Abstract
In order to initiate studies on promoter activities in Bifidobacterium longum and to independently confirm transcriptional data generated by microarray experiments, we have constructed a versatile reporter plasmid based on a B. longum cryptic plasmid and the Escherichia coli gusA gene. The resulting plasmid, pMDY23, has been tested using three B. longum promoters.
Bifidobacteria are natural commensals of the gastrointestinal tract of humans and mammals. Initially isolated from the feces of breast-fed infants by Tissier in 1899, bifidobacteria have been used as a marker to indicate the presence of a healthy microbiota in humans and are today included in a variety of commercial food products accompanied by health-related claims (8).
The determination and analysis of the complete genome sequence of the human infant isolate Bifidobacterium longum NCC2705 (19) has provided insight into the genes that play a role in its metabolic capabilities and its interactions with the host (4). This vast quantity of genomic data cannot be fully exploited at present due to limitations in molecular genetic tools for the analysis of gene function and regulation in the genus Bifidobacterium. In order to initiate studies on promoter activities and to independently confirm data generated by microarray experiments, we constructed the versatile plasmid pMDY23 based on an Escherichia coli origin of replication, a small cryptic B. longum plasmid, and the Escherichia coli β-glucuronidase gene, gusA (5), that has been successfully used as a reporter gene in a wide range of organisms (6, 7, 15, 20). pMDY23 has been designed to function (i) as a reporter for promoter strength or regulation analysis and (ii) for the identification of genomic fragments containing active promoters (promoter probe). Here we demonstrate the use of pMDY23 as a reporter plasmid with the analysis of the activity of three B. longum promoters. Preliminary microarray-based gene expression data were used to identify probable expressed genes, and the regions directly preceding two of them were tested for promoter activity in plasmid pMDY23. These were the potential promoters pr-BL1363 (promoter of gene BL1363) preceding BL1363, encoding the enzyme glyceraldehyde 3-phosphate dehydrogenase, and pr-BL1613, preceding BL1613, encoding a member of the LacI family of bacterial regulator proteins (Pfam no. PF00356). Secondly, a predicted sugar-regulated promoter, pr-BL1518, preceding the gene BL1518, encoding the enzyme α-galactosidase, was also tested.
Analysis of the B. longum NCC2705 genome sequence did not reveal the presence of a complete gene encoding the enzyme β-glucuronidase responsible for the deconjugation of β-d-glucuronides in the gastrointestinal tract; however, NCC2705 contains a truncated, 105-bp open reading frame (BL1077; genome coordinates 1307865 to 1307969) encoding a peptide with similarity to GusA proteins. The absence of β-glucuronidase activity was confirmed using an enzymatic test (Table 1). B. longum NCC2705 obtained from the Nestlé Culture Collection was grown in MRS medium (Becton Dickinson AG, Basel, Switzerland) containing 0.05% (wt/vol) cysteine (MRS-cys) supplemented with 2% (wt/vol) glucose at 37°C. Precultures were incubated in anaerobic conditions using AnaeroGen (Oxoid AG, Basel, Switzerland), while cultures for enzyme activity measurements were grown in a Sixfors fermenter system composed of four individual 500-ml vessels (Infors AG, Bottmingen, Switzerland). Growth medium was sparged overnight with CO2 to produce anaerobic conditions, while fermentations were performed at 37°C with 200 rpm agitation and a constant pH of 6.5 (by addition of 1 M NaOH). All fermentations were performed in duplicate using separate vessels inoculated at 1% (vol/vol) and an approximate starting optical density at 600 nm (OD600) of 0.05.
TABLE 1.
Expression of the E. coli gusA gene in B. longum NCC2705 in plasmid pMDY23 from the selected promoters
| Strain and plasmid | Sugar | Generation timea | GUS activityb | Fold inductionc | Plasmid copy no.d | Plasmid stabilitye (%) |
|---|---|---|---|---|---|---|
| NCC 2705 | Glu | 54.9 ± 0.6 | ND | |||
| NCC 2705 + pMDY23 | Glu | 54.7 ± 0.6 | ND | 12.7 | 100 | |
| NCC 2705 + pGUSA | Glu | 56.1 ± 9.4 | 171.7 ± 36.2 | 10.4 | 98 | |
| NCC 2705 + pGUSB | Glu | 51.5 ± 5.4 | 8.6 ± 0.7 | 11.1 | 99 | |
| NCC 2705 + pGUSC | Glu | 60.8 ± 2.5 | 5.1 ± 3.8 | 100 | ||
| NCC 2705 + pGUSC | Raf | 60.7 ± 7.5 | 62.0 ± 4.9 | 12 | ||
| NCC 2705 + pGUSC | Glu + Raf | 58.6 ± 5.5 | 10.6 ± 2.4 | 2 | 10.2 | 100 |
Generation time in minutes.
