Abstract
I present the results of a culture-independent survey of soil bacterial communities from serpentine soils and adjacent nonserpentine comparator soils using a variety of newly developed phylogenetically based statistical tools. The study design included site-based replication of the serpentine-to-nonserpentine community comparison over a regional scale (∼100 km) in Northern California and Southern Oregon by producing 16S rRNA clone libraries from pairs of samples taken on either side of the serepentine-nonserpentine edaphic boundary at three geographical sites. At the division level, the serpentine and nonserpentine communities were similar to each other and to previous data from forest soils. Comparisons of both richness and Shannon diversity produced no significant differences between any of the libraries, but the vast majority of phylogenetically based tests were significant, even with only 50 sequences per library. These results suggest that most samples were distinct, consisting of a collection of lineages generally not found in other samples. The pattern of results showed that serpentine communities tended to be more similar to each other than they were to nonserpentine communities, and these differences were at a lower taxonomic scale. Comparisons of two nonserpentine communities generally showed differences, and some results suggest that the geographical site may control community composition as well. These results show the power of phylogenetic tests to discern differences between 16S rRNA libraries compared to tests that discard DNA data to bin sequences into operational taxonomic units, and they stress the importance of replication at larger scales for inferences regarding microbial biogeography.
Although the unusual plant communities of serpentine soils have long been the object of extensive investigation as model evolutionary systems, their microbiology remains little known. The combination of high concentrations of potentially toxic heavy metals and low concentrations of calcium and other plant-available nutrients found in serpentine, or ultramafic, soils creates a potent evolutionary force that is responsible for both high levels of endemicity in plant communities and unique physiological traits, such as metal hyperaccumulation (4, 13). We might expect that soil bacteria, being in necessarily intimate contact with both the rhizosphere of these highly endemic plant communities and the unique chemical composition and properties of serpentine soils, would also exhibit strong evolutionary and physiological responses to create microbial communities that are as biologically interesting as the distinctive flora that exist on serpentine soils.
To date, investigations of serpentine soils have been largely culture based and have focused on the heavy-metal resistance of specific strains or the ecology of metal cycling (1). They have revealed that some bacteria resist a range of heavy metals common in serpentine soils by means of plasmid-borne genes (19, 24), and investigators have found that nickel-rich litter from the leaves of nickel-hyperaccumulating plants creates a high-nickel soil microenvironment, from which they isolated several strains of highly nickel-resistant bacteria. The goal of this paper was to compare bacterial communities from serpentine soils to communities from immediately adjacent nonserpentine soils by using 16S rRNA gene clone libraries. Furthermore, I replicated the serpentine-to-nonserpentine community comparison at three different sites over a large-scale landscape (∼100 km).
The question of microbial biogeography in natural environments has become increasingly controversial in recent years. While there is evidence to support the traditional microbiological view that “everything is everywhere” and that the global microbial community consists of a relatively small number of cosmopolitan species (7-9), an increasing number of molecularly based studies have found evidence for biogoegraphical patterns in microbial distribution and diversity (7a, 10, 12, 16, 18, 25). However, studies specifically designed to compare microbial communities across large-scale landscapes and that incorporate replication of these comparisons are rare. Although Fierer and Jackson's results (7a) show the importance of edaphic factors in soil microbial diversity, their use of the coarse-resolution terminal restriction fragment length polymorphism technique provides little detail about the specific lineages that respond to particular edaphic factors. The dramatic chemical differences between serpentine and nonserpentine soils make plausible the existence and detection of consistent differences in the presence and diversity of specific sequence-defined lineages unique to serpentine soils across larger-scale regional geography, should such differences exist.
From an evolutionary perspective, we can state two general hypotheses concerning the bacterial communities of serpentine soils, defined by how they might have adapted to their unique chemical environment. This could either be (i) through long-term evolution of phylogenetically distinct lineages specially adapted to serpentine soils or (ii) through rapid evolution of a small number of adaptive genes, perhaps including horizontal gene transfer or plasmid-borne genes (24). The first hypothesis predicts that a comparison of serpentine to nonserpentine soils will reveal that each community is comprised of different sets of more deeply divergent lineages, while the second hypothesis predicts that essentially the same lineages exist in both communities, with differences being due to a small number of adaptive genes and not likely detectable by comparison of 16S rRNA genes.
