Abstract
Bacillus thuringiensis strains isolated from Latin American soil samples that showed toxicity against three Spodoptera frugiperda populations from different geographical areas (Mexico, Colombia, and Brazil) were characterized on the basis of their insecticidal activity, crystal morphology, sodium dodecyl sulfate-polyacrylamide gel electrophoresis of parasporal crystals, plasmid profiles, and cry gene content. We found that the different S. frugiperda populations display different susceptibilities to the selected B. thuringiensis strains and also to pure preparations of Cry1B, Cry1C, and Cry1D toxins. Binding assays performed with pure toxin demonstrated that the differences in the toxin binding capacities of these insect populations correlated with the observed differences in susceptibility to the three Cry toxins analyzed. Finally, the genetic variability of the three insect populations was analyzed by random amplification of polymorphic DNA-PCR, which showed significant genetic diversity among the three S. frugiperda populations analyzed. The data presented here show that the genetic variability of S. frugiperda populations should be carefully considered in the development of insect pest control strategies, including the deployment of genetically modified maize in different geographical regions.
Fall armyworm, Spodoptera frugiperda (J. E. Smith), is a worldwide pest of economic importance for different crops. This species has a highly polyphagous feeding behavior, which includes the consumption of different cultivated plants, such as maize (Zea mays L.), cotton (Gossypium hirsutum L.), and rice (Oryza sativa L.). To date, the most common method for controlling this pest relies on the use of synthetic insecticides such as methomyl, carbaryl, and cypermethrin (10), in spite of the damage they cause to the environment and to nontarget organisms. Two distinct strains of S. frugiperda, one associated with maize and the other with rice, have been already identified in the United States (17, 19). The detection and characterization of genetic diversity among insect populations is a critical issue for the improvement of pest management strategies, since the evolution of resistance to insecticides among insect populations is dependent on the frequency of resistant alleles, the inheritance of resistance, the relative fitness cost, and the gene flow.
Bioinsecticides are viable alternatives for insect control in agriculture, and among them, Bacillus thuringiensis is the most widely used. B. thuringiensis is compatible with sustainable and environmentally friendly agricultural practices. This bacterium produces insecticidal proteins (Cry protoxins) during sporulation as parasporal crystals, which are highly specific to their target insects; safe for humans, other vertebrates, and plants; and biodegradable (12). Moreover, recombinant DNA technology using cry genes has developed insect-resistant transgenic plants that are used extensively for cotton, corn, and rice production, among others (23).
Information regarding the susceptibility of S. frugiperda to the Cry protein family is limited. The Cry proteins most active against this pest were reported to be the Cry1C and Cry1D toxins (2, 3), with 50% lethal concentrations (LC50) of 31 and 77 ng/cm2, respectively. The Cry1C toxin has also been reported to be toxic against Spodoptera exigua (28), with an LC50 of 68 ng/cm2, and Cry1D has been reported to be slightly active against Spodoptera littoralis, with an LC50 of 423 ng/cm2 (27).
In this study, several B. thuringiensis strains active against larvae of S. frugiperda were identified and characterized, showing different combinations of known cry genes. Differences in susceptibility were found among three S. frugiperda populations from different Latin American countries (Mexico, Colombia, and Brazil) when selected B. thuringiensis strains and pure Cry1B, Cry1C, and Cry1D proteins were analyzed. The three insect strains were collected from fields of maize crops that had never been treated with Bt spray formulations or planted with transgenic crops. These colonies had been reared under laboratory conditions for at least 10 years without exposure to Cry toxins. The differences in susceptibility among the three S. frugiperda populations correlated with differences in the binding of toxin to midgut microvillar membranes from these insect populations and with the molecular variability found by RAPD (random amplification of polymorphic DNA) analysis of their DNA.
MATERIALS AND METHODS
Bacterial strains.
More than 6,000 bacterial strains from different B. thuringiensis strain collections, from Colombia, Mexico, Costa Rica, and Brazil, were analyzed. Strains were isolated from soil samples by the acetate selection method (25). Soil samples were collected from the surface to a depth of 10 cm. All bacterial strains were grown in M-one liquid sporulation medium (20) at 200 rpm and 30 ± 1°C for 48 h until complete autolysis. Lyophilized spore-crystal complexes were used in the bioassays.
Bioassays.
S. frugiperda colonies were maintained on an artificial diet (24) under laboratory conditions at 28 ± 2°C and 65% ± 5% relative humidity, under a 12:12 (light-dark) photoperiod at CIB (Colombia), IBT-UNAM (Mexico), and RGB-EMBRAPA (Brazil). Ten different toxin concentrations were tested, plus a tap water negative control. Twenty-four neonate larvae were assayed per toxin concentration, with four replicates. A constant volume of the sample dilution (35 μl) was applied to the diet surface contained in 24-well polystyrene plates (Cell Wells; Corning Glass Works, NY). One first-instar larva was added per well, and mortality was recorded after 7 days of incubation under laboratory conditions. The concentration at which 50% of the larvae died (i.e., the mean LC50) was estimated by probit analysis (7).
