Abstract
Escherichia coli is the most completely characterized prokaryotic model organism and one of the dominant indicator organisms for food and water quality testing, yet comparatively little is known about the structure of E. coli populations in their various hosts. The diversities of E. coli populations isolated from the feces of three host species (human, cow, and horse) were compared by two subtyping methods: ribotyping (using HindIII) and antibiotic resistance analysis (ARA). The sampling effort required to obtain a representative sample differed by host species, as E. coli diversity was consistently greatest in horses, followed by cattle, and was lowest in humans. The diversity of antibiotic resistance patterns isolated from individuals was consistently greater than the diversity of ribotypes. E. coli populations in individuals sampled monthly, over a 7- to 8-month period, were highly variable in terms of both ribotypes and ARA phenotypes. In contrast, E. coli populations in cattle and humans were stable over an 8-h period. Following the cessation of antibiotic therapy, the E. coli population in the feces of one human experienced a rapid and substantial shift, from a multiply antibiotic-resistant phenotype associated with a particular ribotype to a relatively antibiotic-susceptible phenotype associated with a different ribotype. The high genetic diversity of E. coli populations, differences in diversity among hosts, and temporal variability all indicate complex population dynamics that influence the usefulness of E. coli as a water quality indicator and its use in microbial source tracking studies.
The structure of Escherichia coli populations influences several aspects of public health. Pathogenic subtypes of E. coli are known to cause illness around the world (18), and an increased understanding of the genetic variability of populations in animal reservoirs can inform epidemiological studies. E. coli is also one of the standard indicator organisms for fecal pollution in environmental waters (1). The natural host range of E. coli includes all warm-blooded animals, some cold-blooded animals (12), and environmental reservoirs, such as sediments (2, 32) and free-living strains (24); therefore, the source of fecal pollution in water bodies is often ambiguous. Knowledge of indicator organism source is necessary for risk assessment and remediation of polluted waters, including application, such as total maximum daily load assessment. Consequently, the field of microbial source tracking (MST), which seeks to determine the origin of fecal material in water, has emerged (4, 7, 10, 14, 23, 31, 35).
Many microbial source tracking methods rely on the premise that, in their gastrointestinal tracts, animal species contain distinct subtypes of E. coli that are shed in their feces. Matching the subtypes of E. coli identified in polluted watersheds to those isolated from known sources would hypothetically allow the identification of the source of the pollution (28). Many MST methods require a library of E. coli subtypes isolated from the feces of known animal species in order to act as a predictive tool (13, 14, 17, 22, 23, 29) and are collectively known as library-based methods (30). Methods used to subtype the members of these E. coli subpopulations include, but are not limited to, antibiotic resistance analysis (ARA) (14, 31, 35) and ribotyping (4, 23). Studies of the population structure and dynamics of E. coli within animal species are essential when determining the proper sampling strategy for constructing a representative MST library (30). Natural variability in E. coli populations that could reduce library accuracy may occur at many levels, including inside the gastrointestinal tracts of individual animals (16, 20), over time (36), and over geographic distances (8, 11, 26, 33). Direct comparisons of E. coli population biology in various host animals are rare in the literature, as most studies have focused on one host species (5, 8, 16, 27, 34).
The aim of this study was to investigate the diversity and stability of E. coli populations within the feces of individuals belonging to three mammalian host groups (humans, horses, and beef cattle). These groups have high potentials for impacting surface water quality and are commonly used for MST libraries; furthermore, both human and cattle feces have a high potential for the carriage of human pathogens. Genetic diversity was assessed by ribotyping using one enzyme (HindIII), while phenotypic diversity was assessed by ARA. We hypothesized that subtype diversity would differ from the individual level to the host species level and among different host species. Furthermore, if the basic premise behind MST is true, there should be significantly greater commonality (sharing) of genotypes or phenotypes within host groups than between host groups. The effect of diversity on obtaining a representative sample of E. coli subtypes from hosts was also assessed, as it directly impacts sampling strategies for library construction.
MATERIALS AND METHODS
Sample collection.
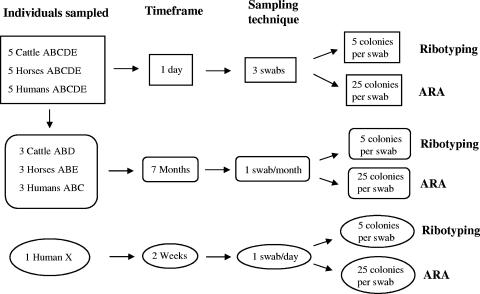
For comparisons of E. coli diversity among host species, fecal samples were obtained from five individuals of humans, horses, and adult beef cows. The sampling scheme is outlined in Fig. 1. Cattle were from the same herd in Tampa, Florida, horses were stabled at the same farm, and humans were coworkers. Fecal samples were collected using sterile swabs that were moisturized with phosphate-buffered saline. Cattle and horse feces were sampled by stabbing one swab multiple times into a fresh fecal mass, while for humans, an anal swab was obtained. Three swab samples per individual from each animal and human were collected in a single day for an experiment in which sampling time points were ∼4 h apart. Three of the five individuals of each animal species were randomly selected for further study. These three individuals were sampled once a month (with a single swab per date) for an additional 7 months. To further study temporal stability of E. coli populations within humans, a single human (human X) who had not previously donated samples was sampled on a daily basis with a single swab for 2 weeks and subsequently on a weekly basis for the next 2 weeks (Fig. 1).