Mean value of β-glucuronidase activity (U) with standard deviations as determined in triplicate performed on two biological replicates. ND indicates enzyme activity not detected.
Fold induction in comparison to the glucose fermentation reference value.
Average plasmid copy number relative to the single-copy genomic gene BL0003.
Percent segregation stability after 10 generations without antibiotic selection.
Samples were taken at regular intervals for optical density, and β-glucuronidase enzyme activity was measured as follows. A 30-ml sample of mid-exponentially growing cells at an OD600 of 0.5 to 0.6 was harvested by centrifugation at 23,000 × g for 15 min at 4°C and suspended in 1 ml of GUS reaction buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 at pH 7.0). The cell suspension was transferred to a screw-top tube containing glass beads (≤10 μm), and the cells were disrupted with a Mini-Bead Beater-8 (Fisher Scientific SA, Wohlen, Switzerland) at 4°C, using three cycles of 1 min with cooling on ice between each cycle. Bacterial debris was removed by centrifugation at 20,000 × g for 10 min, and the cell extracts were transferred to clean tubes. From 20 to 100 μl of cell extract was incubated with 7.5 mM para-nitrophenyl-α-d-glucuronide (Sigma-Aldrich AG, Buchs, Switzerland) at 37°C, and the release of nitrophenol was monitored at 405 nm. Specific β-glucuronidase activity was expressed in nanomolar units of nitrophenol released per minute per milligram of protein. Protein concentrations were determined according to the method of Bradford (2) using the Bio-Rad determination kit (Bio-Rad Laboratories AG, Reinach, Switzerland) and bovine serum albumin as standard. Each enzymatic assay was performed in triplicate, starting from each biological replicate.
As stated above and seen in Table 1, the strain B. longum NCC2705 possesses no β-glucuronidase enzyme activity, hence permitting the use of the gusA gene as a reporter for the analysis of B. longum promoters. DNA manipulations, plasmid isolation, and transformation of E. coli were performed according to standard procedures (18). B. longum strains were screened for plasmids that could be developed into cloning vectors. One microgram of chromosomal DNA isolated according to Germond et al. (3) was separated on a 1% agarose gel, stained with ethidium bromide, and visualized with UV light. B. longum strain NCC293 isolated from infant feces was identified as containing a small extrachromosomal element, further referred to as pNCC293 (data not shown). NCC293 chromosomal DNA was digested with selected enzymes and NarI identified as cutting pNCC293 once, generating a linear plasmid of 1.8 kb in size. The NarI-digested pNCC293 was extracted from the gel, ligated into AccI-digested pUC18 (22), and used to transform E. coli strain TG1 (Stratagene Inc., La Jolla, CA). Transformants were selected on Luria-Bertani (LB) plates supplemented with 100 μg/ml ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and isopropyl-β-d-thiogalactopyranoside and incubated at 37°C. A clone containing pNCC293 was identified and named pDP869. DNA sequence analysis of the cloned plasmid pNCC293 revealed a 1,824-bp circular plasmid with 98.8% identity to the 1,847-bp B. longum plasmid pMB1 (12). The two plasmids differ in the number of 24-bp direct repeats, 3.5 and 4.5 for pNCC293 and pMB1, respectively, and base pair changes in pNCC293 between the end of repB and the repeats that create a 29-bp stem, 46-bp loop hairpin structure with a calculated energy of −42.48 kcal/mol (23). Plasmid pJL74 (pBluescript SK+ carrying a spectinomycin resistance gene) (9) was digested with EcoRI plus BamHI, and the 1.2-kb spectinomycin resistance gene was ligated into pDP869 digested with the same enzymes. Transformants in TG1 were selected on LB plates containing 100 μg/ml spectinomycin. The resulting plasmid was amplified with the primers P1 and P2, listed in Table 2 (MWG-Biotech AG, Ebersberg, Germany), using the Expand High Fidelity DNA polymerase (Roche Diagnostics AG, Rotkreuz, Switzerland), digested with the restriction enzyme AgeI, and ligated to give the E. coli-B. longum shuttle vector pDP870, containing a polylinker and lacking the ampicillin resistance gene. Plasmid pDP870 will be of interest due to its relatively small size and its cloning array of unique restriction sites. The plasmid may also be a combination with other, compatible shuttle vectors derived from B. longum plasmids (10, 11, 13, 14, 17, 21) for the expression of multiple genes in B. longum strains. The 1.85-kb DNA fragment containing the E. coli gusA gene was amplified from plasmid pNZ124-PrecA-GusA (a kind gift from Gabrielle Giliberti) using the primers GUS-F and GUS-R listed in Table 2. The gusA amplicon was digested with XhoI and PstI and ligated into pDP870 digested with XhoI and NsiI to create plasmid pMDY23, shown in Fig. 1.
