Abstract
Diverse rhizobia able to nodulate Biserrula pelecinus evolved following in situ transfer of nodA and nifH from an inoculant to soil bacteria. Transfer of these chromosomal genes and the presence of an identical integrase gene adjacent to a Phe tRNA gene in both the inoculant and recipients indicate that there was lateral transfer of a symbiosis island.
The rates at which rhizobial populations diversify and the evolutionary forces structuring the genetic divergence of rhizobial populations in Australian soils are largely unknown. An opportunity to observe the in situ development of rhizobial diversity arose after the introduction of the pasture legume Biserrula pelecinus L. from the Mediterranean region to Western Australia (W.A.). Numerous field experiments during the past 12 years revealed that the indigenous rhizobial populations in W.A. were not capable of nodulating B. pelecinus. Parallel studies have clearly shown that symbiotically impaired, genetically and phenotypically diverse strains able to nodulate B. pelecinus recently emerged following the sowing of inoculated seed of this legume in W.A. (unpublished data). In 1994 a field site at Northam, W.A., was established using surface-sterilized seeds of B. pelecinus cv. Casbah inoculated with a single mesorhizobial strain, WSM1271, originating from Sardinia (5). The aim of the current study was to determine whether there was in situ transfer of symbiotic genes from WSM1271 to four diverse strains (N17, N18, N45, and N87) isolated in 2000 from nodules on B. pelecinus growing at the Northam site.
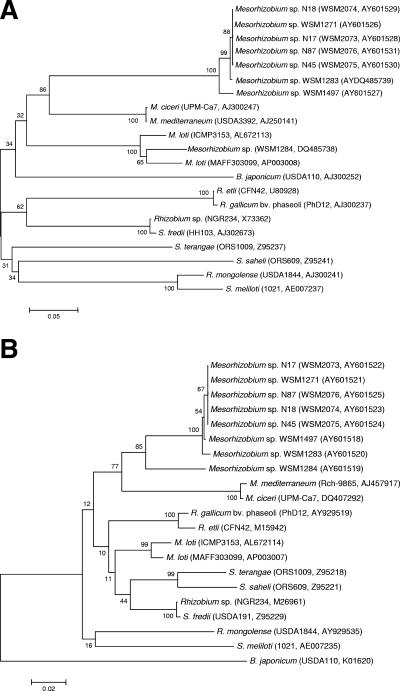
Comparison of the nodA and nifH sequences of Biserrula mesorhizobia.
The nodA nodulation gene determines the type of N-acyl substitution transferred onto the oligosaccharide backbone of Nod factor (1). A 567-bp intragenic nodA fragment was amplified with two primers, nodA-KF (5′-GGTTATGCTGGGAAAATGAGTTGC-3′) and nodA-KR (5′-CATAGCTCTGGACCGTTCC-3′), designed by aligning the nodA sequences of Mesorhizobium loti strains MAFF303099 and ICMP3153. Standard PCR mixture and cycling conditions were used, with five initial cycles (annealing at 52°C), followed by 30 cycles (annealing at 55°C). Two other primers, nodA-midF (5′-CGGGGTGCGTCGGGATCTTG-3′) and nodA-midR (5′-CCGAACCGTGCCGAAAGCGAATGG-3′), were developed from the middle overlapping region of the sequences obtained by using nodA-KF and nodA-KR. All four primers were used to obtain a double-stranded DNA sequence of nodA. Nitrogenase is the key enzyme in nitrogen fixation, and nifH is responsible for the structural development of component II of nitrogenase (11). A 710-bp intragenic nifH fragment was amplified using primers nifH-1 and nifH-2 (2). Two other primers, nifH-midF (5′-AGCCGAACACCGCCCGAATG-3′) and nifH-midR (5′-CTCGGGGACGTGGTTTGCG-3′), were developed in a manner similar to the manner described above for nodA, and all four primers were used to obtain a double-stranded DNA sequence of nifH. Remarkably, the sequences of both genes from the four diverse strains (N17, N18, N45, and N87) exhibited 100% identity to the sequences of the same genes in WSM1271. To put this finding in context, we compared these sequences with the sequences of nodA and nifH genes in three strains of Biserrula mesorhizobia of Mediterranean origin (WSM1283, WSM1284, and WSM1497). There were 2-bp and 5-bp mismatches for nodA and nifH, respectively, between WSM1283 and WSM1271. There were 156-bp and 48-bp mismatches for nodA and nifH, respectively, between WSM1284 and WSM1271, while there were 14-bp and 4-bp mismatches for nodA and nifH, respectively, between WSM1497 and WSM1271. Phylogenetic trees constructed using nodA (Fig. 1A) and nifH (Fig. 1B) clearly show that the identical sequences in WSM1271 and the four diverse strains are very different from those in other strains of Biserrula mesorhizobia and other root nodule bacteria. It is quite common to find intraspecific nucleotide variations in either gene (4, 9). These findings strongly indicate that the diverse strains obtained both nodA and nifH from WSM1271 through lateral transfer.
