Abstract
Fifty-one human glycosyltransferases were expressed in Saccharomyces cerevisiae as immobilized enzymes and were assayed for enzymatic activities. The stem and catalytic regions of sialyl-, fucosyl-, galactosyl-, N-acetylgalactosaminyl-, and N-acetylglucosaminyltransferases were fused with yeast cell wall Pir proteins, which anchor glycosyltransferases at the yeast cell wall glucan. More than 75% of expressed recombinant glycosyltransferases retained their enzymatic activities in the yeast cell wall fraction and will be used as a human glycosyltransferase library. In increasing the enzymatic activities of immobilized glycosyltransferases, several approaches were found to be effective. Additional expression of yeast protein disulfide isomerase increased the expression levels and activities of polypeptide N-acetylgalactosaminyltransferases and other glycosyltransferases. PIR3 and/or PIR4 was more effective than PIR1 as a cell wall anchor when the Pir-glycosyltransferase fusions were expressed under the control of the constitutive glyceraldehyde-3-phosphate dehydrogenase promoter. Oligosaccharides such as Lewis x, Lewis y, and H antigen were successfully synthesized using this immobilized glycosyltransferase library, indicating that the Pir-fused glycosyltransferases are useful for the production of various human oligosaccharides.
Most oligosaccharides exist as glycoproteins and glycolipids, many of which are localized at the cell surface and are involved in biologically important processes such as cellular adhesion, molecular recognition, and signal transduction (58). The oligosaccharides are extremely rich in structural variations, and isomers exhibit different branching structures, anomeric configurations, and linkage positions; these vary widely with species, tissue, and degree of cellular differentiation. To clarify the structure-function relationship of oligosaccharides, the synthesis and remodeling of oligosaccharides are required. In order to synthesize complex oligosaccharides on proteins and lipids or in living cells, enzymatic methods are more suitable than chemical methods, since degradation or denaturation of active proteins, lipids, and living cells can be avoided. Human-derived enzymes are particularly ideal for the synthesis of mammalian oligosaccharides, such as sialylated and branched glycans, because these reactions are difficult to reproduce with either chemical methods or bacterial enzymes (19).
At present, nearly 200 human glycosyltransferase genes are known and are available for use. These are indispensable for enzymatic synthesis of human-type oligosaccharides (41). However, it is important to make these resources suitable for application as enzyme sources. Immobilized enzymes enable the recovery of valuable enzymes for repeated usage and have several advantages for industrial applications; they are usually more stable than free enzymes and can be applied via automated bioreactor-packed columns.
Glycosyltransferases generally contain a catalytic domain at the C-terminal region and a membrane-spanning region at the N terminus, with which glycosyltransferases are localized at the Golgi membrane. It is also known that many glycosyltransferases are secreted into culture media when the membrane-spanning region is truncated (4, 5, 33). Purification of soluble enzymes is laborious, and each enzyme requires a separate protocol for purification from culture broths or cell homogenates. In contrast, immobilization of soluble enzymes to an insoluble carrier by chemical methods is often accompanied by partial loss of enzymatic activity (11, 14, 15). Glycosyltransferases are generally immobilized at the N-terminal region, whereas the C terminus of glycosyltransferases is very sensitive to modification, since it generally includes the catalytic domain. For example, a single-amino-acid replacement of the C terminus of fucosyltransferase III (FUT3) results in a loss of activity (59), while the use of site-directed mutagenesis to eliminate glycosylation sites from some glycosylated human glycosyltransferases, such as FUT3, FUT5, FUT6, α-2,6-sialyltransferase I (ST6Gal I), and core 2 β-1,6-N-acetylglucosaminyltransferase (C2GnT), leads to the loss of enzymatic activity (9, 13, 55). Although some mammalian glycosyltransferases have been expressed in Escherichia coli, they were often in an insoluble form (6, 16). On the other hand, yeasts including Saccharomyces cerevisiae and Pichia pastoris, which are unicellular eukaryotic organisms equipped with a protein glycosylation system, are suitable hosts for producing recombinant human glycosyltransferases, provided that reasonable immobilization protocols are developed (5, 20).
The conventional method for immobilizing proteins on the yeast cell wall was developed using glycosylphosphatidylinositol (GPI) as an anchor to bind proteins onto the β-1,6-glucan of the cell wall (39, 46). However, because the GPI anchor signal sequence is located at the C-terminal end of the precursor proteins (47), this method is not suitable for the immobilization of glycosyltransferases.
It was recently reported that the yeast S. cerevisiae contains another group of cell wall mannoproteins, Pir (protein with internal repeats), which are covalently bound to the β-1,3-glucan of the cell wall in a manner different from that of the GPI anchor (35). The molecular mechanism of Pir protein binding to the cell wall remains unclear (12, 25), but Pir protein sequences possess binding potential irrespective of the fusion site, i.e., at either terminus of the target protein (32, 51). This property is useful for immobilizing glycosyltransferases. In fact, α-1,2-galactosyltransferase from the fission yeast Schizosaccharomyces pombe maintained high enzymatic activity when expressed as a fusion protein with Pir in S. cerevisiae, while the same enzyme completely lost enzymatic activity when fused with the GPI anchor signal sequence at the C terminus (1).
In this study, transmembrane region-truncated forms of various human glycosyltransferases were comprehensively expressed in forms immobilized through the Pir anchor at the yeast cell wall, and their enzymatic activities were analyzed. This analysis indicated that although the expression levels depend on individual enzyme properties, yeast is a suitable host for providing an enzyme source, since more than 75% of the enzymes tested retained their enzyme activities in our yeast cell wall immobilization system.
MATERIALS AND METHODS
Yeast strains, growth media, and genetic methods.
We used S. cerevisiae W303-1A strains (MATa leu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100) (53) in which three of the PIR genes were disrupted: YSF123 (pir1::HisG pir2::HisG pir3::HisG) and YSF124 (pir1::HisG pir2::HisG pir4::HisG). The PDI1 gene was integrated into the HIS3 locus of YSF123 and YSF124 cells and was overexpressed using the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter. SD-Ura medium (49) was used to cultivate yeast cells and to select yeast recombinant transformants. SD medium was maintained at pH 6.0 by addition of HEPES buffer. Yeast transformation was carried out by the lithium acetate method (28).
Cloning of truncated human glycosyltransferase genes.
In order to construct a range of recombinant genes, we used the Gateway system provided by Invitrogen Ltd. (Carlsbad, CA). Truncated glycosyltransferase genes, in which the N-terminal transmembrane region was eliminated, were prepared by PCR using a CACC adaptor sequence at the 5′ end of the forward primer for the glycosyltransferase genes in order to introduce them into the Gateway entry vector pENTER/D-TOPO. The GalNAc α-2,6-sialyltransferase II (ST6GalNAcT II) gene was introduced into the Gateway entry vector pDONOR221. The primer sequences for the truncated glycosyltransferase genes, which were constructed by the authors and M. Nakamura of AIST, are shown in Table 1. Fourteen truncated genes [encoding fucosyltransferases (FUT1, FUT3, FUT6, FUT7, FUT8, FUT9), α-1,3-galactosyltransferases or N-acetylgalactosaminyltransferases (ABO enzymes), β-1,3- and β-1,4-galactosyltransferases (β3GalNAcT1, β4GalT4, and β4GalT5), i-β-1,3-N-acetylglucosaminyltransferase (iGnT), and β-1,3-N-acetylglucosaminyltransferases (β3GnT2 and β3GnT3)], which were subcloned into the Gateway entry vector, were constructed and provided by M. Nakamura. The remaining 20 truncated genes (α-2,3-sialyltransferase III [ST3Gal III], ST6GalNAc I, β4GalT1, UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases [pp-GalNAc-T1, -T2, -T3, -T4, -T6, -T9, -T10, -T12, -T13, -T14, and -T15], core 2 β-1,6-N-acetylglucosaminyltransferases [C2GnT1, C2GnT2, and C2GnT3], IGnT3, β3GnT6, manic fringe [MFNG], and radical fringe [RFNG]), which were subcloned into the Gateway entry vector, were kindly provided by H. Narimatsu (22).
TABLE 1.