An important aspect of this study is that I provide true replication, not pseudoreplication, of the serpentine-to-nonserpentine community comparisons over a geographical extent of ∼100 km. One can argue that true replication is accomplished, for example, by taking several soil samples from a 1-m2 plot and investigating each independently. However, inferences made from these samples are valid only for that 1-m2 plot, in part because of the large potential for soil heterogeneity, but also due to our ignorance of larger-scale microbial biogeography. Even when samples are taken over a larger area, such as an agricultural field or larger study plot, extrapolating the generality of the results to areas outside the study site remains speculation. The general lack of true replication to date in molecular studies is understandable, given the labor-intensiveness and expense of clone library construction and sequencing and the daunting potential for enormous soil heterogeneity over virtually every scale from the microscopic to the regional landscape. The desire to make such general inferences about larger-scale bacterial communities, however, necessitates replication of community profiles over an appropriately larger scale.
In this study, I constructed 16S clone libraries from three different subalpine forest sites in northern California and southern Oregon (Fig. 1). At each of these sites, pairs of soil samples were collected on either side of a sharp boundary between serpentine soil and an immediately adjacent nonserpentine soil, allowing me to make three independent serpentine-to-nonserpentine comparisons. This study design allowed me to (i) test whether there is a consistent pattern of differences in bacterial community composition across a large geographical area, (ii) identify which bacterial groups might be responsible for any observed differences, and (iii) characterize any observed differences in ways that will guide future investigations. The study design allowed independent comparison of the effects of soil type (serpentine versus nonserpentine), as well as the effects of a geographical site, on the microbial community composition.
FIG. 1.
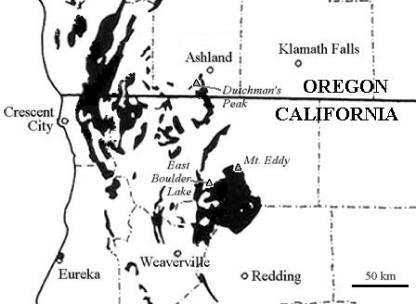
Map of the study area showing the three geographical sampling sites, indicated by triangles, in the context of the serpentine outcrops in Northern California and Southern Oregon, indicated in black. The dashed lines show county borders. A paired set of soil samples was taken at each of these three sites for a total of six clone libraries. The Dutchman's Peak site is at 42.050°N, 122.846°W; Mt. Eddy is at 41.317°N, 122.490°W; and East Boulder Lake at 41.234°N, 122.785°W.
MATERIALS AND METHODS
Study sites and soil sample collection.
I selected sampling sites that exhibited a sharp boundary between serpentine and nonserpentine soils based on both underlying geology and vegetative communities. This enabled me to take paired samples across the boundary separated by short distances (10 to 30 m), which allowed me to control for local microclimate, slope, aspect, hydrology, and physiography as much as possible. The locations of the sampling sites are shown in Fig. 1. All three sites are midelevation (6,500 to 7,500 ft) and are characterized by open subalpine forest vegetation and poorly developed soils, with the serpentine soils dominated by Jeffrey pine (Pinus jeffreyi) at Dutchman's Peak and foxtail pine (Pinus balfouriana) at East Boulder Lake and Mount Eddy. Sampling was done during August 2002, in the middle of the summer dry season. At each of these three sites, I located the precise contact between serpentine and nonserpentine soils and then located a 1-m2 plot on the serpentine side and another 1-m2 plot on the nonserpentine side separated by a short distance ranging from 10 to 30 m. Within each of these plots, I brushed away the surface litter and aseptically collected three 1-cm-diameter by 10-cm soil cores using individually autoclaved soil core tubes, stored them in individually sterilized plastic bags, and kept them on ice until they were returned to the laboratory for homogenization and DNA extraction that same day. The three core samples from a particular soil type at each site were pooled and homogenized prior to DNA extraction and are referred to as a sample. The three geographical sites from which paired samples were taken are referred to as sites. Replication is thus at the site level, not the soil core level, and comparisons of both immediately adjacent and geographically distant sites can be performed.