Protein electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously (22). Concentrated spore-crystal suspensions in Laemmli sample loading buffer 2X (0.125 M Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.01% bromophenol blue) were boiled 5 min, and 5 to 20 μl was loaded in each well. Protein standards were carbonic anhydrase (29 kDa), ovalbumin (45 kDa), bovine serum albumin (66.2 kDa), phosphorylase B (97.4 kDa), β-galactosidase (116.25 kDa), and myosin (205 kDa) (MW-SDS-200; Sigma, St. Louis, MO).
Plasmid patterns.
B. thuringiensis strains were grown to an optical density at 600 nm of 0.8 in Spizizen medium (0.2% NH4SO4, 1.4% K2HPO4, 0.6% KH2PO4, 0.1% sodium citrate, 0.02% MgSO4 · 7H2O) with 0.5% glucose, 0.1% Casamino Acids, and 0.01% yeast extract. Cells were washed in TE buffer (50 mM Tris, 10 mM EDTA, pH 7.8) and incubated for 30 min at 37°C in 10 μg of lysozyme/ml in 0.5 M sucrose, 25 mM Tris, and 10 mM EDTA, pH 8.0. After 10 min at 4°C, lysis buffer (0.2 M NaOH, 1% SDS) was added, and the mixture was incubated for 5 min at 4°C. A solution of 3 M sodium acetate, pH 4.8, was added and stored for 20 min at −20°C. Particles were pelleted at 12,000 rpm for 20 min in a Sorvall SS34 centrifuge. Two volumes of ethanol were added to the supernatant, and the mixture was incubated for 20 min at −80°C to precipitate DNA. DNA was centrifuged as described above, dissolved in distilled water, and visualized in 0.6% agarose gels.
cry gene identification.
The oligonucleotide primers used for detection of the cry1 and cry2 genes have been described previously (5, 6, 13). They were synthesized in a DNA synthesizer (Microsyn 1450A; Systec Inc.) using the reagents and conditions specified by the manufacturer. Novel primers were designed from conserved regions of the related cry1H, cry1I, cry1J, and cry1K genes by using multiple alignments of reported DNA sequences, using ClustalW and GeneWorks 2.3 (Intelligenetics, Inc.). Table 1 shows the specifications of the novel primers. B. thuringiensis strains were grown for 12 h on nutrient medium plates. A loopful of cells was transferred to 0.1 ml H2O, and the mixture was frozen at −70°C for 20 min and boiled for 10 min to lyse the cells. Samples were briefly spun (10 s at 10,000 rpm in an Eppendorf 5415C centrifuge), and 15 μl of supernatant was used as the DNA template in the PCR mixture. PCR mixtures were prepared as described previously (4-6), and PCR was carried out in a Perkin-Elmer model 480 thermal cycler as follows: 2 min at 95°C; 30 cycles of 95°C for 1 min, annealing at a specified temperature (see Table 1) for 1 min, and 72°C for 1 min; and 5 min at 72°C. Samples (15 μl) were electrophoresed in 2% agarose gels.
TABLE 1.
Characteristics of general and specific primers for the cry1H, cry1I, cry1J, and cry1Kgenes
| Primer pair | Positiona | Product size (bp) | Annealing temp (°C) | Sequenceb | Gene recognized | GenBank accession no. |
|---|---|---|---|---|---|---|
| cry1Hgral | 859-1328 | 489 | 51 | 5′-AGTGTATATTGAGTCGCTTAGAGAA (d); 5′-GGTTCAGCAACTGGAGATGT (r) | cry1Ha | Z22513 |
| cry1Hb | U35780 | |||||
| cry1Igral | 1240-1776 | 559 | 51 | 5′-TATACAGACGCAATTGGGAC (d); 5′-GATCCTGAAATGAGTCCTATATG (r) | cry1Ia | X62821 |
| cry1Ib | U07642 | |||||
| cry1Ic | AF056933 | |||||
| cry1d | AF047579 | |||||
| cry1Ie | AF211190 | |||||
| cry1Jgral | 353-1051 | 721 | 53 | 5′-TAGAAGCAACAGTAAGAGCAAAAGCAATC (d); 5′-AGCCGTCATTTCAAGTCCTGACC (r) | cry1Ja | L32019 |
| cry1Jb | U31527 | |||||
| cry1Jc | AX189651 | |||||
| cry1Kgral | 1406-1857 | 474 | 54 | 5′-ACGCAATTATTCGACAACCTCACC (d); 5′-TCTTGAGTCGTTGGACCCATTGA (r) | cry1Ka | U28801 |
Positions at 5′ ends of direct and reverse primers for each PCR primer pair.
d and r, direct and reverse primers, respectively.