FIG. 1.
Experimental design. Letter designations A, B, C, D, E, and X refer to the individual sampled.
All swabs were streaked onto mFC agar plates (100-mm diameter), which were incubated overnight in a water bath at 44.5°C for fecal coliform isolation (1). With sterile toothpicks, 25 blue colonies per swab were transferred into wells of microtiter plates that contained EC broth amended with 4-methylumbelliferyl-β-d-glucuronide (MUG) (50 μg/ml). β-Glucuronidase activity, which is specific to E. coli in the fecal coliform group, was assessed by MUG cleavage and determined by fluorescence in the microtiter plate upon exposure to UV light (3). Only MUG-positive colonies were further analyzed. Ten percent of all MUG-positive colonies were speciated using API 20E biochemical test strips (bioMerieux, Inc.), and all were confirmed as E. coli.
Ribotyping.
A 1-day experiment in which individuals were sampled three times at ∼4-h intervals was conducted. Fifteen E. coli isolates per individual host animal were ribotyped (Fig. 1). These isolates were randomly picked from frozen stocks and were a subset of the 25 E. coli isolates collected at each time point. In the case of the 7-month time course studies, five E. coli isolates per sampling event from each individual were ribotyped. These isolates were also randomly chosen as a subset of the 25 isolates collected from each single swab at each sampling event. All E. coli isolates were ribotyped using HindIII and a modification of the protocol described by Parveen et al. (23), which is detailed in the work by Anderson et al. (2).
ARA.
For the 1-day experiment, approximately 60 E. coli isolates (all of the E. coli isolates collected from all three swabs that were successfully recovered from cryostorage) per host individual were analyzed by antibiotic resistance analysis (Fig. 1) (31, 35). For the time course study, approximately 20 isolates (all isolates collected from each single swab that were successfully processed and recovered) were analyzed by antibiotic resistance analysis for each sampling event for all individuals. Isolates were grown in 96-well microtiter plates filled with EC broth and incubated overnight at 37°C. These cultures were diluted to a McFarland standard of 0.5 (absorbance at 600 nm of ∼0.1) with sterile, nanopure water, and 100 μl of this diluted culture was transferred to a new microtiter plate. Up to 96 E. coli isolates were prepared as inoculum cultures for each microtiter plate. These microtiter plates were used as the inocula for a series of antibiotic Mueller-Hinton plates. Mueller-Hinton agar (Difco, Sparks, MD) plates (150-mm diameter) were each amended with a single antibiotic as described in Table 1.
TABLE 1.
Antibiotics and the concentrations used for ARA and the antibiotic resistance pattern of control strain E. coli ATCC 25922
| Antibiotic | Concn(s) tested (μg ml−1) | Concn (μg ml−1) in E. coli ATCC 25922 |
|---|---|---|
| Amoxicillin | 4.0 and 128.0 | 4.0 |
| Cephalothin sodium salt | 32.0 | 8.0 |
| Chloramphenicol | 4.0 | 0.0 |
| Chlortetracycline hydrochloride | 20.0 and 80.0 | 0.0 |
| Doxycycline hydrochloride | 4.0 | 0.0 |
| Gentamicin sulfate | 1.0 | 0.0 |
| Kanamycin monosulfate | 3.0 | 0.0 |
| Nalidixic acid sodium salt | 3.0 | 0.0 |
| Neomycin | 3.0 | 0.0 |
| Norfloxacin | 0.1 | 0.0 |
| Oxytetracycline hydrochloride | 20.0 | 0.0 |
| Penicillin G | 20.0 and 200.0 | 20.0 |
| Rifampin | 2.0 and 16.0 | 2.0 |
| Streptomycin sulfate | 20.0 and 80.0 | 0.0 |
| Tetracycline hydrochloride | 4.0 and 64.0 | 0.0 |
| Trimethoprim | 0.25 and 1.0 | 0.3 |
| Trimethoprim-sulfamethoxazole (1:19) | 5.0 | 0.0 |
The diluted E. coli inoculum was stamped onto each of the antibiotic plates with a sterile, 96-prong replicator. Each plate was also inoculated with a positive control, E. coli strain ATCC 25922, to assess reproducibility (Table 1). The plates were incubated overnight at 37°C. Any discernible growth on the antibiotic plates was scored as positive, including patchy growth. No attempt was made to distinguish resistance acquired via a recent mutation from preexisting resistance, as the control strain sometimes demonstrated patchy growth in the presence of antibiotics to which it was resistant.
Statistical analysis.
Ribotype similarity was analyzed using BioNumerics software (Applied Maths, Belgium). Similarity dendrograms were constructed using the Dice coefficient algorithm with maximum similarity. The software optimization setting was 0.2, and the position tolerance setting was 0.7. The similarity of replicate analyses of an E. coli control strain (ATCC 9637), starting with cells and proceeding through ribotyping, was 90% similar or greater; therefore, patterns that were at least 90% similar were considered to belong to the same subtype. These relationships were confirmed by eye.
Antibiotic resistance patterns (ARPs) were also analyzed using BioNumerics software. ARPs were compared by constructing a similarity dendrogram using the binary coefficient of simple matching (>50% mean). Simple matching considers the exact antibiotic concentrations when comparing the similarities of different ARPs, giving each concentration equivalent weight, even if the concentrations are geometrically increasing. The similarity of ARPs of an E. coli control (ATCC 25922) using the settings above was at least 94%; therefore, ARPs were considered the same if they were at least 94% similar.