Forward and reverse primer sequences for PCR amplification of glycosyltransferase genes
| Gene | Forward primer sequencea | Reverse primer sequencea |
|---|---|---|
| FUT1 | caccCCACATGGCCTAGGCCTGTCGATCCT | TTCAAGGCTTAGCCAATGTCCAGAGTGG |
| FUT3 | caccGGATCCCCTAGGGCTCCCAGTGGGTC | TTCAGGTGAACCAAGCCGCTATGCTGCG |
| FUT6 | caccTCTCAAGACGATCCCACTGTGTACCC | TTCAGGTGAACCAAGCCGCTATGCCGC |
| FUT7 | caccGGTACCCCGGCACCCCAGCCCACG | TTCAGGCCTGAAACCAACCCTCAAGGTCC |
| FUT8 | caccCGAGATAATGACCATCCTG | GAGCTTTATTTCTCAGCCTCAGGAT |
| FUT9 | caccACCAACAGCTGGATCTTCAGTCCAAT | TTTAATTCCAAAACCATTTCTCTAAATT |
| ST3Gal I | caccAACTACTCCCACACCATGG | TCATCTCCCCTTGAAGATCCGGA |
| ST3Gal III | caccCAGTGGGAGGAGGACTCCAATTCA | TCAGATGCCACTGCTTAGATCAGT |
| ST3Gal IV | caccCGGGAAGACAGTTTTTATT | TCAGAAGGACGTGAGGTTCTTGA |
| ST3Gal VI | caccGAAATGAAACGGAGAAATAAGAT | TCAATCTTGAGTCAAGTTGATTA |
| ST6Gal I | caccTGGAAGGAAAAGAAGAAAGGGAG | TTAGCAGTGAATGGTCCGGAAGC |
| ST6Gal II | caccGACAGCAACCCCGCTGAGCCTGTA | TTAAGAGTGTGGAATGACTGGACTTGG |
| ST6GalNAc II | aaaaagcaggcttcGGAGCCAGGGACACCACATCATT | agaaagctgggtcTCAGCGCTGGTACAGCTGAAGGA |
| ST6GalNAc IV | caccGACCACCACTTCCCCACAGGCTC | CTACTCAGTCCTCCAGGACGGAT |
| Mouse ST8Sia VI | caccCCAGCCGACGCGCCTGCCCGC | TTAAGCCGTTTCACATTTGCT |
| A and B (ABO) | caccATGCCAGGAAGCC | TCACGGGTTCCGGAC |
| β3GalNAcT1 | caccCGCGTGAACTGGATGTACT | AGTTAATAATGGCATGTGGTGTTCCT |
| β3GalT4 | caccGGGTTGGGGGAGGAGCTGCTGAG | GCTCTGAAGCCAGGCTATTGCTC |
| β3GalT5 | caccTACAGTCTAAATCCTTTCAAAGA | GACAGGCGGACAATCTTCCCCCC |
| β4GalT4 | caccGGTGCCATTCAAGAGATTCC | CCAGGGTCATGCACCAAAC |
| β4GalT5 | caccCAAGCCCAAGGCATTCTGATCCG | GTACTCGTTCACCTGAGCCAGCT |
| pp-GalNAc-T7 | caccCTGACCCCGCGGCCGGACGACCC | CTAAACACTATGGATGTTATTCATTTCCC |
| pp-GalNAc-T8 | caccGGGACTTTACAAAACCTGTTTAC | TCACTGGCTGTTGGTCTGACCCC |
| iGnT | caccGCACGGGCAGGAGGA | AAGGGCTCAGCAGCGTC |
| β3GnT2 | caccAGCCAAGAAAAAAATGGAAAAGGGGA | TTTAGCATTTTAAATGAGCACTCTG |
| β3GnT3 | caccGAGCAGCCACCGGCGATCCCCGAG | TTCAGTAGATCTGTGTCTGATTGCCGCAGG |
| MGAT1 | caccACGCGCCCAGCACCTGGCAGGC | CTAATTCCAGCTAGGATCATAGCCCTCCCA |
| MGAT3 | caccCACTTCTTCAAGACCCTGTCCTA | CTAGACTTCCGCCTCGTCCAGTT |
| MGAT4A | caccACTACATGGCAAAATGGGA | TCAGTTGGTGGCTTTTTTAATATGA |
| MGAT5 | caccACCATCCAGCAGCGAACTCAGCC | CTATAGGCAGTCTTTGCAGAGAG |
| MFNG | caccTACCACTTGAACCTGTCCCCGC | AGCAGTTCAGGATTCATCGGGC |
| RFNG | caccGCGCCCGCCCCGGCCCGG | GTCACCGAGAGGTCGGGGCG |
Lowercase letters indicate adaptor sequences.
Construction of expression vectors for Pir-glycosyltransferase fusion genes.
The expression vectors were based on the YEp352GAP II plasmid, having the URA3 selection marker and the GAPDH promoter. Either the PIR1, PIR3, or PIR4 open reading frame, the hemagglutinin (HA) tag sequence, and Gateway Cassette Frame A were ligated in-frame and inserted into the SacI-SalI site of YEp352GAP II. The glycosyltransferase sequences were inserted using the Gateway system (Invitrogen, Carlsbad, CA) according to the supplier's instructions in order to construct each expression plasmid.
Glycosyltransferase assay using pyridylamino (PA)-labeled oligosaccharides or fluorescein isothiocyanate isomer I (FITC)-labeled Muc5AC peptides.
Yeast cells were cultivated at 30°C in 10 ml of SD-Ura medium for 36 to 48 h and were harvested by centrifugation. Yeast cells were homogenized with glass beads, and the cell wall fraction was obtained by centrifugation and used as an enzyme source.
Sialyltransferase and galactosyltransferase activities were measured in a 50-μl reaction mixture containing the cell wall fraction (optical density at 600 nm [OD600], 5), 100 mM HEPES-NaOH (pH 7.2), 5 mM MnCl2, 5 mM CMP-NeuAc or UDP-Gal, and 100 pmol PA-labeled oligosaccharide. In the case of the pp-GalNAc-T assay, the 50-μl reaction mixture contained the cell wall fraction (OD600, 5), 25 mM HEPES-NaOH (pH 7.2), 5 mM MnCl2, 0.25 mM UDP-GalNAc, and 500 pmol FITC-labeled Muc5AC peptide. In the case of α1,2- and α1,3-fucosyltransferases, the reaction mixture contained 50 mM cacodylate buffer (pH 6.8), 12.5 mM MnCl2, 5 mM ATP, 0.4 mM GDP-Fuc, and 100 pmol PA-labeled oligosaccharide. The assay of FUT8 was performed as described by Uozumi et al. (57), and the substrate (GnGn-bi-Asn-PABA) was donated by N. Taniguchi of Osaka University. Reaction mixtures were incubated at 30°C or 37°C for 0.5 to 24 h. The reaction was terminated by adding 100 μl of ice-cold water, and the reaction mixture was centrifuged at 1,000 × g for 5 min. The supernatant was filtered through an Ultrafree-MC membrane (pore size, 0.22 μm; Millipore, Bedford, MA). A 10-μl aliquot of the filtrate was subjected to high-performance liquid chromatography (HPLC) analysis.
HPLC analysis.
PA-labeled oligosaccharide products were analyzed by a Shodex Asahipak NH2P-50 normal-phase column (4.6 mm by 250 mm; Showa Denko, Tokyo, Japan) with solvent A (acetonitrile) and solvent B (20 mM acetic acid-triethylamine, pH 7.0) at a flow rate of 1.0 ml/min. After sample injection, the proportion of solvent B was increased linearly for 35 min from 25% to 46%. p-Nitrophenyl (pNP)-labeled sugars were analyzed by a COSMOSIL 5C18-AR-II reverse-phase column (4.6 mm by 250 mm; Nacalai Tesque Inc., Kyoto, Japan) with solvent A (0.05% trifluoroacetic acid) and solvent B (acetonitrile, 0.05% trifluoroacetic acid) at a flow rate of 1.0 ml/min. After sample injection, the proportion of solvent B was increased linearly for 30 min from 10% to 30%. FITC-labeled Muc5AC peptide was analyzed by a COSMOSIL 5C18-AR-II column (4.6 mm by 250 mm) with solvent A (0.05% trifluoroacetic acid) and solvent B (acetonitrile, 0.05% trifluoroacetic acid) at a flow rate of 1.0 ml/min. After sample injection, the proportion of solvent B was increased linearly for 30 min from 10% to 40%. Glycan structures were confirmed by comparison of retention times with PA-labeled standards after suitable glycosidase digestion. Otherwise, the reaction products were analyzed by matrix-assisted laser desorption ionization-time-of-flight tandem mass spectrometry (MALDI-TOF-MS/MS) (AXIMA-QIT, Shimadzu, Kyoto, Japan) (24).
Immunofluorescence microscopy.
Cells were cultivated for 2 days at 30°C, and a volume of cells equivalent to 1 OD600 unit was collected. Cells were suspended in 250 μl of phosphate-buffered saline (PBS) solution (10 mM KH2PO4, 40 mM K2HPO4, 150 mM NaCl) with bovine serum albumin (BSA) (1 mg/ml) and rat anti-HA antibody 3F10 (Boehringer Mannheim, Mannheim, Germany) and were incubated at 4°C for 30 min. Cells were then washed twice in PBS and were incubated in PBS with BSA (1 mg/ml) and Alexa 488-conjugated goat anti-rat immunoglobulin G (IgG) (Molecular Probes, Eugene, OR) (dilution rate, 1:125) at 0°C for 30 min. After a wash, cells were observed under a model BX50 fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometric analysis.