DNA polymorphism of S. frugiperda populations.
Randomly selected fourth-instar S. frugiperda larvae were collected from each population and maintained in 100% ethanol at −20°C. DNA was extracted according to the procedure of Agusti et al. (1). The selected larvae were placed individually in Eppendorf tubes, homogenized with 500 μl extraction buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 0.3% Triton X-100, and 60 μg/ml DNase-free proteinase K), incubated for 30 min at 65°C, and centrifuged for 10 min at 10,000 × g. The lysate was extracted twice with phenol-chloroform (1:1, vol/vol). The aqueous phase was mixed with the same volume of cold isopropanol and placed at −20°C for 15 min. After centrifugation for 15 min at 12,000 × g, the precipitated DNA was dried, solubilized in 200 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and treated with RNase (20 μg/ml). The DNA concentration was estimated spectrophotometrically at 260 nm, and DNA was stored at −70°C. DNA was amplified in a PTC 100 MJ Research thermal cycler by using five random primers (10 bp each) as described by Operon Technologies, Inc. (OPA-03, OPA-04, OPA-10, OPA-11, and OPA-13) (Table 2). Conditions for each reaction were as follows: 6 mM Tris-HCl (pH 8.8), 2 mM MgCl2, 50 mM KCl, 0.2 mM deoxynucleoside triphosphates, 0.25 U Taq DNA polymerase (Pharmacia), 0.4 μM primers, and 2 ng of insect genomic DNA. Samples were amplified as follows: 3 min at 94°C; 45 cycles of 1 min at 94°C, 1 min at 35°C, and 1 min at 72°C; and, at the end, 5 min at 72°C for final extension. PCR products were analyzed in 1.5% agarose gels stained with ethidium bromide. Finally, the presence and absence of DNA bands were given values of 1 and 0, respectively, and were converted to a binary matrix. The genetic similarity among populations was determined using the Dice similarity coefficient and the SIMQUAL program from the Numerical Taxonomy and Multivariate Analysis System (NTSYS 2.1) (21). Cluster analysis was performed using the unweighted-pair group method with arithmetic means to obtain the dendrogram. In addition, a correspondence analysis was carried out with NTSYS 2.1. Finally, a bootstrap analysis was performed using the TREECON program (version 1.3b) with a 1,000-resample data set.
TABLE 2.
Primers used for analysis of the genetic diversity of the different Spodoptera frugiperdapopulations using RAPD-PCR
| Primer | Sequence | No. of polymorphic bands | Fragment sizes (bp) |
|---|---|---|---|
| OPA-03 | AGT CAG CCA C | 12 | 350, 400, 450, 550, 600, 700, 750, 950, 1,100, 1,200, 1,300, 1,500 |
| OPA-04 | AAT CGG GCT G | 18 | 250, 280, 300, 350, 450, 500, 550, 600, 700, 750, 800, 900, 950, 1,100, 1,250, 1,350, 1,450, 1,800 |
| OPA-10 | GTG ATC GCA G | 12 | 150, 300, 400, 450, 500, 550, 600, 750, 800, 950, 1,200, 1,600 |
| OPA-11 | CAA TCG CCG T | 13 | 250, 350, 400, 450, 500, 600, 650, 800, 850, 900, 950, 1,200, 1,300 |
| OPA-13 | CAG CAC CCA C | 11 | 250, 350, 450, 500, 550, 750, 850, 900, 1,100, 1,300, 1,500 |
Binding assays on isolated BBMV.
The Cry1Ba, Cry1Ca, and Cry1Da toxins were obtained as recombinant proteins expressed in Escherichia coli. Purification of proteins and generation of the toxic trypsin-resistant fragments were performed as described previously (11). All binding assays were performed with activated toxins. Brush border membrane vesicles (BBMV) were prepared as described previously (29). Toxins were biotinylated using biotinyl-N-hydroxysuccinimide ester (RPN28; Amersham) according to the manufacturer's indications. Biotinylated toxins (10 nM) were incubated with 10 μg of BBMV in phosphate-buffered saline buffer, pH 7.6, for 1 h in the presence or absence of a 500-fold excess of unlabeled toxins. Subsequently, unbound toxin was removed by centrifugation (10 min at 14,000 × g), and BBMV were washed twice with 500 μl of the same buffer; BBMV were suspended in 20 μl of phosphate-buffered saline, and an equal volume of Laemmli sample loading buffer 2X was added. Samples were boiled for 5 min, loaded onto an SDS-PAGE gel, and electrotransferred to a nitrocellulose membrane. The biotinylated proteins that were bound to the blotted protein vesicles were visualized by incubation with a streptavidin-peroxidase conjugate (1:4,000 dilution) for 1 h, followed by addition of Supersignal West Pico chemiluminescent substrate (Pierce), as described by the manufacturers.
