiryoiki (C), Kiban (B), (C)] and the Japanese Ministry of Health, Labor and Welfare (H15-Shinko-9, H18-Shinko-11).
We thank Elizabeth Kamei for revising the manuscript.
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Aymerich, T., H. Holo, L. S. Havarstein, M. Hugas, M. Garriga, and I. F. Nes. 1996. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62:1676-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balla, E., and L. M. T. Dicks. 2005. Molecular analysis of the gene cluster involved in the production and secretion of enterocins 1071A and 1071B and of the genes responsible for the replication and transfer of plasmid pEF1071. Int. J. Food Microbiol. 99:33-45. [DOI] [PubMed] [Google Scholar]
- 3.Casaus, P., T. Nilsen, L. M. Cintas, I. F. Nes, P. E. Hernandez, and H. Holo. 1997. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 143:2287-2294. [DOI] [PubMed] [Google Scholar]
- 4.Cintas, L. M., P. Casaus, L. S. Havarstein, P. E. Hernandez, and I. F. Nes. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cintas, L. M., P. Casaus, H. Holo, P. E. Hernandez, I. F. Nes, and L. S. Havarstein. 1998. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J. Bacteriol. 180:1988-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food 71:1-20. [DOI] [PubMed] [Google Scholar]
- 7.Clewell, D. B., and B. L. Brown. 1980. Sex pheromone cAD1 in Streptococcus faecalis: induction of a function related to plasmid transfer. J. Bacteriol. 143:1063-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, C. R., P. S. Coburn, and M. S. Gilmore. 2005. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Protein Pept. Sci. 6:77-84. [DOI] [PubMed] [Google Scholar]
- 10.del Campo, R., C. Tenorio, R. Jiménez-díaz, C. Rubio, R. Gómez-Lui, F. Baquero, and C. Torres. 2001. Bacteriocin production in vancomycin-resistant and vancomycin-susceptible Enterococcus isolates of different origins. Antimicrob. Agents Chemother. 45:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vuyst, L., M. R. Foulquie Moreno, and H. Revets. 2003. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 84:299-318. [DOI] [PubMed] [Google Scholar]
- 12.De Vuyst, L., and E. J. Vandamme. 1994. Antimicrobial potential of lactic acid bacteria, p. 91-142. In L. De Vurst and E. J. Vandamme (ed.), Bacteriocins of lactic acid bacteria: microbiology, genetics and applications. Blackie Academic & Professional, London, United Kingdom.
- 13.Eguchi, T., K. Kaminaka, J. Shima, S. Kawamoto, K. Mori, S.-H. Choi, K. Doi, S. Ohmomo, and S. Ogata. 2001. Isolation and characterization of enterocin SE-K4 produced by thermophilic enterococci, Enterococcus faecalis K-4. Biosci. Biotechnol. Biochem. 65:247-253. [DOI] [PubMed] [Google Scholar]
- 14.Eijsink, V. G. H., L. Axelsson, D. B. Diep, L. S. Håvarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Leeuwenhoek 82:639-654. [DOI] [PubMed] [Google Scholar]
- 15.Floriano, B., J. L. Ruiz-Barba, and R. Jimenez-Diez. 1998. Purification and genetic characterization of enterocin I from Enterococcus faecium 6T1a, a novel antilisterial plasmid-encoded bacteriocin which does not belong to the pediocin family of bacteriocins. Appl. Environ. Microbiol. 64:4883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto, S., H. Hashimoto, and Y. Ike. 1991. Low cost device for electrotransformation and its application to the highly efficient transformation of Escherichia coli and Enterococcus faecalis. Plasmid 26:131-135. [DOI] [PubMed] [Google Scholar]
- 17.Gilmore, M. S., R. A. Segarra, and M. C. Booth. 1990. An HlyB-type function is required for expression of the Enterococcus faecalis hemolysin/bacteriocin. Infect. Immun. 58:3914-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmore, M. S., R. A. Segarra, M. C. Booth, C. P. Bogie, L. R. Hall, and D. B. Clewell. 1994. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176:7335-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giraffa, G. 2002. Enterococci from foods. FEMS Microbiol. Rev. 26:163-171. [DOI] [PubMed] [Google Scholar]
- 20.Giraffa, G. 2003. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 88:215-222. [DOI] [PubMed] [Google Scholar]
- 21.Haas, W., B. D. Shepard, and M. S. Gilmore. 2002. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415:84-87. [DOI] [PubMed] [Google Scholar]