Cells were cultivated in SD medium for 2 days at 30°C, and the cells in 2 ml of culture were washed and resuspended in 0.25 ml of 100 mM PBS (pH 7.4) with 0.1% BSA and 1.25 μl anti-HA antibody, followed by incubation at 4°C for 30 min. After two washes with PBS, cells were resuspended in 0.25 ml PBS with 0.1% BSA and 1.25 μl anti-rat-IgG-antibody labeled with Alexa 488, followed by incubation at 4°C for 30 min. Cells were washed twice with PBS and sonicated for 2 s to dissociate individual cells; they were then subjected to flow cytometric analysis (FACSCalibur; Nippon Becton Dickinson Co., Ltd., Tokyo, Japan).
Western blot analysis of HA-tagged PIR-glycosyltransferase fusion proteins.
Fusion proteins were prepared from cells cultured for 2 days at 30°C. Cells were washed three times in ice-cold wash buffer (10 mM Tris-HCl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride, protease inhibitor Complete [Roche Diagnostics, Mannheim, Germany], EDTA free) and were disrupted by vigorously mixing the cell suspension with glass beads (diameter, 0.45 to 0.5 mm). Cell wall fractions were prepared according to the work of Schreuder et al. (46) with some modifications, after centrifugation of the cell lysates at 1,000 × g for 5 min. Cell wall fractions were washed three times with ice-cold wash buffer, and fusion proteins were extracted according to the method of Mrsa et al. (35), with slight modifications. The cell wall was extracted by heating the cells twice with 100 μl of lysis buffer as described by Masai et al. (31) at 95°C for 10 min, and the remaining cell wall was washed three times with 1 ml of 0.1 M sodium acetate buffer (pH 5.5). Fusion proteins were prepared by extracting the remaining cell wall with 30 mM NaOH overnight at 4°C and were analyzed by Western blotting according to the method of Laemmli (29). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 5-to-20% gradient gel (Atto, Tokyo, Japan) and were transferred to a polyvinylidene difluoride membrane (Pall, East Hills, NJ). HA-tagged fusion proteins were detected with the anti-HA monoclonal antibody HA.11 (Babco, Richmond, CA). Immunoreactive bands were visualized by staining with horseradish-conjugated goat anti-mouse IgG (Cell Signaling Technology, Beverly, MA) and chemiluminescence (NEN Life Science, Boston, MA).
RESULTS
Preparation of yeast cell wall fraction and measurement of glycosyltransferase activity.
We selected more than 50 human glycosyltransferase genes that are well characterized and suitable for the synthesis of typical sugar chains attached to glycoproteins. The truncated cDNA clones encoding these glycosyltransferases lacking the N-terminal transmembrane region were inserted into the entry vector of the Gateway system and were then transferred to the destination vector to be expressed in yeast. The truncated glycosyltransferases were placed after the region consisting of yeast cell wall Pir protein and the HA tag sequence; they were expressed under the control of a strong promoter (GAPDH) in Saccharomyces cerevisiae. Fifty-one human glycosyltransferases, including sialyl-, fucosyl-, galactosyl-, N-acetylgalactosaminyl-, and N-acetylglucosaminyltransferases, were expressed in genetically engineered S. cerevisiae strains in which 3 of the 4 PIR genes were disrupted to increase anchoring of the Pir fusion proteins (2).
Enzymatic activities were measured by HPLC analysis of PA-labeled oligosaccharides, as described in Materials and Methods. Peaks for the acceptor oligosaccharide and reaction products were detected, and the areas of the corresponding peaks were measured. Enzymatic activities were expressed as arbitrary units (1 arbitrary unit = 1 pmol of product/5 OD600 unit of cell wall/24 h). Under these reaction conditions, when all the acceptor was converted to the product, activity was expressed as >100 U, because it is technically difficult to examine each enzymatic reaction profile in more detail, and precise analysis of each enzyme was not the goal of this research. Some exceptions indicating more than 100 U were shown for ST3Gal I incubated for 30 min (Table 2) and ppGalNAcT's measured in the presence of 500 pmol of acceptor (Table 3). The specific activity of each enzyme was not determined, since the measurement of the protein amount was difficult for immobilized glycosyltransferases due to incomplete recovery of fusion proteins and the coexistence of endogenous cell wall mannoproteins.
TABLE 2.
Effects of Pir anchor species on the enzymatic activities of various glycosyltransferases
| Gene | Host | Anchor(s) | Acceptor | Activity (arbitrary units) |
|---|---|---|---|---|
| ST3Gal I | Δpir1,2,3 | PIR1 | Asialo-GM1-tetrasaccharide-PA | 3,917 |
| PIR3 | Asialo-GM1-tetrasaccharide-PA | 4,608 | ||
| PIR4 | Asialo-GM1-tetrasaccharide-PA | 4,032 | ||
| ST3Gal IV | Δpir1,2,3 | PIR1 | LNnT-PA | 0 |
| PIR3 | LNnT-PA | 8 | ||
| PIR1, PIR3 | LNnT-PA | 30 | ||
| ST6Gal I | Δpir1,2,3 | PIR1 | LNnT-PA | 0 |
| PIR3 | LNnT-PA | 70 | ||
| ST6Gal II | Δpir1,2,3 | PIR1 | LNnT-PA | 0 |
| PIR3 | LNnT-PA | 3 | ||
| ST6GalNAc IV | Δpir1,2,3 | PIR1 | NeuAcα2,3Galβ1,3GalNAcβ1,4Galβ1,4Glc-PA (ST3Gal I product) | 11 |
| FUT1 | Δpir1,2,3 | PIR1 | LNnT-PA | 0 |
| LNT-PA | 0 | |||
| PIR3 | LNnT-PA | 48 | ||
| LNT-PA | 74 | |||
| FUT3 | Δpir1,2,3 | PIR1 | LNT-PA | 0 |
| PIR3 | LNT-PA | 16 | ||
| FUT8 | Δpir1,2,3 | PIR3 | N-Biantennary-Asn-PABA | >100 |
TABLE 3.
Glycosyltransferase assays of various pp-GalNAc-T's
| Gene | Host | Anchor | Acceptor | Activity (arbitrary units) |
|---|---|---|---|---|
| pp-GalNAc-T1 | Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC | >500 |
| pp-GalNAc-T2 | Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC | 475 |
| pp-GalNAc-T3 | Δpir1,2,3 PDI1 | PIR3 | FITC-Muc5AC | 0 |
| Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC | 250 | |
| pp-GalNAc-T4 | Δpir1,2,3 PDI1 | PIR3 | FITC-Muc5AC | 0 |
| Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC | 150 | |
| pp-GalNAc-T6 | Δpir1,2,3 | PIR1 | FITC-Muc5AC | 35 |
| Δpir1,2,3 PDI1 | PIR1 | FITC-Muc5AC | 258 | |
| pp-GalNAc-T7 | Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC-GalNAc (pp-GalNAc-T1 product) | 0 |
| PIR1, PIR4 | FITC-Muc5AC-GalNAc (pp-GalNAc-T1 product) | 55 | ||
| pp-GalNAc-T8 | Δpir1,2,3 PDI1 | PIR1, PIR3 | FITC-Muc5AC | 0 |
| pp-GalNAc-T9 | Δpir1,2,3 PDI1 | PIR1 | FITC-Muc5AC-GalNAc (pp-GalNAc-T1 product) | 0 |
| pp-GalNAc-T10 | Δpir1,2,4 | PIR4 | FITC-Muc5AC | 90 |
| Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC | 215 | |
| pp-GalNAc-T12 | Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC | 95 |
| pp-GalNAc-T13 | Δpir1,2,3 | PIR1 | FITC-Muc5AC | 485 |
| pp-GalNAc-T14 | Δpir1,2,3 PD1I | PIR1 | FITC-Muc5AC | 0 |
| pp-GalNAc-T15 | Δpir1,2,4 PDI1 | PIR4 | FITC-Muc5AC | 0 |
Effect of culture medium pH on enzymatic activity.
After yeast cultivation, the pH of the SD-Ura medium generally decreased to below 4. It is possible that yeast proteases, such as GPI-anchored asparagine protease, whose optimal pH is 4.5, are activated and hydrolyze recombinant enzymes, or that the expressed glycosyltransferases are inactivated under acidic conditions. We therefore controlled the pH of the medium during cultivation by addition of HEPES buffer at pH 6. ST3Gal I activity increased from 174 to 706 U (data not shown). The same effect was also observed for ST6Gal I (from 25 to 47 U). Thus, we used these culture conditions for measurement of the remaining glycosyltransferase activities.
Comparison of PIR anchors for immobilization of glycosyltransferases.