RESULTS
Eight B. thuringiensis strains were selected from different Latin American collections based on their high activities against S. frugiperda larvae. Bioassays were performed under identical conditions against first-instar larvae. Strains LBIT27, LBIT193, IB217, and IB412 were isolates from Mexico; S811 came from Brazil, 147-550 and IBUN28 from Colombia, and CIBCM-166 from Costa Rica. Table 3 shows the lethal concentrations of the selected strains assayed against three different S. frugiperda populations. Strains CIBCM-166, S811, IB412, and LBIT27 showed the highest activities against the Mexican population of S. frugiperda, while strains CIBCM-166, S811, and 147-5501 were the most active against the Brazilian and Colombian populations; in addition, strain LBIT27 was also highly toxic to the Brazilian population. Overall, these results suggest that different S. frugiperda populations may have different susceptibilities to the Cry toxins present in the selected strains. The B. thuringiensis strains and the three S. frugiperda populations were further characterized.
TABLE 3.
Mean LC50 estimated for each selected B. thuringiensis strain tested against three populations of S. frugiperdalarvae, and cry gene profiles of selected strains
| B. thuringiensis strain | Mean LC50 (ng/cm2)a of the indicated strain against S. frugiperda populations from:
|
cry gene profile | ||
|---|---|---|---|---|
| Mexico | Brazil | Colombia | ||
| LBIT27 | 288.8 (173.8-479.9) | 300.5 (222.9-405.1) | ND | cry1Ab, cry1Ac, cry1B, cry1E, cry1G, cry1I, cry2 |
| LBIT193 | 329.4 (233.7-464.1) | >2,000 | ND | cry1Ab, cry1Ac, cry1E, cry1G, cry1I |
| IBUN28 | 612.1 (394.2-950.4) | 743.9 (439.3-1,259.6) | 936.1 (460.1-2,832.3) | cry1Aa, cry1Ab, cry1C, cry1D, cry1I |
| S811 | 164.1 (98.4-273.5) | 157.6 (81.1-290.2) | 13.3 (2.4-29.4) | cry1Aa, cry1Ab, cry1B, cry1D, cry1I |
| 147-5501 | 437.7 (280.8-680.5) | 141.8 (84.3-238.4) | 14.2 (2.4-36.3) | cry1Aa, cry1B, cry1D, cry1I, cry2 |
| CIBCM-166 | 95.7 (43.3-208.3) | 294.6 (199.2-435.5) | 12.9 (1.5-32.2) | cry1Aa, cry1Ab, cry1Ac, cry1Ad, cry1B, cry1D, cry1I, cry2 |
| IB217 | 711.1 (480.3-1,052.9) | 928.7 (439.4-1,962.8) | 1,923.9 (1,072.6-6,058.4) | cry1Aa, cry1Ac, cry1Ad, cry1C, cry1D, cry1I |
| IB412 | 200.9 (88.7-455.3) | 863.93 (465.2-1,912) | 358.2 (233.7-597.6) | cry1Ab, cry1Ac, cry1B, cry1E, cry1G, cry1I, cry2 |
| HD137 | 41.8 (12.1-65.7) | 189.6 (136.9-262.5) | 22.9 (10.5-30.9) | cry1Aa, cry1B, cry1C, cry1D |
Values in parentheses are fiducial limits (P = 0.95). ND, not determined.
Characterization of selected B. thuringiensis strains.
The crystal inclusions produced by the selected B. thuringiensis strains were initially observed by phase-contrast microscopy and then by scanning and transmission electron microscopy, showing that all the strains contain bipyramidal crystal inclusions (data not shown). Some strains also contain a small cuboidal crystal (strains 147-5501, CIBCM166, IB412, and LBIT27).
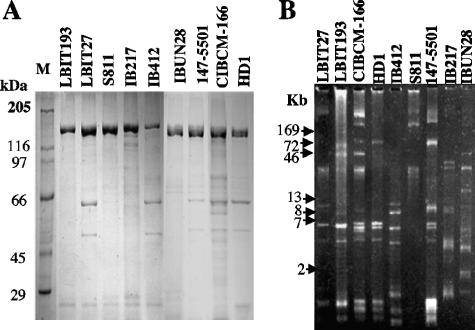
The crystal proteins produced by these strains were characterized by SDS-PAGE of spore-crystal suspensions (Fig. 1A). Strains LBIT27, IB412, 147-5501, and CIBCM-166 showed a protein profile similar to that of the Bacillus thuringiensis serovar kurstaki strain HD1, with major proteins of ca. 130 and 70 kDa. Strains LBIT193, S811, IB217, and IBUN28 showed only proteins of ca. 130 kDa.
FIG. 1.