DNA extraction and PCR.
I performed the DNA extractions using the MoBio UltraClean Soil DNA Kit (MoBio Corp., Carlsbad, CA) according to the manufacturer's protocol and using 90 seconds of bead beating at 4,200/min. I performed PCR for the 16S rRNA gene using the universal bacterial 16S rRNA gene primers 8F (5′-AGRGTTYGATYMTGGCTCAG-3′) and 1492R (5′-CGGCTACCTTGTTACGACTT-3′). PCR conditions were as follows: a buffer of 60 mM Tris-HCl, 15 mM ammonium sulfate, pH 8.5, with 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 0.4 μM of each primer, 1 unit of Taq polymerase, and 0.4 μl of a 10−1 dilution of the pooled DNA extract as the template in 20-μl reaction volumes. For each DNA extract, I performed 16 replicate PCRs and pooled the product prior to clone library construction in order to minimize the effect of any PCR drift in individual reactions during amplification. The PCR cycling conditions were 94°C for a 1-minute initial denaturation, followed by 35 cycles of 94°C for 30 seconds, 52°C for 1 minute, and 72°C for 1 minute and 30 seconds, with a final extension of 72°C for 10 min. The PCR products were run out on a 1% low-melting-point agarose gel, and the ∼1.5-kb fragments were excised from the gel and purified with the QIAEXII Gel Extraction Kit (QIAGEN Corp., Valencia, CA) in preparation for cloning.
Clone library construction and sequencing.
I used the pGEM-T Easy cloning vector system (Promega Corp., Madison, WI) according to the manufacturer's protocol to clone the pooled PCR products. I chose 50 random clones from each library for sequencing using the primer 926F (5′-AAACTYAAAKGAATTGACGG-3′) and the ABI BigDye v3.0 cycle-sequencing kit (Applied Biosystems, Foster City, CA). I used an ABI 310 Genetic Analyzer for the actual sequencing. I manually checked and edited all sequencer chromatograms prior to alignment. The risk of sequencing chimeric sequences was reduced by the fact that, while I cloned almost the entire length of the 16S rRNA gene, I sequenced only the last third of it, thus missing a large percentage of potential chimeric breakpoints. Sequences were checked for possible chimeric origin by using the Ribosomal Database Project's CheckChimera program and the Mallard program, which is based on the Pintail algorithm by Ashelford et al. (2); I did not use the Bellerophon chimera detection software due to the relatively short sequence lengths. A few potentially suspicious sequences were discarded, and additional clones were sequenced until each library consisted of 50 nonchimeric sequences. The final data for each clone consisted of approximately 580 bases at the 3′ end of the 16S rRNA gene, which contained the V6, V7, V8, and V9 hypervariable regions.
Phylogenetic analyses.
I performed manual sequence alignments based on the 16S rRNA structural diagrams of Cannone et al. (4a), including 38 known sequences representative of major bacterial divisions. I used a conservative sequence mask that resulted in an alignment of 568 base positions for phylogenetic analyses. Exploratory phylogenetic analyses revealed that Aquifex and Thermotoga were basal to all new sequences, so I used these two as outgroups in all further phylogenetic analyses rather than archaeal sequences. In order to compare two different libraries, I combined the 50 sequences from each library, added Aquifex and Thermotoga, and estimated their phylogeny with maximum likelihood using fastDNAml. This was repeated for all 15 possible pairwise comparisons of the six libraries. In order to directly compare sites, I combined the serpentine and nonserpentine sequences from both samples at a site and then estimated the phylogeny rooted with Aquifex and Thermotoga using fastDNAml. This was repeated for the three possible site-versus-site comparisons. Finally, to make an overall comparison of the pooled serpentine-versus-nonserpentine sequences, all 300 sequences were combined, and an enormous computational effort was devoted to estimating a maximum likelihood phylogeny rooted with Aquifex and Thermotoga using fastDNAml. I similarly constructed Jukes-Cantor distance matrices for all levels of hierarchical pairwise comparisons (sample versus sample, site versus site, and serpentine versus nonserpentine) by using DNADIST from the PHYLIP package.