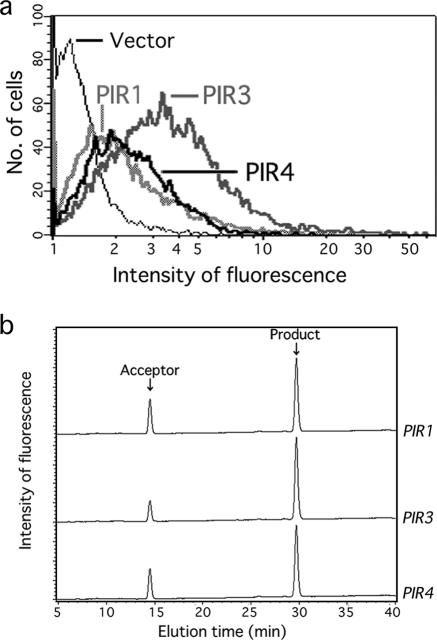
In our previous study, human glycosyltransferase (FUT6) was fused with PIR1 as an anchor protein (2). However, the PIR family comprises PIR1 to PIR4, with different numbers of 17- to 18-amino-acid repeated sequences, different gene expression regulation mechanisms (36, 54), and different localizations in the cell wall. Pir1 and Pir2 localize at bud scar regions, whereas Pir3 and Pir4 exist throughout the cell wall (51). To specify a suitable Pir protein for immobilization of glycosyltransferases, glycosyltransferase activities anchored by different Pir sequences were compared. ST3Gal I, anchored by PIR1, PIR3, and PIR4, exhibited activity at 3,917, 4,608, and 4,032 U, respectively (Fig. 1b; Table 2). The amounts of immobilized glycosyltransferases were monitored by flow cytometry after HA tag staining with an HA tag-specific antibody coupled with fluorescence, which revealed much stronger signals for PIR3-anchored cells than for cells anchored by PIR1 or PIR4 (Fig. 1a). Similarly, FUT1 anchored by PIR3 showed the strongest activity (40 U) compared with PIR1 and PIR4 (28 and 35 U, respectively) (see Table 5). Furthermore, FUT3, ST3Gal IV, ST6Gal I, and ST6Gal II did not show any activity with the PIR1 fusion, but the PIR3 fusions exhibited enzymatic activity (Table 2). On the other hand, pp-GalNAc-T3 and -T4 and β3GnT2 showed activities only when fused with the PIR4 anchor (Tables 3 and 4), suggesting that PIR4 is a suitable anchor protein for the first trial of the immobilized enzyme assay. Therefore, PIR4 was used as an anchor for the expression of further glycosyltransferases.
FIG. 1.
Effect of Pir anchor on amount of cell wall immobilization and enzymatic activity. (a) HA-tagged PIR1, PIR3, and PIR4 fusion proteins at the cell surface were monitored by flow cytometry. (b) Enzymatic activities of PIR1-, PIR3-, and PIR4-anchored ST3Gal I at the cell wall were measured and compared by HPLC analysis under the conditions described in Materials and Methods. Enzymatic activities were expressed as arbitrary units (1 arbitrary unit = 1 pmol of product/5 OD600 units of cell wall/24 h). PIR1-HA-ST3Gal I, PIR3-HA-ST3Gal I, and PIR4-HA-ST3Gal I (labeled PIR1, PIR3, and PIR4, respectively) showed enzymatic activities of 3,917, 4,608, and 4,032 U, respectively.
TABLE 5.
Effect of SED1 or OCH1 disruption on enzymatic activity
| Gene | Host | Anchor | Acceptor | Activity (arbitrary units) |
|---|---|---|---|---|
| FUT9 | Δpir1,2,3 | PIR3 | LNnT-PA | 5 |
| Δpir1,2,4 PDI1 | PIR4 | LNnT-PA | 29 | |
| Δpir1,2,4 PDI1Δsed1 | PIR4 | LNnT-PA | 45 | |
| FUT1 | Δpir1,2,4 PDI1 | PIR1 | LNnT-PA | 28 |
| Δpir1,2,4 PDI1Δsed1 | PIR1 | LNnT-PA | 74 | |
| Δpir1,2,4 PDI1 | PIR3 | LNnT-PA | 40 | |
| Δpir1,2,4 PDI1Δsed1 | PIR3 | LNnT-PA | 71 | |
| Δpir1,2,4 PDI1 | PIR4 | LNnT-PA | 35 | |
| Δpir1,2,4 PDI1Δsed1 | PIR4 | LNnT-PA | 62 | |
| ST3Gal VI | Δpir1,2,3 | PIR1 | LNnT-PA | 2.2 |
| Δpir1,2,3Δoch1 | PIR1 | LNnT-PA | 4.1 |
TABLE 4.
Enzymatic activities of yeast cell wall-immobilized human β3GnT2 containing native and variant N-glycosylation sites
| Genea | Host | Anchor | Acceptor | Activity (arbitrary units) |
|---|---|---|---|---|
| β3GnT2 (NLS) | Δpir1,2,3 PDI1 | PIR1 | LNnT-PA | 0 |
| Δpir1,2,3 PDI1 | PIR3 | 0 | ||
| Δpir1,2,4 PDI1 | PIR4 | 2 | ||
| β3GnT2 (NLT) | Δpir1,2,4 PDI1 | PIR4 | LNnT-PA | 6 |
NLS, native N-glycosylation sites; NLT, variant N-glycosylation sites.
Two types of Pir anchor were used simultaneously for ST3Gal IV, which showed no detectable activity with the PIR1 anchor and low activity (8 U) with the PIR3 anchor (Table 2). However, when PIR1 and PIR3 fusion proteins were expressed simultaneously, the enzyme activity increased to 30 U, which was much higher than the combined individual activities (Table 2). This result suggests that different kinds of Pir proteins may interact with one another and bind to form a mannoprotein network in the cell wall, resulting in increased immobilization of recombinant enzymes in the cell wall. The PIR gene family is known to have two binding regions for the cell wall. One is the repetitive sequence, which is necessary for binding with β1,3-glucan in an alkaline-sensitive manner; the other is the cysteine residue-rich C-terminal region, which is necessary for binding with an unidentified component of the cell wall through the disulfide bond (12, 34, 51).
Expression of pp-GalNAc-T's and effect of PDI on activity.
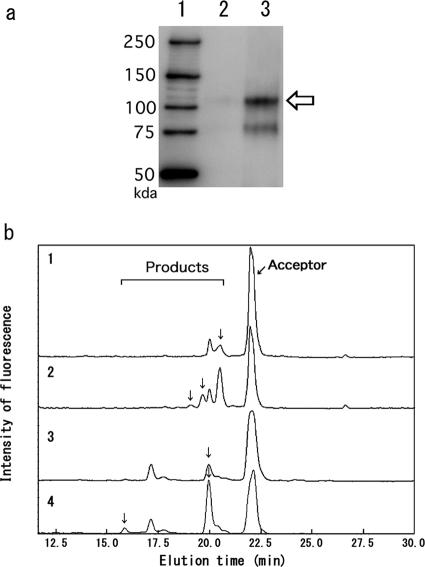
Many pp-GalNAc-T's expressed in mammalian cells, including pp-GalNAc-T2, -T6, and -T10, have strong activities (8, 22). However, these enzymes initially showed very low activity when expressed in yeast (Table 3). When the expression of these immobilized enzymes at the yeast cell wall was examined, the amounts of fusion protein expressed were significantly smaller than those of other glycosyltransferases, as judged from fluorescence microscopic intensity (data not shown) and Western blot analysis (Fig. 2a). To address the causes of the poor expression of the pp-GalNAc-T family in yeast, we examined the coding sequences of glycosyltransferases. The amino acid sequences of these glycosyltransferases contain about 16 cysteine residues, almost three times as many as other glycosyltransferases. It is possible that the numerous disulfide bonds in pp-GalNAc-T's hinder prompt folding of the proteins, and subsequent production of active enzymes, because yeast cells may lack the capacity to make sufficient disulfide bond. Therefore, protein disulfide isomerase (PDI), a molecular chaperone, was overexpressed in yeast cells. pp-GalNAc-T6 showed a fourfold increase in the protein amount, as judged from the band intensity after probing with an antibody raised against the HA tag, and a comparable increase in activity, as judged from HPLC analysis (Fig. 2b; Table 3). The same effect was also observed for other glycosyltransferases, such as FUT9 (Table 5) and ST3Gal IV (data not shown), suggesting that the Pir protein-mediated immobilization of human glycosyltransferases may be adversely affected by protein folding difficulties in the yeast endoplasmic reticulum.
FIG. 2.
Effect of PDI on amount of Pir fusion protein in the cell wall. (a) An HA-tagged fusion protein (HA-pp-GalNAc-T10) was detected by Western blotting from YSF124 cells (lane 2) and from YSF124 cells with PDI overexpression (lane 3). Lane 1, molecular markers. Arrow indicates the fusion protein. (b) The enzymatic activities of pp-GalNAc-T6 and -T10 were analyzed by HPLC as described in Materials and Methods. Enzymatic activities were expressed as arbitrary units (1 arbitrary unit = 1 pmol of product/5 OD600 units of cell wall/24 h). Arrows indicate the peaks for acceptor and product oligosaccharides. Activities were as follows: for pp-GalNAc-T6 expressed in YSF123 cells (curve 1), 35 U; for pp-GalNAc-T6 expressed in YSF123 cells with PDI overexpression (curve 2), 150 U; for pp-GalNAc-T10 expressed in YSF124 cells (curve 3), 90 U; for pp- GalNAc-T10 expressed in YSF124 cells with PDI overexpression (curve 4), 230 U.