Characterization of the selected Bacillus thuringiensis strains. (A) SDS-PAGE of spore-crystal suspensions of selected Bacillus thuringiensis strains. (B) Agarose gel electrophoresis of the plasmid profile present in selected Bacillus thuringiensis strains.
Figure 1B shows the plasmid profiles of the selected B. thuringiensis strains. All the strains showed different plasmid profiles, indicating the high diversity of these strains.
Identification of cry genes in the selected B. thuringiensis isolates.
The cry1 and cry2 gene contents of the selected strains were determined by PCR analyses (Table 3). None of the native strains showed the same cry gene profile as the control Bacillus thuringiensis subsp. aizawai strain HD137. The native strains most active against the three S. frugiperda populations contain a combination of cry1Aa, cry1B, cry1D, and cry1I genes. Interestingly, the native strains that harbor the cry1C and cry1D genes (IBUN28 and IB217) did not show the highest insecticidal activity.
Susceptibilities of S. frugiperda populations to single Cry toxins.
In order to determine the susceptibilities of the three insect populations to single Cry toxins and correlate their toxicities with specific binding, susceptibilities to pure preparations of some Cry proteins were evaluated. We selected for the assay two Cry toxins reported to be toxic to S. frugiperda larvae (Cry1Ca and Cry1Da) and one toxin reported to be nontoxic to these larvae (Cry1Ba) (2). Table 4 shows that the three insect populations differ in their susceptibilities to these toxins. The Mexican population was susceptible to the Cry1Ca and Cry1Da toxins but not to Cry1Ba. In contrast, the Brazilian population showed susceptibility to Cry1Ca, moderate susceptibility to Cry1Ba, and no susceptibility to Cry1Da. Finally, the Colombian insect population showed high susceptibilities to all three Cry toxins analyzed (Table 4).
TABLE 4.
Mean LC50 estimated for different Cry toxins tested against three populations of S. frugiperdalarvae
| Cry toxin | Mean LC50 (ng/cm2)a of the indicated Cry toxin against S. frugiperda populations from:
|
||
|---|---|---|---|
| Mexico | Brazil | Colombia | |
| Cry1B | >2,000 | 403 (198-690) | 74 (31-148) |
| Cry1C | 42 (27-55) | 84 (61-129) | 21 (6-48) |
| Cry1D | 80 (66-128) | >2,000 | 7 (2-22) |
Values in parentheses are fiducial limits (P = 0.95).
Analysis of binding of Cry toxins to BBMV from S. frugiperda populations.
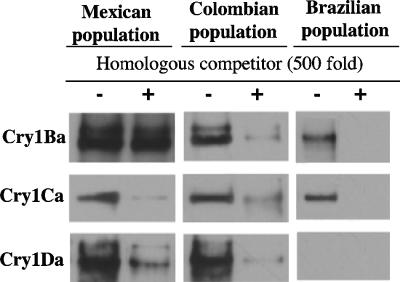
Trypsin-activated Cry1Ba, Cry1Ca, and Cry1Da toxins were labeled with biotin, and homologous competition binding assays were performed on BBMV of S. frugiperda populations. The homologous competition with the Mexican population showed that both the Cry1Ca and Cry1Da toxins were able to bind and that this interaction was specific, as binding was competed by a 500-fold excess of unlabeled Cry1Ca or Cry1D toxin, respectively (Fig. 2). The Cry1Ba toxin showed nonspecific binding; there was no competition in the presence of unlabeled toxin. In contrast, the homologous competition assays carried out with the Brazilian population showed that the Cry1Ba and Cry1Ca toxins bound specifically to BBMV, whereas Cry1Da did not bind to the membranes (Fig. 2). Finally, in the case of the Colombian population, the three toxins bound specifically, since all of them were competed in the homologous competition experiments by their corresponding unlabeled toxins (Fig. 2).
FIG. 2.
Homologous competition binding assays on BBMV isolated from Manduca sexta larvae. Biotinylated trypsin-activated Cry toxins were incubated with the BBMV in the absence or in the presence of a 500-fold excess of unlabeled toxin. After 1 h of incubation, unbound toxins were removed, and vesicles containing bound toxins were loaded onto an SDS-PAGE gel and blotted onto a nitrocellulose membrane. Labeled proteins were visualized by means of a streptavidin-peroxidase conjugate.
Genetic variability among the three S. frugiperda populations.