Statistical analyses.
I performed the phylogenetic P tests described by Martin (15) at all levels of comparison (sample versus sample, site versus site, and serpentine versus nonserpentine) using the maximum likelihood trees. In these tests, the soil type was coded as a character on the tree, and the minimum number of changes between soil types required to explain the tree was computed by MacClade (14). The number of changes was compared to the distribution of changes on 1,0000 random trees in order to produce a P value for the likelihood of the observed number of changes. These P values were used to determine if phylogeny significantly covaried with soil type. I used Arlequin (22) to perform the F test as described by Martin (15). In this test, FST values between sets of sequences are based on the average pairwise distance of two sequences within a set compared to the average pairwise distance of two sequences from different sets. Significance is tested by randomly permuting the sequences into two sets 10,000 times and computing a null distribution of FST values to which the observed FST value can be compared for significance by calculating a P value. I used S-LIBSHUFF (21, 23) to make pairwise comparisons between clone libraries at all levels (sample versus sample, site versus site, and serpentine versus nonserpentine) by using the Cramer-von Mies statistic. These tests use distance matrices to estimate the coverage of a clone library over a range of taxonomic levels, enabling the detection of significantly different microbial communities (as defined by nonoverlapping coverage), as well as whether one library is a subset of another (defined by differences in reciprocal coverage). S-LIBSHUFF calculates P values by using a random-permutation procedure to establish the statistical significance of overlapping coverage.
To estimate diversity statistics, I used DOTUR (20) to compute the Shannon diversity and the Chao and ACE richness estimates at a range of taxonomic levels based on the Jukes-Cantor distance matrices. This program uses the furthest-neighbor method to collapse similar sequences into groups at arbitrary levels of taxonomic similarity and then computes the Shannon, Chao, and ACE statistics for that taxonomic level. I computed these statistics for each sample separately, for each site separately, and also for the overall serpentine and nonserpentine sequences. Bootstrapping procedures within the DOTUR program assess the confidence limits of these estimators.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rRNA gene sequences from each of the six samples are DQ457699 through DQ457998.
RESULTS
I obtained 50 16S rRNA gene sequences from each of the six samples. The large phylogenetic analysis with representatives of known groups was used in conjunction with BLAST search results to place as many as possible of the sequences into known divisions. The 300 sequences collectively were broadly similar to other 16S rRNA gene clone libraries from other forest soils (3, 11); grouped by soil type, the serpentine and the nonserpentine collections were similar as well (Table 1). Though not statistically significant, there were several minor differences between the serpentine and nonserpentine communities—representatives of the OP10 division were exclusively in nonserpentine soils, while representatives of the OP8 division were exclusively in serpentine soils; also, there were four sequences from the nonserpentine community which could not be placed in any known or candidate division, while there were none from the serpentine community that could not be placed. The four sequences that could not be placed were all from a single library, the Dutchman's Peak nonserpentine sample. Among the 44 alphaproteobacterial sequences, many were very closely related to Rhizobium and Bradyrhizobium, suggesting that in these poorly developed soils, nitrogen fixation is an important process.
TABLE 1.
Numbers of clones allied to known and candidate divisions from the 16S rRNA gene libraries from serpentine and nonserpentine soils
| Phylum or divisiona | No. of clones
|
|
|---|---|---|
| Serpentine | Nonserpentine | |
| Actinobacteria | 26 | 32 |
| Acidobacteria | 25 | 21 |
| Alphaproteobacteria | 17 | 27 |
| Verrucomicrobia | 13 | 5 |
| Green-nonsulfur-bacterium related | 13 | 4 |
| Gemmatimonadetes | 12 | 4 |
| Planctomycetes | 11 | 8 |
| Bacteriodetes | 10 | 8 |
| Betaproteobacteria | 8 | 11 |
| Deltaproteobacteria | 7 | 8 |
| OP8 | 5 | 0 |
| Gammaproteobacteria | 2 | 6 |
| TM7 | 1 | 2 |
| OP10 | 0 | 5 |
| Other | 0 | 4 |
| Firmicutes | 0 | 3 |
| Nitrospirae | 0 | 1 |
| Total | 150 | 150 |
Phyla and divisions are listed according to their abundance in the serpentine soil samples.