Effect of SED1 or OCH1 gene disruption.
SED1 encodes a major GPI-cell wall mannoprotein in stationary-phase yeast cells. Gene disruption of SED1 causes zymolyase sensitivity (50). Many yeast cell wall proteins, including the Pir family, increase their expression levels during perturbation of cell wall integrity (7, 23). To investigate whether the elimination of one of the major cell wall mannoproteins induces a compensatory response leading to greater incorporation of Pir proteins into the cell wall, we examined immobilized glycosyltransferase activities in SED1-disrupted cells. As shown in Table 5, FUT9 and FUT1 activities increased about 1.5- to 2.6-fold as a result of SED1 disruption.
OCH1 encodes α-1,6-mannosyltransferase, which is required for the initiation of yeast-specific mannose outer chain elongation of N-glycans (40). To address whether the large mannose outer chain inhibits glycosyltransferase activities, we examined ST3Gal VI activity. The ST3Gal VI activity of the Δoch1 cells reproducibly increased about twofold compared with wild-type cells (Table 5). However, other enzymes, including ST3Gal I, showed decreased activity in Δoch1 cells (data not shown). Thus, it is difficult to generalize the effect of yeast-specific polymannosylation on the enzymatic activities of recombinant proteins.
Comprehensive expression of various glycosyltransferases, including N-acetylglucosaminyltransferases and galactosyltransferases.
In total, 51 human glycosyltransferase genes were expressed as Pir fusion proteins to mediate localization in the yeast cell wall, and we were successful in detecting enzymatic activity for 40 (see Table 7). All fucosyltransferases expressed showed significant activity.
TABLE 7.
Summary of the expression of immobilized human glycosyltransferase genes in S. cerevisiae
| Enzyme | No. of entry clones | No. of expressed transferases | No. of active enzymes |
|---|---|---|---|
| FucT | 11 | 6 | 6 |
| SiaT | 20 | 10 | 8 |
| pp-GalNAc-T | 20 | 13 | 9 |
| GalT and GalNAcT | 20 | 8 | 5 |
| GnT | 26 | 14 | 12 |
| Total | 97 | 51 | 40 |
Among the 14 N-acetylglucosaminyltransferases (GnT's), including β3GnT2, C2GnT1, N-acetylglucosaminyltransferase V (MGAT5), and MFNG, 12 showed enzymatic activity (Tables 4 and 6). β3GnT2 is an important enzyme in the synthesis of polylactosamine; however, the enzymatic activity was not very strong in our system (Table 4). To obtain increased activity, we constructed a variant form of the β3GnT2 gene, in which the N-glycosylation site of the C-terminal end was changed from Asn-Leu-Ser (NLS) to Asn-Leu-Thr (NLT), a change reported to increase glycosylation frequency in insect cells (26). As shown in Table 4, β3GnT2 activity increased about threefold, indicating that this amino acid exchange is useful in improving glycoprotein productivity in both insect cells and yeast cells.
TABLE 6.
Enzymatic activities of yeast cell wall-immobilized human glycosyltransferases
| Gene | Host | Anchor | Acceptor | Activity (arbitrary units) |
|---|---|---|---|---|
| β3GalT4 | Δpir1,2,4 PDI1 | PIR4 | GM2-tetrasaccharide-PA | 0 |
| β3GalT5 | Δpir1,2,4 PDI1 | PIR4 | Agalacto-LNnT-PA | 0 |
| β4GalT1 | Δpir1,2,4 PDI1 | PIR4 | Agalacto-LNnT-PA | >100 |
| β4GalT4 | Δpir1,2,4 PDI1 | PIR4 | Agalacto-LNnT-PA | 2 |
| β4GalT5 | Δpir1,2,4 PDI1 | PIR4 | Core 1-pNP | 27 |
| A(ABO) | Δpir1,2,4 PDI1 | PIR4 | Lacto-N-fucopentaose-PA | 99 |
| B(ABO) | Δpir1,2,4 PDI1 | PIR4 | Lacto-N-fucopentaose-PA | 90 |
| β3GalNAc-T1 | Δpir1,2,4 PDI1 | PIR4 | Globoriose-PA | 0 |
| β3GnT3 | Δpir1,2,4 PDI1 | PIR4 | Core 1-pNP | 0.4 |
| β3GnT6 | Δpir1,2,4 PDI1 | PIR4 | GalNAc-pNP | 6 |
| C2GnT1 | Δpir1,2,4 PDI1 | PIR4 | Core 1-pNP | 29 |
| C2GnT2 | Δpir1,2,4 PDI1 | PIR4 | Core 1-pNP | 0.7 |
| C2GnT3 | Δpir1,2,4 PDI1 | PIR4 | Core 1-pNP | 0.6 |
| IGnT3 | Δpir1,2,4 PDI1 | PIR4 | LNnT-PA | 6 |
| iGnT | Δpir1,2,4 PDI1 | PIR4 | LNnT-PA | 0 |
| MGAT1 | Δpir1,2,4 PDI1 | PIR4 | M5-Gn2-PA | 57 |
| MGAT3 | Δpir1,2,4 PDI1 | PIR4 | Agalacto-N-biantennary-PA | 0 |
| MGAT4A | Δpir1,2,4 PDI1 | PIR4 | Agalacto-N-biantennary-PA | 12 |
| MGAT5 | Δpir1,2,4 PDI1 | PIR4 | Agalacto-N-triantennary-PA | 7 |
| MFNG | Δpir1,2,4 PDI1 | PIR4 | Fucose-pNP | 0.3 |
| RFNG | Δpir1,2,4 PDI1 | PIR4 | Fucose-pNP | 0.2 |
| ST3Gal III | Δpir1,2,4 PDI1 | PIR4 | LNnT-PA | >100 |
| ST3Gal III | Δpir1,2,4 PDI1 | PIR4 | LNT-PA | 70 |
| ST3Gal VI | Δpir1,2,3 PDI1 | PIR3 | LNnT-PA | 34 |
| ST6GalNAc I | Δpir1,2,4 PDI1 | PIR4 | H-[Ala-Thr(GlcNAc)-Ala]2-7-OH, Muc1 peptide-GlcNAc | 0 |
| ST6GalNAc II | Δpir1,2,4 PDI1 | PIR4 | Asialofetuin, Muc1 peptide-GlcNAc | <0.1 |
| ST8Sia VI | Δpir1,2,4 PDI1 | PIR4 | NeuAcα2,3Galβ1,3GalNAc-PA | 0 |
| FUT6 | Δpir1,2,4 PDI1 | PIR4 | LNnT-PA | 81 |
| FUT7 | Δpir1,2,3 PDI1 | PIR2ΔCT | Sia-LNnT-PA(ST3Gal IV product) | 0.3 |
Nine of 13 pp-GalNAc-T's showed some activity (Tables 6 and 7), although the activity level was relatively low compared with that of other families, probably due to the requirement of more complex disulfide bond formation.
Among the eight galactosyltransferase family proteins (GalT's), β4GalT1, α-1,3-N-acetylgalactosaminyltransferase [A(ABO)], and α-1,3-galactosyltransferase [B(ABO)] showed high activity, low activity was seen for β4GalT4 and β4GalT5, and no activity was seen for β3GalNAcT1, β3GalT4, and β3GalT5 while these fusion proteins were expressed at the cell wall (Table 6). Because these glycosyltransferases use glycolipids or glycopeptides as in vivo acceptors, it is likely that PA-labeled oligosaccharides are not suitable acceptors.
Eight of 10 sialyltransferases showed some activity (Tables 6 and 7). Pir-mediated ST6GalNAc IV and ST3Gal I fusions had significant activity, indicating that our system has advantages for expressing glycosyltransferases compared with previous reports, in which the soluble form of ST6GalNAc IV was unstable (18, 21) and inactive ST3Gal I was secreted from the yeast Pichia pastoris (48). ST6GalNAc I and ST6GalNAc II did not show any activity by HPLC analysis when Muc1 peptide-O-GalNAc was used as an acceptor, but ST6GalNAc II showed slight activity when tritium-labeled CMP-sialic acid and asialofetuin were used as the donor and acceptor in an enzyme assay (Table 6). However, no activity was detected for ST6GalNAc I, even under these assay conditions.
ST8Sia VI also showed no enzymatic activity (Table 6), whereas relatively strong signals were detected for the HA-tagged fusion construct (data not shown), suggesting that an unknown cofactor or protein modification may be required for expression of enzymatic activity.
Oligosaccharide synthesis using PIR-mediated fusions of human glycosyltransferases.