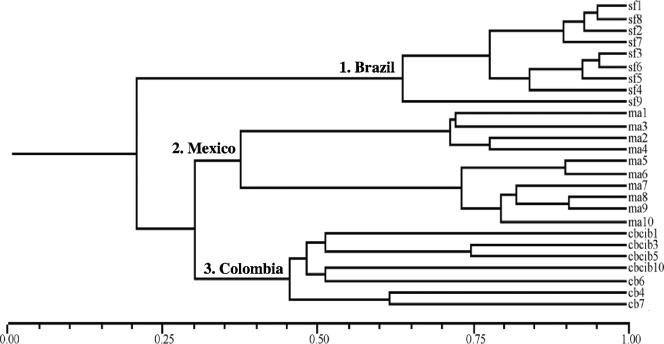
The RAPD-PCR methodology was used to analyze fourth-instar larvae from each S. frugiperda population. The commercially available RAPD primers produced different bands in the three populations. Table 2 shows the sum of polymorphic markers produced by each primer when the three populations were analyzed. As stated in Materials and Methods, the presence and absence of DNA bands were given values of 1 and 0, respectively, and converted to a binary matrix. Binary data were used to generate a dendrogram that shows the genetic relationship among these insect populations (Fig. 3). Three main clusters were clearly discriminated, assembling larvae from each population. The level of variability found within each group was lower than that between two different geographical populations. One group corresponds to the larvae collected from the Brazilian population, which showed high similarity (65 to 95%) within their members. In contrast, the Brazilian group shared only 22% similarity with the groups from Mexico and Colombia. The Mexican and Colombian groups shared only 31% similarity between them. The Mexican population clearly splits into two subgroups that shared 37% similarity. Each of the clusters present in the Mexican population shared high similarity, ranging from 73 to 90%, among their members. The third group integrates larvae from the Colombian population, which showed 45 to 75% similarity among their members. These data show that the diversity of S. frugiperda larvae clearly correlates with their geographical origin.
FIG. 3.
Genetic variability among the three S. frugiperda populations. Shown is a dendrogram obtained from RAPD-PCR analysis of 10 fourth-instar larvae from each S. frugiperda population.
DISCUSSION
A great diversity among B. thuringiensis strains active against S. frugiperda larvae, in terms of plasmid profiles, cry gene content, and insecticidal activity against three Latin American S. frugiperda populations, was found. In this work we also analyzed the genetic variability among the three different S. frugiperda populations; our data indicated that these insect populations are genetically different, suggesting that the independent evolution of these populations generated genetic divergence. It is noteworthy that these populations showed different responses to intoxication with Cry toxins. The correlation of this genetic divergence with mutations in specific toxicity-related genes remains to be determined.
Morphological characterization of the crystal inclusions of the selected B. thuringiensis strains showed typical bipyramidal and cuboidal crystals common to most lepidopteran-active strains. These data are in agreement with the expected protein composition observed in the SDS-PAGE analyses. However, a high diversity of plasmid profiles was observed, suggesting important variability among these B. thuringiensis strains, which agrees with the different cry gene contents found in these strains. The toxicities of these strains were tested against the three S. frugiperda populations, showing significant differences in susceptibility. The Colombian population showed the highest sensitivity to some B. thuringiensis strains (S811, 147-5501, CIBCM-166, and IB292), in contrast to the Mexican and Brazilian populations, which showed low to moderate susceptibility. All these strains have in common the presence of the cry1Aa, cry1Ba, and cry1Da genes.
To date, the reported toxicity data concerning the susceptibility of S. frugiperda to pure Cry proteins seem to be contradictory. Cry1A and Cry1Ba toxins have been reported to have low toxicities against this pest, while Cry1C and Cry1D showed the highest toxicities (2, 3, 27). However, another report indicated that Cry1Bb was toxic to this pest while Cry1C was not (15). Furthermore, attempts to correlate the cry gene contents of a large number of B. thuringiensis strains and their toxicities to S. exigua were unsuccessful (16). The selected strains used in this work that harbored the cry1C and cry1D genes but not cry1B (IBUN28 and IB217) were not the most active. Our data are in agreement with those of a previous report (14), which showed that the presence of the cry1C and cry1D genes does not necessarily correlate with high toxicity to S. frugiperda larvae and suggested that other proteins present in the B. thuringiensis strains analyzed might be more important for toxicity. We found that the most active native B. thuringiensis strains have a combination of cry1Aa, cry1B, and cry1D genes. However, because a great variability of cry1B genes has been reported in recent years (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/index.html), it will be worthwhile to identify the specific cry1B genes present in the more active strains and test the individual proteins in bioassays. Additionally, other factors, such as differences in toxin expression or synergism effects between some Cry toxins, could account for the variability in toxicity of B. thuringiensis strains harboring the same set of cry genes. The protein concentration of each Cry toxin within the crystal could influence the final toxicity of a particular B. thuringiensis strain. We did not quantify the amount of each Cry protein in the crystal inclusion of the selected B. thuringiensis strains, but we tested the toxicity of pure preparations of three Cry toxins and demonstrated that there is a difference in susceptibility to these Cry toxins among the three insect populations (Table 4). These data indicate that the insect populations used in this study have evolved differently regarding their susceptibilities to specific Cry toxins. A previous report indicated that different populations of Plutella xylostella also showed differences in susceptibility to Cry toxins (8).