Diversity indices were used to assess the structures of E. coli populations. The observed frequency and distribution of E. coli subtypes, determined by ribotype or ARP, were compared within and between three host categories. Four different indices from the Hill family of diversity indices were used: (i) a richness estimator (S) was calculated the number of different subtypes; (ii) Shannon's index (H′) was calculated as H′ = −Σpiln(pi) (where pi is the number of isolates with pattern [i]/total isolates); (iii) Simpson's index (λ) was calculated as Σ[ni(ni − 1)]/[n(n − 1)] (where ni is the number of isolates with subtype [i] and n is the total number of isolates); and (iv) Pielou's evenness (J′) was calculated as H′/lnS (19). Diversity measurements were calculated separately for each individual, and means were calculated for each host category. One-way analysis of variance with Dunnett's posttest was performed using GraphPad InStat, version 3.00 (GraphPad Software, San Diego, CA), on all diversity measurements to compare E. coli population structures among the different source categories. Significance was determined at a P value of <0.05. Accumulation curves were created using the statistical software EcoSim 700 (9). The values for these curves were obtained using the accumulation curve calculation with 1,000 iterations within the species diversity function of EcoSim 700. These values were imported into Microsoft Excel for graph production.
RESULTS AND DISCUSSION
E. coli diversity in individuals over 1 day.
The diversity measurements of E. coli ribotypes sampled during the 1-day experiment were calculated for each of five individual host animals and were averaged in order to compare the diversities of E. coli populations of cattle, horses, and humans (Table 2). Ribotype diversity in the feces of individual horses was significantly greater than that in either cow or human individuals based on all diversity measurements (P < 0.05), although evenness (J′) measurements were not significantly different among hosts (Table 2). The ribotype diversity of E. coli populations of individual cattle was not significantly different from those in humans, although diversity tended to be higher in cattle.
TABLE 2.
Average diversity measurements of E. coli subtypesa
| Subtyping method and source | Richness estimator (S) | Shannon index (H′) | Pielou's evenness (J′) | Simpson index (1/λ) |
|---|---|---|---|---|
| Ribotyping | ||||
| Beef cow | 3.4 (± 2.1) a | 0.67 (± 0.58) a | 0.69 (± 0.28) a | 2.2 (± 1.4) a, b |
| Horse | 9.2 (± 3.2) b | 2.00 (± 0.46) b | 0.90 (± 0.09) a | 21.1 (± 19.0) b |
| Human | 2.2 (± 2.0) a | 0.43 (± 0.69) a | 0.95 (± 0.08) a | 2.1 (± 2.0) a |
| Antibiotic resistance analysis | ||||
| Beef cow | 10.2 (± 2.9) a | 1.40 (± 0.38) a | 0.61 (± 0.13) a | 3.0 (± 1.1) a |
| Horse | 10.5 (± 1.7) a | 1.86 (± 0.25) a | 0.79 (± 0.06) a | 5.5 (± 2.7) b |
| Human | 4.4 (± 2.3) b | 0.59 (± 0.44) b | 0.55 (± 0.28) a | 1.6 (± 0.6) c |
Subtypes were isolated from five individuals per host species during the 1-day study. Values that share the same letter within columns are not significantly different.
Diversity measurements of ARPs of E. coli populations were also significantly different among host species (Table 2). ARPs of E. coli populations in individual humans were significantly less diverse than in the E. coli populations of both cattle and horse individuals based on all diversity measurements. However, there was no significant difference noted between the evenness values (J′) among host species. There was no significant difference in the diversity measurements obtained for the E. coli populations of cow individuals compared to those of horse individuals according to the richness estimator and the Shannon index. However, the E. coli populations of horse individuals demonstrated a significantly higher Simpson value (1/λ = 5.5) than did the E. coli populations of cattle (1/λ = 3.0).
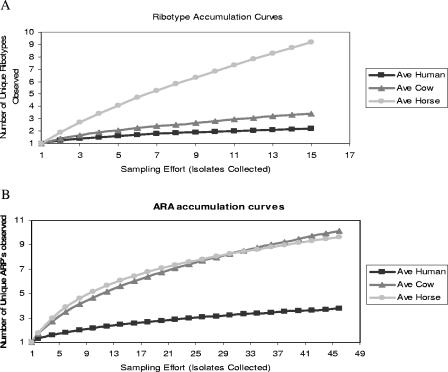
Accumulation curves were constructed for E. coli ribotypes sampled from each host species during the 1-day study. The graphs demonstrate the average number of new E. coli subtypes that were observed with increased sampling effort for each host species (Fig. 2A and B). Ribotype accumulation curves demonstrated that, on average, the dominant E. coli populations within individual humans required the fewest isolates to obtain a representative sample (Fig. 2A). Thus, in the case of humans and beef cattle, collecting several isolates (e.g., five) per individual could adequately represent the ribotype diversity of the dominant E. coli populations. Furthermore, the same set of ribotypes was generally sampled from an individual at each of the three times over the 1-day period, demonstrating temporal stability of the population over an 8-h period. In contrast, the slope of the accumulation curve for E. coli ribotypes from horses did not approach an asymptote and it is clear that more than 15 isolates per individual were required to adequately sample the populations in horses (Fig. 2A). Temporal stability of sampled E. coli ribotypes was not observed in horses; however, from the data collected, it is not possible to determine whether the E. coli population in horses was turning over on a <12-h time scale or whether the apparent lack of temporal stability was due entirely to undersampling of the population.