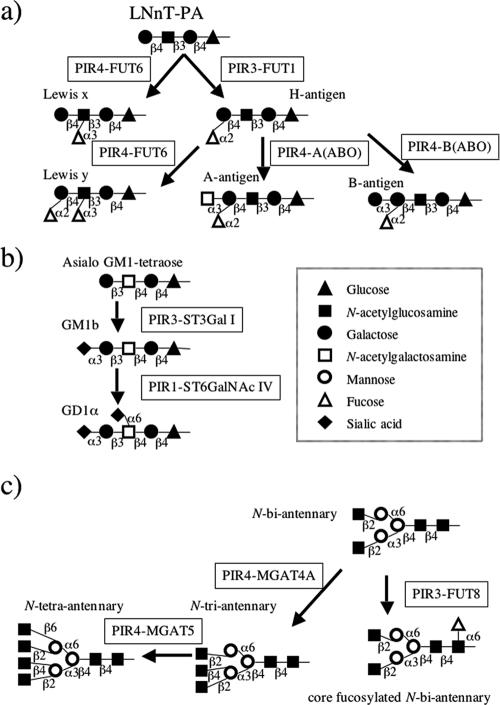
With the 40-member immobilized human glycosyltransferase library, various types of oligosaccharides could be synthesized. Because FUT1 and ST3Gal III can recognize both lacto-N-tetraose-PA (LNT-PA) and lacto-N-neotetraose-PA (LNnT-PA) as acceptors (Tables 2 and 6), these enzymes are suitable for the synthesis of several useful oligosaccharides. For example, FUT1 synthesized type 1 and type 2 H antigen from LNnT-PA and LNT-PA, respectively. By using type 2 H antigen as an acceptor, Lewis y oligosaccharide was further synthesized by FUT6 in microgram quantities (Fig. 3). In another experiment, ST3Gal III synthesized Sia α-2,3-Gal β1,3-Gn-R and Sia α-2,3-Gal β1,4-Gn-R from LNnT-PA and LNT-PA, respectively (Table 6). MGAT4A and MGAT5 synthesized agalacto-N-triantennary-PA and agalacto-N-tetra-antennary-PA oligosaccharides, respectively (Fig. 3), and C2GnT1 and β3GnT6 synthesized core 2 and core 3 oligosaccharides, respectively (Table 6).
FIG. 3.
Synthesis of oligosaccharides using immobilized glycosyltransferases. (a) Synthesis of Lewis x, Lewis y, H antigen, A antigen, and B antigen from LNnT-PA using immobilized FUT1, FUT6, and A(ABO) or B(ABO). (b) Synthesis of GM1b and GD1α from asialo-GM1 tetraose-PA using immobilized ST3Gal I and ST6GalNAc IV. (c) Synthesis of fucosylated and branched N-glycans using FUT8, MGAT4A, and MGAT5.
For more practical purposes, acceptors for some glycosyltransferases were synthesized using our immobilized glycosyltransferases. ST6GalNAc IV uses the Sia α2,3-Gal β1,3-GalNAc-R structure as an acceptor, but this structure is not commercially available. Therefore, we synthesized Sia α2,3-Gal β1,3-GalNAc β1,4-Gal β1,4-Glc-PA oligosaccharide from asialo-GM1-tetraose-PA with the immobilized ST3Gal I (Table 2; Fig. 3). In addition, pp-GalNAc-T1 shows strong primary activity that transfers a GalNAc residue(s) to peptides, whereas pp-GalNAc-T7 and pp-GalNAc-T9 mainly show secondary activity that transfers a GalNAc residue(s) only to peptides already containing a GalNAc residue(s). Therefore, we first produced Muc5AC-GalNAc by using pp-GalNAc-T1, and then we used Muc5AC-GalNAc purified by HPLC as an acceptor for pp-GalNAc-T7 and pp-GalNAc-T9 (Table 3).
DISCUSSION
Factors affecting expression of human glycosyltransferases in the yeast cell wall.
We attempted to immobilize glycosyltransferases in vivo at the yeast cell surface by fusion with cell wall Pir proteins. After construction of the genetically engineered yeast strains, which express glycosyltransferases that were covalently linked to cell wall glucans, simple cultivation of the yeast cells and centrifugation of the culture broth provided large amounts of immobilized enzymes. A total of 51 human glycosyltransferases were expressed in this study as immobilized proteins, and 40 showed enzymatic activity; more than 75% of human enzymes expressed in yeast were active (Table 7), indicating that the yeast expression system is useful for the production of large numbers of human glycosyltransferases.
In this project, we identified several important factors that affect the amount of immobilized human glycosyltransferase at the yeast cell wall: the choice of Pir anchor proteins, the use of the molecular chaperone PDI, and the codons used, in addition to other factors described above. Comparison of several Pir fusion proteins expressed under the control of the same GAPDH promoter showed higher activities for the PIR3 fusion than for the PIR1 and PIR4 fusions, confirming the more efficient and homogenous localization of PIR3 fusions in the cell wall (51). Interestingly, a synergistic effect was observed when PIR1 and PIR3 fusions were expressed simultaneously; higher levels of glycosyltransferase activity were observed than the combined activities with PIR1 and PIR3 alone. This suggests that different Pir proteins interact in the yeast cell wall, although the mechanism responsible remains unclear. Furthermore, perturbation of the cell wall architecture by deletion of the major GPI-anchored mannoprotein SED1 (Table 5) or of endogenous Pir proteins (2) increased the amounts of Pir-glycosyltransferase fusion proteins.
Eleven glycosyltransferases that we attempted to express in this study showed no detectable activity. Four of these included the pp-GalNAc-T family, in which the presence of many disulfide bonds may inhibit proper protein folding, while some pp-GalNAc-T's retain very weak activity, even in mammalian cells (22). Among the GalT and GalNAc-T groups, no activity was detected for three enzymes, probably because the PA-labeled oligosaccharides used as acceptors were not suitable for these enzymes. They may recognize glycolipids or glycopeptides in vivo, even if these enzymes are reported to be able to transfer some amounts of sugar to oligosaccharide acceptors in vitro. No activity was detected for ST6GalNAc I or ST8Sia VI, while significant amounts of protein were detected by Western blot analysis. It was reported that unique disulfide bond structures are required for ST8Sia IV activity (3, 10), suggesting that our PIR4-ST8Sia VI fusion construct disturbs the correct disulfide bond formation of ST8Sia VI protein. For ST8Sia II and ST8Sia IV, it was reported that autopolysialylation is required for activation of the polysialyltransferases (37), suggesting that ST8Sia VI is also activated by autopolysialylation. Alternatively, it is possible that unknown subunits or partners are required for expression of enzymatic activity. In fact, POMT1 and POMT2 are known to form a complex to retain their activities (30).
The HA tag signal intensity of PIR-HA-glycosyltransferase was often much weaker than that of the PIR-HA anchor, suggesting that the presence of human-derived sequences may inhibit the production of fusion proteins (data not shown). This may be caused by differences in codon usage between humans and yeast. For example, the CGG codon for arginine is the rarest in yeast S. cerevisiae, with a frequency of only 0.17% among all codons (Codon Usage Database [http://www.kazusa.or.jp/codon/]), and is known to inhibit protein translation in yeast (43), whereas it is more common in humans (frequency, 1.16%). Therefore, it is possible that the low levels of human protein production in yeast are partly due to the presence of rare codons, like CGG, in human genes. The five glycosyltransferases showing strong activity—FUT8, ST3Gal I, ST3Gal III, β4GalT1, and pp-GalNAc-T1—contain 0, 5, 6, 6, and 3 CGG codons, respectively, whereas the eight glycosyltransferases showing no activity—ST6GalNAc I, β3GalT4, β3GalT5, pp-GalNAc-T8, -T9, -T14, and -T15, and MGAT3, contain 9, 8, 3, 9, 17, 12, 7, and 3 CGG codons, respectively. This suggests that the increase in rare yeast codons in human genes may decrease the protein productivity and activity of these enzymes.
Although ST3Gal VI increased its enzymatic activity by eliminating hypermannosylation, which may cause the steric hindrance of catalytic sites (Table 5), other glycosyltransferases, including ST3Gal I, did not increase their activities (data not shown). The och1 gene disruptants generally showed slow growth and weakened cell wall strength (40), which may reduce amounts of the immobilized Pir fusion proteins.
Oligosaccharide synthesis by recombinant glycosyltransferases.