- 22.Hechard, Y., and H. G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from gram-positive bacteria. Biochimie 84:545-557. [DOI] [PubMed] [Google Scholar]
- 23.Ike, Y., and D. B. Clewell. 1984. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J. Bacteriol. 158:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ike, Y., D. B. Clewell, R. A. Segarra, and M. S. Gilmore. 1990. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J. Bacteriol. 172:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue, T., H. Tomita, and Y. Ike. 2006. Bac 32, a novel bacteriocin widely disseminated among clinical isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 50:1202-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalmokoff, M. L., S. K. Banerjee, T. Cry, M. A. Hefford, and T. Gleeson. 2001. Identification of a new plasmid-encoded sec-dependent bacteriocin produced by Listeria innocua 743. Appl. Environ. Microbiol. 67:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leclercq, R., S. Dutka-Malen, J. Duval, and P. Courvalin. 1992. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob. Agents Chemother. 36:2005-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maqueda, M., A. Galvez, M. M. Bueno, M. J. Sanchez-Barrena, C. Gonzalez, A. Albert, M. Rico, and E. Valdivia. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein Pept. Sci. 5:399-416. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Bueno, M., A. Galbez, E. Valdivia, and M. Maqueda. 1990. A transferable plasmid associated with AS-48 production in Enterococcus faecalis. J. Bacteriol. 172:2817-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Métivier, A., M.-F. Pilet, X. Dousset, O. Sorokine, P. Anglade, M. Zagorec, J.-C. Piaed, D. Marion, Y. Cenatiempo, and C. Fremaux. 1998. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergence V41: primary structure and genomic organization. Microbiology 144:2837-2844. [DOI] [PubMed] [Google Scholar]
- 31.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, T., N. Yamazaki, H. Tamiguchi, and S. Fujimura. 1983. Production, purification, and properties of a bacteriocin from Staphylococcus aureus isolated from saliva. Infect. Immun. 39:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 34.Nes, I. F., and H. Holo. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 55:50-61. [DOI] [PubMed] [Google Scholar]
- 35.Netz, D. J. A., R. Pohl, A. G. Beck-Sickinger, T. Selmer, A. J. Pierik, M. C. F. Bactos, and H. G. Sahl. 2002. Biochemical characterization and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 319:745-756. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solioz, M., and M. Waser. 1990. Efficient electrotransformation of Enterococcus hirae with a new Enterococcus-Escherichia coli shuttle vector. Biochimie 72:279-283. [DOI] [PubMed] [Google Scholar]
- 39.Tanimoto, K., and D. B. Clewell. 1993. Regulation of the pAD1-encoded sex pheromone response in Enterococcus faecalis: expression of the positive regular TraE1. J. Bacteriol. 175:1008-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanimoto, K., and T. Iino. 1985. Additional genes essential for replication of the mini-F plasmid from origin I. Mol. Gen. Genet. 198:358-359. [DOI] [PubMed] [Google Scholar]
- 41.Tomich, P. K., F. Y. An, and D. B. Clewell. 1980. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1996. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J. Bacteriol. 178:3585-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1997. Cloning and genetic and sequence analysis of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 179:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomita, H., C. Pierson, S. K. Lim, D. B. Clewell, and Y. Ike. 2002. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J. Clin. Microbiol. 40:3326-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomita, H., K. Tanimoto, S. Hayakawa, K. Morinaga, K. Ezaki, H. Oshima, and Y. Ike. 2003. Highly conjugative pMG1-like plasmids carrying Tn1546-like transposons that encode vancomycin resistance in Enterococcus faecium. J. Bacteriol. 185:7024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yagi, Y., R. E. Kessler, J. H. Shaw, D. E. Lopatin, F. An, and S. B. Clewell. 1983. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J. Gen. Microbiol. 129:1207-1215. [DOI] [PubMed] [Google Scholar]