FIG. 2.
Accumulation curves representing the average number of unique patterns observed per host individual by ribotyping (A) and antibiotic resistance analysis (B) during the 1-day experiment.
Accumulation curves based on ARPs demonstrated that feces from human individuals contained the least diverse E. coli population, requiring ∼15 isolates to represent the dominant ARPs in their E. coli populations (Fig. 2B). In contrast to ribotyping results, the accumulation curves of ARPs indicate that E. coli populations of individual cows would require a sampling effort similar to that for horses (∼45 to 50 isolates per individual). The slopes of the ARP accumulation curves for all host species, particularly cattle, were steeper than the slopes of ribotyping accumulation curves. The graphs demonstrate that the sampling efforts required to represent population diversity differed based on both the host source of the E. coli population (i.e., cattle, humans, or horses) and the typing technique used.
This is the first study to systematically compare the diversities of E. coli populations of individuals from different animal hosts using both a phenotypic and a genotypic subtyping method. The diversity of E. coli populations was determined by using several different measurements because each measurement has advantages and disadvantages, i.e., richness captures the presence of rare subtypes by giving all subtypes equal value regardless of abundance, the Simpson's index gives more weight to the most abundant, dominant subtypes over rare subtypes, and the Shannon index falls in between, accounting for both the frequency and abundance of each subtype (15). In the host groups sampled, E. coli populations of individual horses were the most diverse according to all indices by both genotypic and phenotypic assessments, followed by those of cattle and humans. It is probable that the diversity of E. coli ribotypes in horses was underestimated in this study as well as the diversity of ARPs in cattle and horses, as illustrated by the species accumulation curves (Fig. 2A and B). However, since the E. coli ribotypes of human and cow individuals and the ARPs of human individuals appear to have been adequately sampled, the general trend is still informative.
These results have several important biological and public health implications. In general, they suggest that the conclusions made about E. coli population structure and dynamics in a single host species should not be generalized to other host species. The observed differences in diversity particularly impact MST methods, as an MST library should adequately represent the diversity of the indicator organism population for it to be an effective predictor of the source of fecal pollution (25). Previous MST studies have generally used arbitrary criteria to select the number of isolates to analyze per individual fecal sample (4, 10, 14, 31, 35), i.e., a set number for all source types or all of the available isolates from a fecal sample. Our data indicate that preliminary studies of E. coli diversity in various animal hosts should be carried out as a prelude to library development and should be conducted separately using individual methods if a method comparison is planned. Such strategy would ensure that the genetic diversity of E. coli populations in host animals is captured, while avoiding unnecessary, expensive processing of isolates that would cause redundancy in the library (17). Furthermore, this strategy is generally applicable to studies of E. coli population biology.
Diversity measurements for all host species were significantly higher by ARA than by ribotyping, and accumulation curves demonstrated that, for representative sampling, greater sampling effort is required with ARA than with ribotyping. Almost four times as many isolates were processed using ARA compared to that using ribotyping, which was a logistically driven decision based on the relative costs of the analyses. Although most published MST studies have relied mainly on logistical factors to determine the number of isolates typed per animal, this work demonstrates the pitfalls associated with such an approach. Comparing ribotype diversity data with ARP diversity data from this study must be done with caution because of the discrepancy in the number of isolates typed; however, comparisons can be made in the cases of host species whose fecal flora was adequately represented by the sampling effort. E. coli ribotypes in cattle, which were adequately sampled (Fig. 2A), were far less diverse than ARPs in cattle. Although cattle ARPs were undersampled, the comparison is still informative because, if anything, the undersampling of cattle ARPs underestimated the phenotypic diversity. Likewise, E. coli ribotypes from human individuals were less diverse than ARPs and both of these parameters were adequately sampled (Fig. 2A and B).
Subtype sharing within and among host species on a limited temporal scale.
Individuals were sampled three times in one day to determine the observed overlap of E. coli subtypes within individuals, among individuals of the same host species, and among individuals belonging to different species. The individuals belonging to each species were not from independent populations, as horses were from one herd, cattle were from one herd, and humans worked in the same laboratory. We hypothesized that extensive sharing of subtypes within a host group would occur, particularly among horse individuals and among cow individuals; however, almost 47% of E. coli ribotypes sampled from horses were unique (observed only once in horses during the study) (Table 3). This observation reflects the high diversity of the populations in horse feces and is probably also influenced by the fact that they were undersampled. Ribotypes of E. coli isolated from cattle and humans, which were adequately sampled, were much less likely to be unique within their respective groups, but were most likely to be sampled from only one individual (50.7 and 95.9%, respectively). Only cattle demonstrated a tendency to share ribotypes between two individuals (46.7%), in contrast to results reported in a previous study (16) that found that any given E. coli ribotype was infrequently sampled from more than one animal in a herd. Although the authors did not report diversity measurements in their work, a rough comparison with our results is possible by comparing estimated richness values. The average richness of E. coli ribotypes from cows was 3.4 in this study. Twenty-four cows sampled once each in the previous study (16) yielded 240 different ribotypes, for a rough richness measurement of 10 within each animal. The greater richness estimate obtained from the previous study could well be due to a difference in the ribotyping method, as that study used two enzymes for ribotyping, while our study used one enzyme. One possible interpretation of this comparison is that the ability of the two-enzyme ribotyping method to discriminate between closely related E. coli subtypes obscured the relatedness of the E. coli isolates within the cattle herds.