We believe that our system provides a promising approach to synthesizing a variety of oligosaccharides using immobilized human glycosyltransferases. As described above, some useful oligosaccharides, such as H antigen, Lewis x, and Lewis y, were synthesized from LNnT. A and B antigens were further synthesized from H antigen by A(ABO) and B(ABO) enzymes, respectively (Fig. 3a). ST3Gal I added sialic acid at the terminal galactose of asialo-GM1-tetraoligosaccharide, and the reaction product was further used as an acceptor for ST6GalNAc IV (Fig. 3b). MGAT4A and MGAT5 synthesized tri- and tetra-antennary complex N-glycans, respectively (Fig. 3c), which are useful in the production of N-glycan-engineered erythropoietin (52). Oligosaccharides or oligosaccharide conjugates inhibit infection by pathogens; for example, GM3 trisaccharide inhibits influenza virus infection (42), and O-glycans with terminal α1,4-linked N-acetylglucosamine inhibit Helicobacter pylori infection (27). Sialyl Lewis x inhibits inflammation (17, 38, 44, 45), and GQ1b affects cellular differentiation (56). A greater variety of oligosaccharides is required for studies on infection, the immune system, cellular differentiation, and cancer. Some of these oligosaccharides will be useful when large-scale production systems are established for the food and medical industries.
Acknowledgments
H. Narimatsu donated 18 human glycosyltransferase genes. The ST6Gal I, ST6Gal II, and ST8Sia VI genes were kindly donated by O. Takashima at Riken. MGAT4A and MGAT5 genes were provided by A. Yoshida of the Kirin brewery. An acceptor, biantennary-Asn-PABA, was kindly donated by N. Taniguchi. Some oligosaccharide products were confirmed using mass spectrometry performed by A. Kameyama. Muc1 peptides with GalNAc were kindly provided by S. Nishimura. We are grateful to S. Tsuji, H. Abe, A. Togayachi, T. Sato, and K. Tachibana for technical advice.
This work was supported by the Research and Development of Structural Glycoproteomics Project from the New Energy and Industrial Technology Development Organization of Japan (NEDO).
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Abe, H., Y. Shimma, and Y. Jigami. 2003. In vitro oligosaccharide synthesis using intact yeast cells that display glycosyltransferases at the cell surface through cell wall-anchored protein Pir. Glycobiology 13:87-95. [DOI] [PubMed] [Google Scholar]
- 2.Abe, H., M. Ohba, Y. Shimma, and Y. Jigami. 2004. Yeast cells harboring human α-1,3-fucosyltransferase at the cell surface engineered using Pir, a cell wall-anchored protein. FEMS Yeast Res. 4:417-425. [DOI] [PubMed] [Google Scholar]
- 3.Angata, K., T.-Y. Yen, A. ElBattari, B. A. Macher, and M. Fukuda. 2001. Unique disulfide bond structures found in ST8Sia IV polysialyltransferase are required for its activity. J. Biol. Chem. 276:15369-15377. [DOI] [PubMed] [Google Scholar]
- 4.Babad, H., and W. Z. Hassid. 1966. Soluble uridine diphosphate d-galactose: d-glucose β-4-d-galactosyltransferase from bovine milk. J. Biol. Chem. 241:2672-2678. [PubMed] [Google Scholar]
- 5.Bencurova, M., D. Rendic, G. Fabini, E.-M. Kopecky, F. Altmann, and I. B. H. Wilson. 2003. Expression of eukaryotic glycosyltransferases in the yeast Pichia pastoris. Biochimie 85:413-422. [DOI] [PubMed] [Google Scholar]
- 6.Boeggeman, E. E., P. V. Balaji, N. Sethi, A. S. Masibay, and P. K. Qasba. 1993. Expression of deletion constructs of bovine β-1,4-galactosyltransferase in Escherichia coli: importance of Cys134 for its activity. Protein Eng. 6:779-785. [DOI] [PubMed] [Google Scholar]
- 7.Boorsma, A., H. de Novel, B. Riet, B. Bargmann, S. Brul, K. J. Hellingwerf, and F. M. Klis. 2004. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21:413-427. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, L., K. Tachibana, Y. Zhang, J.-M. Guo, K. Tachibana, A. Kameyama, H. Wang, T. Hiruma, H. Iwasaki, A. Togayachi, T. Kudo, and H. Narimatsu. 2002. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T10. FEBS Lett. 531:115-121. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, L. L., U. B. Jensen, P. Bross, and T. F. Orntoft. 2000. The C-terminal N-glycosylation sites of the human α1,3/4-fucosyltransferase III, -V, and -VI (hFucTIII, -V, and -VI) are necessary for the expression of full enzyme activity, Glycobiology 10:931-939. [DOI] [PubMed] [Google Scholar]
- 10.de Vries, T., T.-Y. Yen, R. K. Joshi, J. Storm, D. H. Eijnden, R. M. Knegtel, H. Bunschoten, D. H. Joziasse, and B. A. Macher. 2001. Neighboring cysteine residues in human fucosyltransferase VII are engaged in disulfide bridges, forming small loop structures. Glycobiology 11:423-432. [DOI] [PubMed] [Google Scholar]
- 11.Drobnik, J., V. Saudek, F. Svec, J. Kalal, V. Vojtisec, and M. Barta. 1979. Enzyme immobilization techniques on poly(glycidyl methacrylate-co-ethylene dimethacrylate) carrier with penicillin amidase as model. Biotechnol. Bioeng. 21:1317-1332. [DOI] [PubMed] [Google Scholar]
- 12.Ecker, M., R. Deutzmann, L. Lehle, V. Mrsa, and W. Tanner. 2006. Pir proteins of Saccharomyces cerevisiae are attached to β-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 281:11523-11529. [DOI] [PubMed] [Google Scholar]
- 13.Fast, D. G., J. C. Jamieson, and G. McCaffrey. 1993. The role of the carbohydrate chains of Galβ-1,4-GlcNAcα2,6-sialyltransferase for enzyme activity. Biochim. Biophys. Acta 1202:325-330. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira, J. P., R. Sasisekharan, O. Louie, and R. Langer. 1993. Influence of chemistry in immobilization of cobra venom phospholipase A2: implications as to mechanism. Biochemistry 17:8098-8102. [DOI] [PubMed] [Google Scholar]
- 15.Gerasimas, V. B., V. M. Chernoglazov, and A. A. Klesov. 1980. Effect of progressive chemical modification of the activity and thermal stability of soluble and immobilized glucoamylase. Biokhimiya 45:1086-1092. [PubMed] [Google Scholar]
- 16.Hamamoto, T., Y.-C. Lee, N. Kurosawa, T. Nakaoka, N. Kojima, and S. Tsuji. 1994. Expression of mouse Galβ1,4GlcNAc α1,6-sialyltransferase in an insoluble form in Escherichia coli and partial renaturation. Bioorg. Med. Chem. 2:79-84. [DOI] [PubMed] [Google Scholar]
- 17.Han, K. T., S. R. Sharar, M. L. Phillips, J. M. Harlan, and R. K. Winn. 1995. Sialyl Lewis(x) oligosaccharide reduces ischemia reperfusion injury in the rabbit ear. J. Immunol. 155:4011-4015. [PubMed] [Google Scholar]
- 18.Harduin-Lepers, A., D. C. Stokes, W. F. A. Steelant, B. Samyn-Petit, M. A. Krzewinski-Recchi, V. Vallejo-Ruiz, J. P. Zanetta, C. Auge, and P. Delannoy. 2000. Cloning, expression and gene organization of a human Neu5Acα2-3Galβ1-3GalNAc α2,6-sialyltransferase: hST6GalNAc IV. Biochem. J. 352:37-48. [PMC free article] [PubMed] [Google Scholar]
- 19.Hidari, K. I. P. J., N. Horie, T. Murata, D. Miyamoto, T. Suzuki, T. Usui, and Y. Suzuki. 2005. Purification and characterization of a soluble recombinant human ST6Gal I functionally expressed in Escherichia coli. Glycoconj. J. 22:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, E. H., T.-Y. Yen, S. Thomas, R. Joshi, A. Nguyen, T. Long, F. Gallet, A. Maftah, R. Julien, and B. A. Macher. 2000. Human α1,3/4 fucosyltransferases. Characterization of highly conserved cysteine residues and N-linked glycosylation sites. J. Biol. Chem. 275:24237-24245. [DOI] [PubMed] [Google Scholar]
- 21.Ivannikova, T., F. Bintein, A. Malleron, S. Juliant, M. Cerutti, A. Harduin-Lepers, P. Delannoy, C. Auge, and A. Lubineau. 2003. Recombinant (2-3)-α-sialyltransferase immobilized on nickel-agarose for preparative synthesis of sialyl Lewisx and Lewisa precursor oligosaccharides. Carbohydr. Res. 338:1153-1161. [DOI] [PubMed] [Google Scholar]
- 22.Iwasaki, H., Y. Zhang, K. Tachibana, M. Gotoh, N. Kikuchi, Y.-D. Kwon, A. Togayachi, T. Kudo, T. Kubota, and H. Narimatsu. 2003. Initiation of O-glycan synthesis in IgA1 hinge region is determined by a single enzyme, UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2. J. Biol. Chem. 278:5613-5621. [DOI] [PubMed] [Google Scholar]