The binding analyses performed with individual toxins indicated differences among the toxin-receptor interactions within the three insect populations. We propose that the Mexican and Colombian populations contain a functional Cry1Da receptor, in contrast to the Brazilian population, where this binding site is missing. In addition, the Colombian and Brazilian populations bind the Cry1Ba toxin, while the Mexican population showed nonspecific binding of this toxin. These data correlated with the susceptibilities of the different S. frugiperda colonies to Cry toxins: the Brazilian population was not susceptible to the Cry1D toxin, while the Mexican population was not affected by the Cry1Ba toxin.
The RAPD-PCR technique has been used at different levels in the molecular characterization of different insect pests and other related species. Monnerat et al. (18) showed that there is no intrapopulation genetic variability for three species of Diadegma (Hymenoptera: Ichneumonidae), an important parasitoid of Plutella xylostella. Tsai et al. (26) demonstrated the effectiveness of this technique in the phylogenetic analysis of the microsporidian Nosema isolated from different lepidopteran larvae, such as Spodoptera littura, S. exigua, Helicoverpa armigera, P. xylostella, and Pieris spp., showing that isolates from Pieris spp., S. exigua, and H. armigera were phylogenetically related.
In this work we used RAPD-PCR to evaluate the molecular variability in the different S. frugiperda populations. These studies allowed us to obtain preliminary information about the genetic variability among these populations. It is known that genetic differences may evolve if the physiological adaptation of an insect to a certain host plant entails a decrease in performance on the alternative host (9). According to this principle, the rice strain of S. frugiperda is associated with rice plants, whereas the maize strain occurs in maize (17). The three S. frugiperda populations characterized here are associated with maize. However, we found genetic variability among the three colonies of S. frugiperda, and we found that these populations were clustered according to their geographical origin, suggesting that other factors besides the host plant have influenced the selection of genetic differences. We also found that the populations from Colombia and Mexico are slightly closer than the Brazilian population. However, one important observation is that these populations showed different susceptibilities to B. thuringiensis Cry toxins. Therefore, the susceptibilities of S. frugiperda populations to different Cry toxins should be carefully evaluated in the development of insect pest control strategies, including the deployment of genetically modified maize, in different geographical regions.
Acknowledgments
We thank Regina Basurto and Eleazar Urbina for excellent technical assistance.
This work was supported in part by USDA grant 2002-35302-12539, CYTED III.5, and FAO.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Agusti, N., M. C. De Vicente, and R. Gabarra. 1999. Development of sequence amplified characterized region (SCAR) markers of Helicoverpa armigera: a new polymerase chain reaction-based technique for predator gut analysis. Mol. Ecol. 8:1467-1474. [DOI] [PubMed] [Google Scholar]
- 2.Aranda, E., J. Sánchez, M. Peferoen, L. Guereca, and A. Bravo. 1996. Interaction of Bacillus thuringiensis crystal protein with the midgut epithelial cells of Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Invertebr. Pathol. 68:203-212. [DOI] [PubMed] [Google Scholar]
- 3.Bohorova, N., M. Cabrera, C. Abarca, R. Quintero, A. M. Maciel, R. M. Brito, D. Hoisington, and A. Bravo. 1997. Susceptibility of four tropical lepidopteran maize pest to Bacillus thuringiensis CryI type insecticidal toxins. J. Econ. Entomol. 90:412-415. [Google Scholar]
- 4.Bravo, A., S. Sarabia, L. Lopez, H. Ontiveros, C. Abarca, A. Ortiz, M. Ortiz, L. Lina, F. J. Villalobos, G. Peña, M. E. Nuñez-Valdez, M. Soberón, and R. Quintero. 1998. Characterization of cry genes in a Mexican Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 64:4965-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerón, J., L. Covarrubias, R. Quintero, A. Ortíz, M. Ortíz, E. Aranda, L. Lina, and A. Bravo. 1994. PCR analysis of the cryI insecticidal crystal family genes from Bacillus thuringiensis. Appl. Environ. Microbiol. 60:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerón, J., A. Ortíz, R. Quintero, L. Güereca, and A. Bravo. 1995. Specific PCR primers directed to identify cryI and cryIII genes within a Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 61:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finney, D. J. 1971. Probit analysis. Cambridge University Press, Cambridge, United Kingdom.
- 8.Gonzalez-Cabrera, J., S. Herrero, A. H. Sayyed, B. Escriche, Y. B. Liu, S. K. Meyer, D. J. Wright, B. E. Tabashnik, and J. Ferré. 2001. Variations in susceptibility to Bacillus thuringiensis toxins among unselected strains of Plutella xylostella. Appl. Environ. Microbiol. 67:4610-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould, F. 1979. Rapid host range evolution in a population of the phytophagous mite Tetramychus urticae Koch. Evolution 33:791-802. [DOI] [PubMed] [Google Scholar]
- 10.Hill, D. S. 1983. Agricultural insect pests of the tropics and their control. Cambridge University Press, Cambridge, United Kingdom.