TABLE 3.
Sharing of E. coli subtypes within and among individuals belonging to the same host speciesa
| Subtyping method and pattern | Cattle (%) | Horse (%) | Human (%) |
|---|---|---|---|
| Ribotyping | |||
| Unique (observed once) | 10.6 | 46.6 | 4.1 |
| Multiple within individual | 42.7 | 50.7 | 95.9 |
| Shared by two individuals | 46.7 | 2.7 | 0.0 |
| Shared by three individuals | 0.0 | 0.0 | 0.0 |
| Shared by four individuals | 0.0 | 0.0 | 0.0 |
| Total | 100 | 100 | 100 |
| ARA | |||
| Unique (observed once) | 9.5 | 2.7 | 3.5 |
| Multiple within individual | 13.5 | 5.5 | 21.1 |
| Shared by two individuals | 52.8 | 23.1 | 42.2 |
| Shared by three individuals | 12.1 | 40.2 | 0.0 |
| Shared by four individuals | 12.1 | 28.5 | 33.2 |
| Total | 100 | 100 | 100 |
The categories are mutually exclusive, e.g., a subtype shared by three individuals would be counted only in that category. For ribotyping, n was 75, 72, and 75 for cattle, horses, and humans, respectively; for ARA, n was 226, 302, and 355 for cattle, horses, and humans, respectively.
ARPs were generally shared among many individuals, in contrast to ribotypes. The majority of E. coli isolates from cattle, horses, and humans had an ARP found in two or more individuals, and a number of ARPs were observed in as many as four different individuals from the same host species (Table 3). Horses demonstrated the greatest frequency of ARP sharing (91.8% of ARPs shared among multiple individuals), while humans demonstrated the lowest frequency of ARP sharing among multiple individuals (75.4%), although this value was still very high compared to that for ribotype-sharing frequency. The ARP sharing among horses stands in contrast to ribotype sharing and may be due, in part, to greater sampling effort in the case of ARA.
Ribotype and ARP sharing among different host species were also assessed (Table 4). Most of the E. coli isolates (70.9%) had a ribotype that was observed in only one host species (Table 4). More than 22% of E. coli isolates shared a ribotype that was observed in both horses and cattle, and 6.7% of the E. coli isolates had a ribotype that was shared by cattle, horses, and humans. It is possible that a greater extent of interspecies ribotypes sharing would have been observed if E. coli populations in horses had been adequately sampled.
TABLE 4.
E. coli subtype distribution among host speciesa
| Host species | % Ribotypes (n = 222) | % ARPs (n = 857) |
|---|---|---|
| Human only | 28.8 | 2.4 |
| Cow only | 10.9 | 9.0 |
| Horse only | 31.1 | 11.6 |
| Human and horse | 0 | 3.4 |
| Human and cow | 0 | 0.6 |
| Horse and cow | 22.5 | 14.8 |
| Human, horse, and cow | 6.7 | 59.2 |
The categories are mutually exclusive, e.g., a subtype shared by three host species would be counted only in that category.
A high degree of interspecies sharing of E. coli ARPs was observed (Table 4) in spite of the fact that E. coli populations in both horses and cattle were undersampled. One would expect that the extent of interspecies sharing would increase, if anything, if representative sampling were attained for all host species. The majority of E. coli isolates (77.0%) had an ARP that was observed in two or more host species. In fact, 59% of the E. coli isolates analyzed by ARA had an ARP observed in all three host species. A very small proportion of E. coli isolates had an ARP that was shared only by horses and humans or by cattle and humans.
Few studies have directly compared E. coli subtype distributions within different host species. One study subtyped 655 E. coli isolates by multilocus enzyme electrophoresis from 34 different individuals (23 humans, 6 dogs, and 5 cats) (5). Subtypes were frequently observed multiple times within single individuals (85% of E. coli isolates), and only a small proportion of subtypes (7%) were shared between individuals (5). Comparatively greater E. coli subtype sharing between individuals was observed in our study, particularly in the cases of ribotypes from cattle and in general for ARPs. The extent of subtype sharing was probably influenced both by the host species sampled and by the typing methods used.
We originally hypothesized that subtype sharing would be hierarchical, i.e., the greatest frequency of subtype sharing would occur within individual animals, followed by sharing between different individuals in the same host category, and finally between animals in different host categories (the last two are major assumptions of library-based MST methods). Horses and cattle tended to share ribotypes, and the majority of E. coli ARPs were observed in all three source categories. These observations are not consistent with the “host specificity” characteristic of an ideal source identifier, which states that subtypes be restricted to a particular host species or group (30). Limited sharing among host groups can still allow differential distribution of E. coli subtypes (13, 15), which is a characteristic of a useful source identifier (30); however, library-based methods using ribotyping (21, 22, 26, 29) and ARA (13, 21) of E. coli have demonstrated incomplete discrimination among isolates from various host species. The sharing of subtypes among various host groups is doubtless a major contributor to the inaccurate source classifications observed in these studies.
Temporal variability of E. coli populations within individuals.