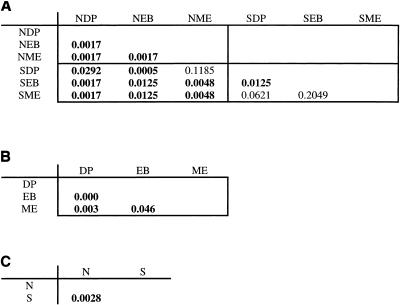
The results of Martin's phylogenetic P tests are presented in Fig. 2. I did not do any Bonferroni correction with these P values, because I wished to focus, not on the detection of absolute significant differences, but rather on the pattern of differences between the different soil types. All three of the nonserpentine-versus-nonserpentine comparisons were significant (P < 0.05), and eight out of the nine serpentine-versus-nonserpentine comparisons were significant, but only one of the three serpentine-versus-serpentine comparisons was significant (Fig. 2A). The results of the site-versus-site comparisons (with serpentine and nonserpentine sequences from the same site pooled) were all significant, as was the overall comparison by soil type (Fig. 2B and C).
FIG. 2.
Results of phylogenetic P tests. (A) Matrix of P values comparing maximum likelihood phylogenetic trees from individual clone libraries using Martin's phylogenetic P test. The first letter of each sample code is either S for serpentine or N for nonserpentine, and the next two letters encode the specific site (DP for Dutchman's Peak, EB for East Boulder Lake, or ME for Mount Eddy). The four quadrants of the matrix are groupings of like tests (nonserpentine versus nonserpentine in the upper left, serpentine versus nonserpentine in the lower left, and serpentine versus serpentine in the lower right). Significant P values (those less than 0.05) are in boldface; nonsignificant results indicate that 16S phylogeny does not covary with the sampling site. (B) Comparisons of one site to another, with serpentine and nonserpentine samples from each site pooled. (C) Overall serpentine-nonserpentine comparison.
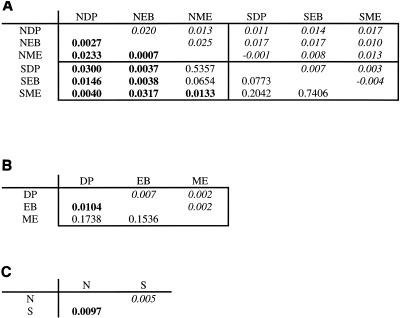
Figure 3A shows FST values comparing the different samples above the diagonal and the P values from significance testing below the diagonal. All three of the nonserpentine-versus- nonserpentine comparisons were significant, seven out of the nine serpentine-versus-nonserpentine comparisons were significant, and none of the three serpentine-versus-serpentine values was significant. Figure 3B shows that only the Dutchman's Peak-versus-East Boulder Lake site comparison was significant, while Fig. 3C shows that the overall comparison by soil type was significant.
FIG. 3.
FST values based on average pairwise differences within and between 16S clone libraries and results of significance tests. (See the legend to Fig. 2 for site codes.) (A) FST values based on average pairwise differences within and between 16S clone libraries are given above the diagonal; below the diagonal are P values from significance testing by permutation tests using Arlequin. The four quadrants of the matrix are groupings of like tests (nonserpentine versus nonserpentine in the upper left, serpentine versus nonserpentine in the lower left, and serpentine versus serpentine in the lower right). P values of less than 0.05 are in boldface; nonsignificant results indicate that there is not significant population differentiation based on a matrix of pairwise distances. (B) Comparisons of one site to another, with serpentine and nonserpentine samples from each site pooled. (C) Overall serpentine-nonserpentine comparison.