- 11.Hofmann, C., H. Vanderbruggen, H. Höfte, J. Van Rie, S. Jansens, and H. Van Mellaert. 1988. Specificity of Bacillus thuringiensis δ-endotoxins is correlated with the presence of high affinity binding sites in the brush border membrane of target insect midguts. Proc. Natl. Acad. Sci. USA 85:7844-7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Höfte, H., and H. R. Whiteley. 1989. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53:242-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibarra, J. E., M. C. del Rincón, S. Ordúz, D. Noriega, G. Benintende, R. Monnerat, L. Regis, C. M. F. de Oliveira, H. Lanz, M. H. Rodriguez, J. Sánchez, G. Peña, and A. Bravo. 2003. Diversity of Bacillus thuringiensis strains from Latin America with insecticidal activity against different mosquito species. Appl. Environ. Microbiol. 69:5269-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loguercio, L. L., C. G. Santos, M. R. Barreto, C. T. Guimaraes, and E. Paiva. 2001. Association of PCR and feeding bioassays as large scale method to screen tropical Bacillus thuringiensis isolates for a cry constitution with higher insecticidal effect against Spodoptera frugiperda (Lepidoptera: Noctuidae) larvae. Lett. Appl. Microbiol. 32:362-367. [DOI] [PubMed] [Google Scholar]
- 15.Luo, K., D. Banks, and M. J. Adang. 1999. Toxicity, binding, and permeability analyses of four Bacillus thuringiensis Cry1 δ-endotoxins using brush border membrane vesicles of Spodoptera exigua and Spodoptera frugiperda. Appl. Environ. Microbiol. 65:457-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez, C., J. E. Ibarra, and P. Caballero. 2005. Association analysis between serotype, cry gene content, and toxicity to Helicoverpa armigera larvae among Bacillus thuringiensis isolates native to Spain. J. Invertebr. Pathol. 90:91-97. [DOI] [PubMed] [Google Scholar]
- 17.McMichael, M., and D. P. Prowell. 1994. Differences in amplified fragment-length polymorphisms in fall armyworm (Lepidoptera: Noctuidae) host strains. Ann. Entomol. Soc. Am. 92:175-181. [Google Scholar]
- 18.Monnerat, R. G., S. Leal-Bertioli, D. Bertioli, T. Butt, and D. Bordat. 2004. Variabilidade genética do parasitóide Diadegma sp. através de RAPD-PCR. Horticultura Brasileira 22:90-92. [Google Scholar]
- 19.Pashley, D. P. 1986. Host-associated genetic differentiation in fall armyworm (Lepidoptera: Noctuidae): a sibling species complex. Ann. Entomol. Soc. Am. 79:898-904. [Google Scholar]
- 20.Restrepo, N., D. Gutierrez, M. M. Patiño, I. Thiery, A. Delecluse, and S. Ordúz. 1997. Cloning, expression and toxicity of a mosquitocidal toxin gene from Bacillus thuringiensis subsp. medellin. Mem. Inst. Oswaldo Cruz 92:257-262. [DOI] [PubMed] [Google Scholar]
- 21.Rohlf, F. J. 1993. NTSYS-pc: Numerical Taxonomy and Multivariate System. Version 2.9. Applied Biostatistics, New York, N.Y.
- 22.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 23.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shorey, H. H., and R. L. Hale. 1965. Mass-rearing of the larvae of nine Noctuid species on a simple artificial medium. J. Econ. Entomol. 58:522-524. [Google Scholar]
- 25.Travers, R. S., P. A. W. Martin, and C. F. Reichelderfer. 1987. Selective process for efficient isolation of soil Bacillus spp. Appl. Environ. Microbiol. 53:1263-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai, S. J., C. F. Lo, Y. Soichi, and C. H. Wang. 2003. The characterization of microsporidian isolates (Nosematidae: Nosema) from five important lepidopteran pests in Taiwan. J. Invertebr. Pathol. 83:51-59. [DOI] [PubMed] [Google Scholar]
- 27.Van Rie, J., S. Jansens, H. Höfte, D. Degheele, and H. Van Mellaert. 1990. Receptors on the brush border membrane of the insect midgut as determinants of the specificity of Bacillus thuringiensis δ-endotoxins. Appl. Environ. Microbiol. 56:1378-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visser, B., T. Van der Salm, W. Van der Brink, and G. Folkers. 1988. Genes from Bacillus thuringiensis entomocidus 60.5 coding for insect-specific crystal proteins. Mol. Gen. Genet. 212:219-224. [Google Scholar]
- 29.Wolfersberger, M., P. Lüthy, A. Maurer, F. Parenti, V. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A:301-308. [Google Scholar]