- 23.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 24.Kameyama, A., N. Kikuchi, S. Nakaya, H. Ito, T. Sato, T. Shikanai, Y. Takahashi, K. Takahashi, and H. Narimatsu. 2005. A strategy for identification of oligosaccharide structures using observational multistage mass spectral library. Anal. Chem. 77:4719-4725. [DOI] [PubMed] [Google Scholar]
- 25.Kapteyn, J. C., P. Van Egmond, E. Sievi, H. Van Den Ende, M. Makarow, and F. M. Klis. 1999. The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and β1,6-glucan-deficient mutants. Mol. Microbiol. 31:1835-1844. [DOI] [PubMed] [Google Scholar]
- 26.Kato, T., M. Suzuki, T. Murata, and E. Y. Park. 2005. The effects of N-glycosylation sites and the N-terminal region on the biological function of β1,3-N-acetylglucosaminyltransferase 2 and its secretion. Biochem. Biophys. Res. Commun. 329:699-705. [DOI] [PubMed] [Google Scholar]
- 27.Kawakubo, M., Y. Ito, Y. Okimura, M. Kobayashi, K. Sakura, S. Kasama, M. N. Fukuda, M. Fukuda, T. Katsuyama, and J. Nakayama. 2004. Natural antibiotic function of a human gastric mucin against Helicobacter pylori infection. Science 305:1003-1006. [DOI] [PubMed] [Google Scholar]
- 28.Klebe, R. J., J. V. Harriss, Z. D. Sharp, and M. G. Douglas. 1983. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene 25:333-341. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Manya, H., A. Chiba, A. Yoshida, X. Wang, Y. Chiba, Y. Jigami, R. U. Margolis, and T. Endo. 2004. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. USA 101:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masai, H., T. Miyake, and K. Arai. 1995. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 14:3094-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattila, P., V. Joutsjoki, E. Kaitera, M.-L. Majuri, J. Niittymaki, N. Saris, H. Maaheimo, O. Renkonen, R. Renkonen, and M. Makarow. 1996. Targeting of active rat α2,3-sialyltransferase to the yeast cell wall by the aid of the hsp 150Δ-carrier: toward synthesis of sLex-decorated l-selectin ligands. Glycobiology 6:851-859. [DOI] [PubMed] [Google Scholar]
- 33.Mollicone, R., A. Gibaud, A. Francois, M. Ratcliffe, and R. Oriol. 1990. Acceptor specificity and tissue distribution of three human α-3-fucosyltransferases. Eur. J. Biochem. 191:169-176. [DOI] [PubMed] [Google Scholar]
- 34.Moukadiri, I., and J. Zueco. 2001. Evidence for the attachment of Hsp150/Pir2 to the cell wall of Saccharomyces cerevisiae through disulfide bridges. FEMS Yeast Res. 1:241-245. [DOI] [PubMed] [Google Scholar]
- 35.Mrsa, V., T. Seidl, M. Gentzsch, and W. Tanner. 1997. Specific labeling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast 13:1145-1154. [DOI] [PubMed] [Google Scholar]
- 36.Mrsa, V., and W. Tanner. 1999. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of Pir protein family) in stability of the Saccharomyces cerevisiae cell wall. Yeast 15:813-820. [DOI] [PubMed] [Google Scholar]
- 37.Muhlenhoff, M., A. Manegold, M. Windfuhr, B. Gotza, and R. Gerardy-Schahn. 2001. The impact of N-glycosylation on the functions of polysialyltransferases. J. Biol. Chem. 276:34066-34073. [DOI] [PubMed] [Google Scholar]
- 38.Mulligan, M. S., J. C. Paulson, S. de Frees, Z. L. Zheng, J. B. Lowe, and P. A. Ward. 1993. Protective effects of oligosaccharides in P-selectin-dependent lung injury. Nature 364:149-151. [DOI] [PubMed] [Google Scholar]
- 39.Murai, T., M. Ueda, M. Yamamura, H. Atomi, Y. Shibasaki, N. Kamasawa, M. Osumi, T. Amachi, and A. Tanaka. 1997. Construction of a starch-utilizing yeast by cell surface engineering. Appl. Environ. Microbiol. 63:1362-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama, K., T. Nagasu, Y. Shimma, J. Kuromitsu, and Y. Jigami. 1992. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 11:2511-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narimatsu, H. 2004. Construction of a human glycogene library and comprehensive functional analysis. Glycoconj. J. 21:17-24. [DOI] [PubMed] [Google Scholar]
- 42.Ohta, T., N. Miura, N. Fujitani, F. Nakajima, K. Niikura, R. Sadamoto, C.-T. Guo, T. Suzuki, Y. Suzuki, K. Monde, and S. Nishimura. 2003. Glycotentacles: synthesis of cyclic glycopeptides, toward a tailored blocker of influenza virus hemagglutinin. Angew. Chem. Int. Ed. 42:5186-5189. [DOI] [PubMed] [Google Scholar]
- 43.Percudani, R., A. Pavesi, and S. Ottonello. 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268:322-330. [DOI] [PubMed] [Google Scholar]
- 44.Renkonen, O., S. Toppila, L. Penttila, H. Salmine, J. Helin, H. Maaheimo, C. E. Costello, J. P. Turunen, and R. Renkonen. 1997. Synthesis of a new nanomolar saccharide inhibitor of lymphocyte adhesion: different polylactosamine backbones present multiple sialyl Lewis x determinants to l-selectin in high-affinity mode. Glycobiology 7:453-461. [DOI] [PubMed] [Google Scholar]
- 45.Salo, H., E. Sieve, T. Suntio, M. Mechlin, P. Mattila, R. Renkonen, and M. Makarow. 2005. Co-expression of two mammalian glycosyltransferases in the yeast cell wall allows synthesis of sLex. FEMS Yeast Res. 5:341-350. [DOI] [PubMed] [Google Scholar]
- 46.Schreuder, M. P., S. Brekelmans, H. van den Ende, and F. M. Klis. 1993. Targeting of a heterologous protein to the cell wall of Saccharomyces cerevisiae. Yeast 9:399-409. [DOI] [PubMed] [Google Scholar]
- 47.Schreuder, M. P., A. T. Mooren, H. Y. Toschka, C. T. Verrips, and F. M. Klis. 1996. Immobilizing proteins on the surface of yeast cells. Trends Biotechnol. 14:115-120. [DOI] [PubMed] [Google Scholar]
- 48.Shang, J., R. Qiu, J. Wang, J. Liu, R. Zhou, H. Ding, S. Yang, S. Zhang, and C. Jin. 1999. Molecular cloning and expression of Galβ1,3GalNAc α2,3-sialyltransferase from human fetal liver. Eur. J. Biochem. 265:580-588. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 50.Shimoi, H., H. Kitagaki, H. Ohmori, Y. Iimura, and K. Ito. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180:3381-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sumita, T., T. Yoko-o, Y. Shimma, and Y. Jigami. 2005. Comparison of cell wall localization among Pir family proteins and functional dissection of the region required for cell wall binding and bud scar recruitment of Pir1p. Eukaryot. Cell 4:1872-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeuchi, M., N. Inoue, T. W. Strickland, M. Kubota, M. Wada, R. Shimizu, S. Hoshi, H. Kozutsumi, S. Takasaki, and A. Kobata. 1989. Relationship between sugar chain structure and biological activity of recombinant human erythropoietin produced in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 86:7819-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 54.Toh-e, A., S. Yasunaga, H. Nisogi, K. Tanaka, T. Oguchi, and Y. Matsui. 1993. Three yeast genes, PIR1, PIR2 and PIR3, containing internal tandem repeats, are related to each other, and PIR1 and PIR2 are required for tolerance to heat shock. Yeast 9:481-494. [DOI] [PubMed] [Google Scholar]
- 55.Toki, D., M. Sarkar, B. Yip, F. Reck, D. Joziasse, M. Fukuda, H. Schachter, and I. Brockhausen. 1997. Expression of stable human O-glycan core 2 β-1,6-N-acetylglucosaminyltransferase in Sf9 insect cells. Biochem. J. 325:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsuji, S., M. Arita, and Y. Nagai. 1983. GQ1b, a bioactive ganglioside that exhibits novel nerve growth factor (NGF)-like activities in the two neuroblastoma cell lines. J. Biochem. 94:303-306. [DOI] [PubMed] [Google Scholar]
- 57.Uozumi, N., T. Teshima, T. Yamamoto, A. Nishikawa, Y. E. Gao, E. Miyoshi, C. X. Gao, K. Noda, K. N. Islam, Y. Ihara, S. Fujii, T. Shiba, and N. Taniguchi. 1996. A fluorescent assay method for GDP-l-Fuc:N-acetyl-β-d-glucosaminide α1-6 fucosyltransferase activity, involving high performance liquid chromatography. J. Biochem. 120:385-392. [DOI] [PubMed] [Google Scholar]
- 58.Varki, A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3:97-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, Z., L. Vo, and B. A. Macher. 1996. Structure-function analysis of human α1,3-fucosyltransferase. Amino acids involved in acceptor substrate specificity. J. Biol. Chem. 271:8818-8823. [DOI] [PubMed] [Google Scholar]