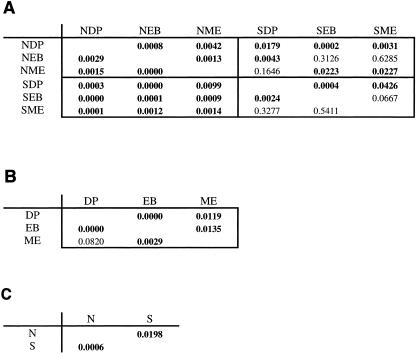
Figure 4A shows the matrix of P values produced by S-LIBSHUFF comparisons of clone library distance matrices. Values below the diagonal test the coverage of the first library by the second, while values above the diagonal test the coverage of the second library by the first. Three out of the 6 serpentine-versus-serpentine comparisons were significant, while 15 out of the 18 comparisons between serpentine and nonserpentine and all 6 of the nonserpentine-versus-nonserpentine comparisons were significant. Figure 4B and C shows that five out of six of the site-versus-site comparisons were significant and that the overall serpentine-versus-nonserpentine comparison was significant.
FIG. 4.
Results of S-LIBSHUFF clone library comparisons. (See the legend to Fig. 2 for site codes.) (A) Matrix of P values produced by S-LIBSHUFF comparisons of clone library distance matrices. The values test the heterologous coverage CROW,COLUMN versus the homologous coverage, CROW. The four quadrants of the matrix are groupings of like tests (nonserpentine versus nonserpentine in the upper left, serpentine versus nonserpentine in the lower left, and serpentine versus serpentine in the lower right). P values of less than 0.05 are in boldface; nonsignificant results indicate overlapping coverage of the second library by the first. For cases in which CXY is not significant and CYX is, library X is considered to be a subset of library Y. The matrix suggests that SME and NME are both separate subsets of SDP and that NEB is a subset of both SEB and SME. (B) Comparisons of one site to another, with serpentine and nonserpentine samples from each site pooled. These results suggest that the Mount Eddy community is a subset of the Dutchman's Peak community. (C) Overall serpentine-nonserpentine comparison.
The results for the diversity statistics produced by the DOTUR program were not informative. Although large differences in the ACE (from 110 to 301) and Chao (from 97 to 197) diversity estimators for individual samples were calculated at the 97% similarity level, there were no significant differences, as the bootstrapped 95% confidence intervals were very large, resulting in complete overlap between all samples for both estimators. Similarly, comparisons of ACE and Chao diversity estimators at the site level and the overall serpentine-versus-nonserpentine level were not significant (data not shown). The Shannon diversities of individual library samples ranged from 3.5 to 3.8, and there were no significant differences at the sample, site, or soil type level (data not shown). Because the DOTUR program can produce these comparisons at arbitrary levels of sequence similarity, I explored these statistics at other levels of similarity (95%, 90%, and 80%) with similar results—large 95% confidence intervals that precluded the detection of any significant differences among samples, sites, or soil types with my sample sizes.
DISCUSSION
Comparisons of individual samples.
With six different clone libraries, there are 15 different pairwise comparisons that can be performed for each of the phylogenetic tests; this allows independent replication of serpentine-to-nonserpentine comparisons. Before discussing particular results, it is important to note that the vast majority of P tests, F tests, and library-shuffling tests between individual libraries returned significant results at the 95% confidence level (Fig. 2A, 3A, and 4A). This points to a general distinctiveness of each soil sample, consisting of a collection of lineages generally not found in the other samples. The detection of this many significant differences among soil sample communities is remarkable, considering that the number of clones I sequenced from each sample (50) was tiny compared to the vast diversity of the community sampled. These results suggest that these statistical tests (Martin's P and F tests and Schloss and Handelsman's library-shuffling tests) are very powerful tools to enable the discernment of community differences at finer taxonomic scales by making use of the full DNA sequence information produced by these libraries. Merely classifying 16S rRNA gene clones into divisions discards much of the data upon which these distinctions are based.