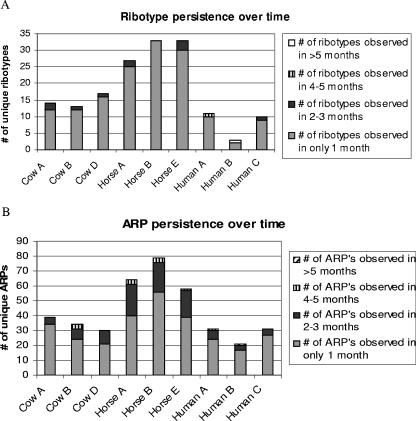
Three individuals of each host species were sampled once a month for 6 to 8 months. The number of months during which each specific ribotype was observed in each individual is shown in Fig. 3. Each bar represents the number of different subtypes observed within one individual, and the pattern coding represents the number of times it was observed. For example, samples from cow A yielded 14 different ribotypes over the time course (Fig. 3A); 12 were observed in only one sample (one month), and 2 were observed during 2 to 3 months. The trends in genotypic diversity observed in this 8-month study were similar to those observed in the 1-day experiment discussed above, i.e., E. coli subtype diversity was highest in horse feces, followed by cattle, and then humans. This observation is apparent from the height of each bar in Fig. 3A; although roughly the same number of isolates (35 to 40) were typed per individual, the number of ribotypes observed over time in horses was much greater than that observed in cows and humans. E. coli subtype richness values were similar (and not significantly different) among individuals belonging to the same host species. The vast majority of ribotypes were observed at only a single sampling event in a given animal (Fig. 3A). There was little difference between the persistence of E. coli ribotypes from different source categories, although the most persistent ribotype was isolated from human B and was observed in samples from six different months (resulting in a very short bar in Fig. 3A).
FIG. 3.
The temporal persistence of E. coli subtypes isolated from individual cattle, horses, and humans as determined by ribotyping (A) and ARA (B). Each bar represents the number of different subtypes sampled from one individual over 7 to 8 months, and the shading designates the number of months in which the subtypes were observed.
ARA subtyping of E. coli yielded similar results (Fig. 3B). The measured diversity for all individuals at any given month was similar to that observed at the one-time sampling event; horses harbored the most diverse E. coli population and humans the least diverse. Most ARPs were observed at only a single sampling event, but the most persistent ARP was observed in human B and was sampled at six different months, which coincides with ribotyping results (Fig. 3B). These results suggest that the E. coli populations in feces of individual humans, horses, and cows are not temporally stable and experience significant turnover on a monthly time scale but also that these population characteristics can differ among host individuals of the same species.
Subtype sharing among individuals within a host species was assessed across the 6- to 8-month sampling period (Table 5). For this analysis, subtypes were considered “shared” if E. coli strains with the same subtype were isolated from two different individuals, regardless of the sampling date. Trends in subtype sharing were similar to those in the 1-day experiment, as a small proportion of ribotypes and ARPs were shared between individuals within each source category. Humans demonstrated the lowest frequency of ribotype sharing among individuals. Only one ribotype (4.5% of the different ribotypes identified) was shared by two individuals, and no ribotypes were shared by three individuals (Table 5). Cattle demonstrated a higher frequency of ribotype sharing. Of the different ribotypes identified in cow feces, 18.9% were shared by two individuals, and none were shared by three. Horses also tended to share ribotypes over time and were the only species in which one ribotype (8.5%) was shared by three individuals. Similar trends were noted for ARA (Table 5), although the frequency of sharing was generally greater than that observed for ribotyping. The frequency of ARP sharing was lowest among humans (17.6%) and slightly higher among cattle and among horses. In contrast with ribotyping results, shared ARPs were frequently observed among all three individuals of each host species (53% for cattle, 34% for horses, and 17% for humans).
TABLE 5.
Subtype sharing over a 6- to 8-month temporal scalea
| Subtyping method (no. of individuals) | No. of different ribotypes or ARPs | No. (%) of ribotypes or ARPs shared within a host species | % Sharing prevalence inb:
|
|
|---|---|---|---|---|
| Two individuals | Three individuals | |||
| Ribotypes | ||||
| Cow (3) | 37 | 7 (18.9) | 100.0 | 0 |
| Horse (3) | 80 | 11 (13.8) | 91.5 | 8.5 |
| Human (3) | 22 | 1 (4.5) | 100.0 | 0 |
| ARPs | ||||
| Cow (3) | 76 | 17 (22.4) | 47.0 | 53.0 |
| Horse (3) | 155 | 32 (20.6) | 66.0 | 34.0 |
| Human (3) | 68 | 12 (17.6) | 83.0 | 17.0 |
Results show the number of different subtypes observed in three individuals of each host species and the frequency of subtype sharing among individuals within a host species. Prevalence represents the number of host individuals that shared these subtypes.
The percentage of shared subtypes observed in two or three individuals from a host species.
The distributions of E. coli subtypes in the feces of individuals were not temporally stable and changed on a monthly basis, although they were stable over a 1-day time scale (discussed above). Furthermore, the majority of E. coli subtypes observed at any given time point within an individual did not persist for longer than 1 month. These results support the findings from a previous study on changes in the E. coli population isolated from a human over time (6), in which typing by multilocus enzyme electrophoresis determined that most subtypes were observed at only one sampling event, and the previously discussed study on E. coli populations in cattle (16). Our study extends these findings to multiple individuals of three host species. These findings indicate that an MST library of E. coli subtypes isolated from known sources is unlikely to be representative of the E. coli population impacting a water body unless known-source fecal samples are collected concurrently with water samples, which is not the current practice in MST studies (30).