FIG. 1.
Phylogenetic trees showing the relationships of novel isolates N17, N18, N45, and N87 and Biserrula mesorhizobial strains WSM1271, WSM1283, WSM1284, and WSM1497 to some members of the root nodule bacteria in the α-Proteobacteria based on aligned sequences of the nodA (A) and nifH (B) genes. Phylogenetic analyses were performed using MEGA, version 3.1 (8). Kimura two-parameter distances were derived from the aligned sequences (7) and a bootstrap analysis (3) was performed with 500 replicates in order to construct a consensus unrooted tree using the neighbor-joining method (12) for each gene alignment separately. B., Bradyrhizobium; M., Mesorhizobium; R., Rhizobium; S., Sinorhizobium.
Localization of nodA and nifH in Biserrula mesorhizobia.
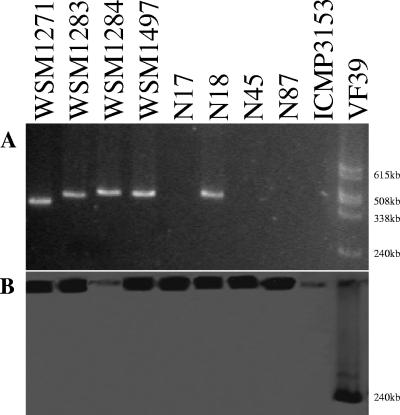
To determine the location of nodA and nifH in Biserrula mesorhizobia, we searched for plasmid-borne symbiotic genes by first separating plasmids by Eckhardt gel electrophoresis (6) and then hybridizing blotted plasmid DNA to nodA and nifH probes using standard procedures (Boehringer Mannheim). WSM1271 contained a single approximately 500-kb plasmid. WSM1283, WSM1284, WSM1497, and N18 each had a single plasmid that was slightly larger than the plasmid of WSM1271, while N17, N45, and N87 lacked a plasmid (Fig. 2A). The Eckhardt gel procedure was repeated three times with multiple replicates of each strain to confirm the absence of a plasmid in N17, N45, and N87. The symbiotic probes did not hybridize with the plasmid DNA of any strain, while hybridization was detected in wells containing chromosomal DNA from WSM1271, WSM1283, WSM1497, N17, N18, N45, and N87 (Fig. 2B), suggesting that nodA and nifH are located on the chromosome in these strains. Biserrula mesorhizobia are closely related to M. loti (10). Sullivan and Ronson (13) demonstrated that symbiotic genes of M. loti strain ICMP3153 are present on a chromosomally located mobile symbiosis island. Therefore, we investigated whether the symbiotic genes of Biserrula mesorhizobia are located on a similar mobile genetic element.
FIG. 2.
(A) Eckhardt gel displaying plasmid profiles of strains WSM1271, WSM1283, WSM1284, WSM1497, N17, N18, N45, and N87; M. loti strain ICMP3153 (negative control); and Rhizobium leguminosarum strain VF39 (positive control used to estimate the approximate sizes of plasmids). (B) Southern blot of the Eckhardt gel hybridized with a probe mixture consisting of nifH and nodA of Biserrula mesorhizobia and nodC of R. leguminosarum. Note the hybridization of the nodC probe to the 240-kb plasmid of strain VF39 and the hybridization of the nifH and nodA probes to the chromosomal DNA in the wells.
Symbiosis island insertion site of Biserrula mesorhizobia.