The combined results of the P tests, F tests, and library shuffling do suggest that serpentine communities are more similar to each other than serpentine communities are to nonserpentine communities. The pattern of significant results in the P tests (Fig. 2A), F tests (Fig. 3A), and library shuffling (Fig. 4A) all show a bias toward nonsignificant results in the quadrant comparing serpentine communities to other serpentine communities, with most of the serpentine-to-nonserpentine comparisons and nonserpentine-to-nonserpentine comparisons producing significant results. This suggests that the soil type or, more accurately, the parent rock type does help to determine microbial community composition across a broad geographic region at my sampling sites, which are all moderate-elevation forested sites with poorly developed soils. The fact that all of the nonserpentine-to-nonserpentine comparisons in all three tests were significant may be partially explained by the fact that I have used “nonserpentine” as a negative definition, which obscures a diversity of parent rock types among the nonserpentine sites. The geology ranges from quartz biotite schist at Dutchman's Peak to diorite at Mt. Eddy to quartz diorite at East Boulder Lake. If this edaphic diversity helps to determine community composition in these samples, then we might expect some significant differences among the comparisons (7a).
Although large differences in the ACE and Chao richness estimators were calculated from individual samples, there were no significant differences at the sample, site, or soil type level. Shannon diversities estimated by DOTUR produced a much narrower range among samples (from 3.5 to 3.8), with no significant differences. These results are consistent with those of studies that have determined that very large numbers of sequences must be obtained from clone libraries of complex environments in order to accurately estimate a number of parameters, such as species richness (5, 6, 17).
Comparisons by geographical site.
To make site-based comparisons, we pooled the serpentine and nonserpentine libraries at each site to create a larger sequence set that represented the community at that geographical site. The P tests, F tests, and library-shuffling tests based on site-versus-site comparisons showed conflicting results. The Dutchman's Peak-versus-East Boulder Lake comparison produced significant results in all of the statistical tests, while the other two comparisons produced a mix of significant and nonsignificant results (Fig. 2B, 3B, and 4B). These results suggest that the geographical site can play a role in determining soil bacterial communities. However, in doing the site-based tests, frequently two very distinctive communities were combined and compared to a combination of two other very distinctive communities, a process that could tend to produce spurious significant results and reduce the effective power of these statistical tests.
Overall comparison by soil type.
The relative abundances of different bacterial divisions from serpentine and nonserpentine communities were not significantly different from each other (Table 1) and are generally similar to those in other culture-independent studies of forest soils (3, 11). At the broadest taxonomic scale, there is little evidence for a marked change in community composition or unique dominant groups on serpentine soils (Table 1). However, the P tests, F tests, and library-shuffling results (Fig. 2C, 3C, and 4C) comparing overall serpentine to overall nonserpentine communities did all have significant results, suggesting that differences in the communities do exist at a finer taxonomic scale than the division level. In order to discover which phylogenetic groups might be contributing to the significant overall serpentine-to-nonserpentine P-test result, I attempted to remove the significance by pruning branches off the overall tree and recalculating the P test until the result was nonsignificant (data not shown). It took at least four separate prunings of a minimum of 57 sequences in order to remove the overall serpentine-to-nonserpentine significance, supporting the idea that there is no single major group that is largely responsible for this difference. There is some suggestion of higher-level groups that are unique to serpentine soils, such as a clade within the green nonsulfur bacterium-related bacteria and those related to the OP8 division (Table 1), but they are minor components of the overall communities, and the differences are not significant with my sample sizes.
Conclusions.
One hypothesis is that the chemistry of serpentine soils constrains the community composition to be drawn from a limited number of bacterial groups. This would predict a pattern similar to the one I observed, with most serpentine-to-serpentine results being nonsignificant, while the larger pool of groups from which other soil type communities may draw from would result in a larger number of significant differences. The library-shuffling data do not support the idea that serpentine communities are generally subsets of other communities—the overall serpentine-to-nonserpentine comparison (Fig. 4C) is significant in both directions, and of those comparisons which did identify one library as a subset of another (Fig. 4A), there are actually more cases of nonserpentine communities being subsets of serpentine communities than vice versa. The general pattern of more differences between serpentine and nonserpentine communities at the lower taxonomic levels is consistent with horizontal gene transfer of relatively few genes allowing adaptation to the serpentine environment.