Temporal variability of the E. coli population of one human over 1 month.
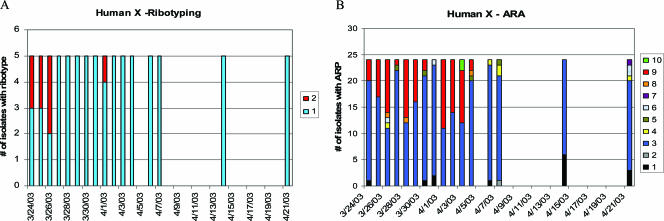
The preceding experiments demonstrated that within individuals, the dominant E. coli subtypes that are shed in feces changes greatly in a single month. In order to assess the turnover of an E. coli population within an individual on a finer time scale, daily samples were taken from a human (human X) who was not sampled during the previous experiments (Fig. 4A and B). Human X had been on an antibiotic treatment of trimethoprim-sulfamethoxazole for 1 week, which ended 5 days prior to the first sampling. Both ribotyping and ARA results demonstrated similar trends: the E. coli population was dominated by two distinct subtypes. Ribotype 2 and ARP 9 were observed in only the first few days of sampling, while ribotype 1 and ARP 3 persisted throughout the entire month. Ribotype 2 was always associated with ARP 9. E. coli with ribotype 1 usually had ARP 3 (94%), but some ribotype 1/ARP 9 isolates were sampled during the second week of the experiment (6%). ARP 9 represents an E. coli subtype that was relatively antibiotic resistant, i.e., ARP 9 denotes resistance to amoxicillin (32 μg ml−1), cephalothin (8 μg ml−1), chlortetracycline (80 μg ml−1), doxycycline (4 μg ml−1), oxytetracycline (20 μg ml−1), penicillin (200 μg ml−1), streptomycin (20 μg ml−1), tetracycline (64 μg ml−1), trimethoprim (1 μg ml−1), and trimethoprim/sulfonamide (5 μg ml−1) and could also grow on low levels of chloramphenicol (4 μg ml−1) and rifampin (2 μg ml−1). ARP 3 was susceptible to low levels of all antibiotics, tolerating only chloramphenicol (4 μg ml−1), chlortetracycline (20 μg ml−1), penicillin (20 μg ml−1), rifampin (2 μg ml−1), and trimethoprim (0.25 μg ml−1). Although isolates with ARP 9 were assessed for plasmid presence, none were found and the possibility of integron presence was not explored.
FIG. 4.
The abundance and persistence of subtypes within human X over 1 month as determined by ribotyping (A) and ARA (B). Each bar represents the number of isolates typed for each sampling date. Each different subtype is designated by a number and a color.
The dominance of ribotype 2/ARP 9 in the first days of the study, followed by the turnover and dominance by ribotype 1/ARP 3 (Fig. 4), suggests that the E. coli population experienced selective pressure due to antibiotic therapy prior to the sample cycle. The ribotype 2/ARP 9 strain may have been outcompeted by ribotype 1/ARP 3 as the selective pressure diminished. In spite of the presumably disruptive influence of antibiotic therapy on the microbial population of the gastrointestinal tract, the E. coli population shed by human X demonstrated far less turnover in 1 month than did populations sampled over the 6- to 8-month time period, with the exception of the loss of ribotype 2/ARP 9. This experiment detailed the succession of one dominant subtype over another and included a possible mechanistic explanation (antibiotic treatment, followed by withdrawal). Other selective pressures that may contribute to temporal variability of E. coli subtypes in individuals include diet, exposure to novel strains through travel or contact with other individuals, and the health of the individual.
Concluding remarks.
E. coli populations of beef cattle, horses, and humans exhibited complex structures and were temporally dynamic. Although the observed diversity of E. coli populations in this study was dependent upon the typing method, the trend in diversity of horses > cattle > humans was consistent. The sample size used to characterize population diversity in this study was arbitrarily chosen, and the data presented here illustrate the importance of using preliminary data to estimate an adequate sampling effort. The unstable and diverse nature of E. coli populations observed in this study has several practical implications with respect to its use as an indicator of water quality. The survival rates of E. coli subtypes (strains) in environmental waters can differ significantly according to strain, and some subtypes exhibit prolonged persistence in the culturable state in environmental waters (2). Such “survivor” E. coli strains can generate a false indication of recent fecal contamination and may well be present in the absence of pathogens. The results of this study suggest that it will be very difficult or impossible to generalize findings on E. coli persistence from one situation to the next, as the makeup of E. coli populations entering the water will be highly variable. With regard to MST methods, the dependence of E. coli population diversity on the host species, the great sampling effort required to obtain a representative sample in some host species, the temporal variability observed in all host types, and the lack of host specificity suggest that library-based MST methods utilizing E. coli experience major logistical limitations.
Acknowledgments
This study was funded by a grant from the U.S. Department of Agriculture, NRI grant 2001-35102-10790.
We thank Miriam Brownell, Kimberly Anderson, Mariya Dontchev, Joshua Day, and Sonia Magana for logistical and technical help.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.American Public Health Association. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, D.C.