The symbiosis islands in M. loti strains ICMP3153 and MAFF303099 integrate into a phenylalanine tRNA (Phe tRNA) gene (13). These islands contain a gene that codes for a phage P4-like integrase (intS) that is responsible for the excision and insertion of symbiosis islands (13). This integrase gene is located at one end of the island and is next to the Phe tRNA gene (13). Therefore, a forward primer, phetRNAf (5′-GCCCAGATAGCTCAGTTGGT-3′), was developed by aligning Phe tRNA gene sequences of the two strains described above and 10 other diverse bacterial genera. A reverse primer, intS522r (5′-ATTGCATATCGAAACACG-3′), was developed to anneal in the midregion of intS by aligning intS sequences of ICMP3153 and MAFF303099. An 800-bp fragment was successfully amplified for the four Mediterranean Biserrula mesorhizobia and the four diverse strains using standard PCR procedures with annealing at 50°C. A 51-bp region of the Phe tRNA gene, which did not include the forward primer binding site, was identical in all eight strains. Furthermore, identical sequences were present in N17, N18, N45, N87, and WSM1271 for both intS and the intergenic region (198 bp) between the Phe tRNA gene and intS (Fig. 3). There was considerable variation in the sequences of intS and the intergenic region among WSM1271, WSM1283, WSM1284, and WSM1497 (Fig. 3). However, the intS sequences (1,070 bp) of the Biserrula mesorhizobia exhibited >93.5% similarity to the intS sequence of M. loti (data not shown).
FIG. 3.
Sequence alignment for intergenic region between the Phe tRNA gene and intS for Biserrula mesorhizobia and M. loti strains ICMP3153 and MAFF303099. The start of intS is indicated by shading. Sequences were aligned using Genetool lite (BioTools Inc., Edmonton, Canada).
The evidence that the Mediterranean Biserrula mesorhizobia and the four diverse strains isolated from Northam contain a P4-like integrase gene similar to that found in the symbiosis islands of M. loti and the location of this gene adjacent to a Phe tRNA gene led us to propose that the symbiotic genes of Biserrula mesorhizobia are located on a mobile symbiosis island which integrates into a Phe tRNA gene. These findings confirm that the diverse and ineffective strains isolated in Northam, W.A., that are able to nodulate B. pelecinus evolved through in situ transfer of symbiotic genes on a symbiosis island from WSM1271 to resident soil bacteria.
Acknowledgments
This work was supported by a postdoctoral research-funded fellowship from Grains Research and Development Corporation (GRDC) and Australian Wool Innovation Ltd. (AWI) within the national Rhizobium program.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Downie, J. A. 1998. Functions of rhizobial nodulation genes, p. 387-402. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 2.Eardly, B. W., J. P. W. Young, and R. K. Selander. 1992. Phylogenetic position of Rhizobium sp. strain Or 191, a symbiont of both Medicago sativa and Phaseolus vulgaris, based on partial sequences of the 16S rRNA and nifH genes. Appl. Environ. Microbiol. 58:1809-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 4.Haukka, K., K. Lindstrom, and J. P. W. Young. 1998. Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl. Environ. Microbiol. 64:419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howieson, J. G., A. Loi, and S. J. Carr. 1995. Biserrula pelecinus L.—a legume pasture species with potential for acid, duplex soils which is nodulated by unique root-nodule bacteria. Aust. J. Agric. Res. 46:997-1009. [Google Scholar]
- 6.Hynes, M. F., R. Simon, and A. Pühler. 1985. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid 13:99-105. [DOI] [PubMed] [Google Scholar]
- 7.Kimura, M. 1980. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 8.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefs Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 9.Laguerre, G., S. M. Nour, V. Macheret, J. Sanjuan, P. Drouin, and N. Amarger. 2001. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981-993. [DOI] [PubMed] [Google Scholar]
- 10.Nandasena, K. G., G. W. O'Hara, R. P. Tiwari, R. J. Yates, and J. G. Howieson. 2001. Phylogenetic relationships of three bacterial strains isolated from the pasture legume Biserrula pelecinus L. Int. J. Syst. Evol. Microbiol. 51:1983-1986. [DOI] [PubMed] [Google Scholar]
- 11.Rubio, L. M., and P. W. Ludden. 2000. The gene products of the nif regulon, p. 101-130. In G. J. Leigh (ed.), Nitrogen fixation at the millennium. Elsevier, Amsterdam, The Netherlands.
- 12.Saitou, N., and M. Nei. 1987. Reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]