Despite having only a tiny sample size of three geographical sites and a relatively small number of 50 sequences per library, a pattern does emerge from the results of the P tests, F tests, and library-shuffling tests that suggests that serpentine communities are phylogenetically more similar to each other than they are to nonserpentine communities, but at a lower taxonomic level than the division. I have controlled my samples to be from moderate-elevation poorly developed forest soils in southern Oregon and northern California, so I cannot infer similarities among serpentine soils of different types and from other geographical locations. However, my results suggest that future work devoted to expanding the range of soil types (e.g., low-elevation serpentine grasslands or Darlingtonia bogs) within the Klamath Mountains region, as well as expanding the geographical scope outside the region to similar elevations and qualities of forest soils, will be a fruitful avenue for defining the range of serpentine environments over which such a pattern of similarity may hold. Using true replication to determine the geographical and soil type limits over which bacterial communities in natural soil environments are phylogenetically similar would be an important advance in microbial biogeography.
Acknowledgments
This research was performed with equipment purchased by National Science Foundation grant DBI 0115892, which funded the creation of the Southern Oregon University Biotechnology Center.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Amir, H., and R. Pineau. 1998. Influence of plants and cropping on microbiological characteristics of some New Caledonian ultramafic soils. Aust. J. Soil Res. 36:457-471. [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axelrood, P. E., M. L. Chow, C. C. Radomski, J. M. McDermott, and J. Davies. 2002. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48:655-674. [DOI] [PubMed] [Google Scholar]
- 4.Brady, K. U., A. R. Kruckeberg, and H. D. Bradshaw, Jr. 2005. Evolutionary ecology of plant adaptation to serpentine soils. Annu. Rev. Ecol. Syst. 36:243-266. [Google Scholar]
- 4a.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Guttell. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. (Erratum, 3:15.) [DOI] [PMC free article] [PubMed]
- 5.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenchel, T., and B. J. Finlay. 2003. Is microbial diversity fundamentally different from biodiversity of larger animals and plants? Eur. J. Protist 39:486-490. [Google Scholar]
- 7a.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, B. J., and K. J. Clarke. 1999. Ubiquitous dispersal of microbial species. Nature 400:828. [Google Scholar]
- 10.Green, J. L., A. J. Holmes, M. Westoby, I. Oliver, D. Briscoe, M. Dangerfield, M. Gillings, and A. J. Beattie. 2004. Spatial scaling of microbial eukaryote diversity. Nature 432:747-750. [DOI] [PubMed] [Google Scholar]
- 11.Hackl, E., S. Zeckmeister-Boltenstern, L. Bodrossy, and A. Sessitch. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horner-Devine, M. C., M. Lage, J. B. Hughes, and B. J. Bohannan. 2004. A taxa-area relationship for bacteria. Nature 432:750-753. [DOI] [PubMed] [Google Scholar]
- 13.Kruckeberg, A. R. 1985. California serpentines: flora, vegetation, geology, soils, and management problems. University of California Press, Berkeley.
- 14.Maddison, W. P., and J. R. Maddison. 1992. MacClade: analysis of phylogeny and character evolution. Sinauer, Sunderland, Mass.
- 15.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martiny, J. B., B. J. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A. L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 17.Narang, R., and J. Dunbar. 2003. Modeling bacterial species abundance from small community surveys. Microb. Ecol. 47:396-406. [DOI] [PubMed] [Google Scholar]
- 18.Papke, R. T., N. B. Ramsing, M. M. Bateson, and D. M. Ward. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650-659. [DOI] [PubMed] [Google Scholar]
- 19.Schlegel, H. G., J.-P. Cosson, and J. M. Baker. 1991. Nickel-hyperaccumulating plants provide a niche for nickel-resistant bacteria. Bot. Acta 104:18-25. [Google Scholar]
- 20.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider, S., D. Roessli, and L. Excofier. 2000. Arlequin: a software for population genetics data analysis, version 2.000. University of Geneva, Geneva, Switzerland.
- 23.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoppel, R.-D., and H. G. Schlegel. 1995. Nickel-resistant bacteria from anthropogenically nickel-polluted and naturally nickel-percolated ecosystems. Appl. Environ. Microbiol. 61:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic Archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]