- 2.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bitton, G., B. Koopman, and K. Jung. 1995. An assay for the enumeration of total coliforms and Escherichia coli in water and wastewater. Water Environ. Res. 67:906-909. [Google Scholar]
- 4.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caugant, D. A., B. R. Levin, and R. K. Selander. 1984. Distribution of multilocus genotypes of Escherichia coli within and between host families. J. Hyg. Camb. 92:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon, D. M. 1997. The genetic structure of Escherichia coli populations in feral house mice. Microbiology 143:2039-2046. [DOI] [PubMed] [Google Scholar]
- 9.Gotelli, N. J., and G. L. Entsminger. 2004. EcoSim: null models software for ecology, version 7. Acquired Intelligence, Inc. and Kesey-Bear, Burlington, Vt.
- 10.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartel, P. G., J. D. Summer, J. L. Hill, J. V. Collins, J. A. Entry, and W. I. Segars. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 12.Harwood, V. J., J. Butler, D. Parrish, and V. Wagner. 1999. Isolation of fecal coliform bacteria from the diamondback terrapin (Malaclemys terrapin centrata). Appl. Environ. Microbiol. 65:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harwood, V. J., B. Wiggins, C. Hagedorn, R. D. Ellender, J. Gooch, J. Kern, M. Samadpour, A. C. H. Chapman, B. J. Robinson, and B. C. Thompson. 2003. Phenotypic library-based microbial source tracking methods: efficacy in the California collaborative study. J. Water Health 1:153-166. [PubMed] [Google Scholar]
- 14.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins, M. B., P. G. Hartel, T. J. Olexa, and J. A. Stuedemann. 2003. Putative temporal variability of Escherichia coli ribotypes from yearling steers. J. Environ. Qual. 32:305-309. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leclerc, H., D. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201-234. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig, J. A., and J. F. Reynolds. 1988. Statistical ecology. A primer on methods and computing. John Wiley and Sons, New York, N.Y.
- 20.McLellan, S. L., A. D. Daniels, and A. L. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, D. F., V. J. Harwood, D. M. Ferguson, J. Lukasik, P. Hannah, M. Getrich, and M. Brownell. 2005. Evaluation of antibiotic resistance analysis and ribotyping for identification of faecal pollution sources in an urban watershed. J. Appl. Microbiol. 99:618-628. [DOI] [PubMed] [Google Scholar]
- 22.Myoda, S. P., C. A. Carson, J. J. Fuhrmann, B.-K. Hahm, P. G. Hartel, H. Yampara-Iquise, L. Johnson, R. L. Kuntz, C. H. Nakatsu, M. J. Sadowsky, and M. Samadpour. 2003. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 1:167-180. [PubMed] [Google Scholar]
- 23.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Power, M. L., J. Littlefield-Wyer, D. M. Gordon, D. A. Veal, and M. B. Slade. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631-640. [DOI] [PubMed] [Google Scholar]
- 25.Ritter, L. L., E. Carruthers, C. A. Carson, R. D. Ellender, V. J. Harwood, K. Kingsley, C. Nakatsu, M. Sadowsky, B. Shear, B. West, J. E. Whitlock, B. A. Wiggins, and J. D. Wilbur. 2004. Assessment of statistical methods used in library-based approaches to microbial source tracking. J. Water Health 1:209-223. [PubMed] [Google Scholar]
- 26.Scott, T. M., S. Parveen, K. M. Portier, J. B. Rose, M. L. Tamplin, S. R. Farrah, A. Koo, and J. Lukasik. 2003. Geographical variation in ribotype profiles of Escherichia coli isolates from humans, swine, poultry, beef, and dairy cattle in Florida. Appl. Environ. Microbiol. 69:1089-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selander, R. K., T. K. Korhonen, V. Vaisanen-Rhen, P. H. Williams, P. E. Pattison, and D. A. Caugant. 1986. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect. Immun. 52:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 29.Stoeckel, D. M., M. V. Mathes, K. E. Hyer, C. Hagedorn, H. Kator, J. Lukasik, T. L. O'Brien, T. W. Fenger, M. Samadpour, K. M. Strickler, and B. A. Wiggins. 2004. Comparison of seven protocols to identify fecal contamination sources using Escherichia coli. Environ. Sci. Technol. 38:6109-6117. [DOI] [PubMed] [Google Scholar]
- 30.U.S. Environmental Protection Agency. 2005. Microbial source tracking guide document. EPA/600/R-05/064. U.S. Environmental Protection Agency, Washington, D.C.
- 31.Whitlock, J. E., D. T. Jones, and V. J. Harwood. 2002. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 36:4273-4282. [DOI] [PubMed] [Google Scholar]
- 32.Whitman, R. L., and M. B. Nevers. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 69:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Geographic components of linkage disequilibrium in natural populations of Escherichia coli. Mol. Biol. Evol. 1:67-83. [DOI] [PubMed] [Google Scholar]
- 34.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Multilocus genetic structure in natural populations of Escherichia coli. Proc. Natl. Acad. Sci. USA. 80:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiggins, B. A., P. W. Cash, W. S. Creamer, S. E. Dart, P. P. Garcia, T. M. Gerecke, J. Han, B. L. Henry, K. B. Hoover, E. L. Johnson, K. C. Jones, J. G. McCarthy, J. A. McDonough, S. A. Mercer, M. J. Noto, H. Park, M. S. Phillips, S. M. Purner, B. M. Smith, E. N. Stevens, and A. K. Varner. 2003. Use of antibiotic resistance analysis for representativeness testing of multiwatershed libraries. Appl. Environ. Microbiol. 69:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]