TABLE 2.
List of primers used in this worka
| Primer name | Primer sequence, 5′-3′ |
|---|---|
| P1 | ATTCAACCGGTCCCGGGAGATCTGAGCTCACTAGTCATATGCTGTCAGACCAAGTTTAC |
| P2 | TATTACCGGTCTCGAGATGCATAAGCTTGCATGCCTGCAG |
| GUS-F | TCTTCCTCGAGATGCATGTCGACTTACGTCCTGTAGAACC |
| GUS-R | TCTAATGTCACTAACCTGC |
| pr-BL1363-5′ | CCTAGAGATCTTCGCTGACTTGCATGCCG |
| pr-BL1363-3′ | TGATTCTCGAGCATGTAATTCTCCCTTGTAG |
| pr-BL1518-5′ | CAGGAAGATCTATATCGGCACCATCGCGTTC |
| pr-BL1518-3′ | GCTGCCCTCGAGCATATTGTCTCCTTCGAC |
| pr-BL1613-5′ | CCTAGAGATCTTCTTATGGTTGCTGTGCC |
| pr-BL1613-3′ | TGATTCTCGAGCATTGTTCGAATCCATTTCTGC |
| qPCR-pDP870-5′ | CATCGGACATGGCTACAGCAT |
| qPCR-pDP870-3′ | TGACTTGGTTCCCGCGTATT |
| qPCR-BL0003-5′ | TTCCGAGCGAATCCAATCAT |
| qPCR-BL0003-3′ | CCTGATTTGCGAGATCGAAGT |
| BL0003-5′ | CCGGTATTGGCAATCGGC |
| BL0003-3′ | ACTAGCTGGCAAACAGGC |
Introduced restriction sites are underlined.
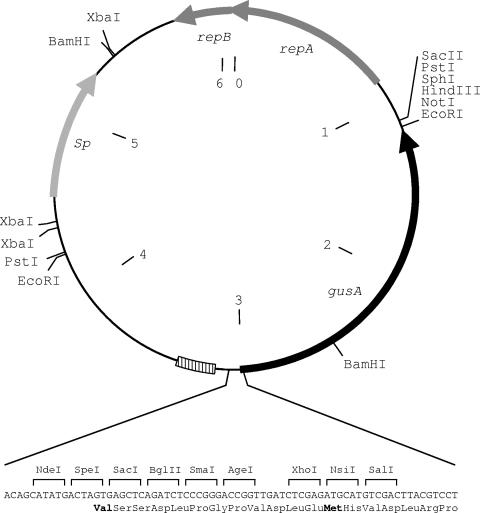
FIG. 1.
Schematic map of the 6,092-bp plasmid pMDY23 containing the E. coli origin of replication (hatched box). The gusA reporter gene is in black, the spectinomycin resistance gene (Sp) is light gray, and the B. longum replicon pNCC293 predicted replication proteins Rep1 and Rep2 are dark gray. The polylinker at the start of the gusA reporter gene is expanded and annotated with restriction sites plus translation products below the plasmid. Potential GTG and ATG initiation codons are marked in boldface type, and the size markers (in kilobases) are within the circle.
Microarray cDNA hybridization intensities of exponentially growing cultures of NCC2705 (data not shown) indicated high and low expression levels for the genes BL1363 and BL1613, respectively. While the preceding genes are oriented in the same orientation, an operon structure was judged unlikely, as the distances of 240 and 532 bp, respectively, for BL1363 and BL1613 were predicted to be excessive for efficient translational coupling. The DNA sequences preceding these genes were therefore selected for functional testing of pMDY23 in B. longum NCC2705. The promoter pr-BL1363 was amplified from NCC2705 chromosomal DNA using Expand High Fidelity DNA polymerase and the primers pr-BL1363-5′ and pr-BL1363-3′ listed in Table 2. The 250-bp pr-BL1363 amplicon (genome coordinates 266282 to 266033) was digested with the restriction enzymes BglII and PstI and ligated into plasmid pMDY23, digested with BglII plus NsiI to give plasmid pGUSA. Similarly, the 500-bp pr-BL1613 amplicon (genome coordinates 2018040 to 2018540) amplified with primers pr-BL1613-5′ and pr-BL1613-3′ was cloned into pMDY23 to give plasmid pGUSB. The DNA sequences of the promoters were verified, and plasmids pMDY23, pGUSA, and pGUSB were used to transform B. longum NCC2705 (containing the small cryptic plasmid pBL01 [accession number AF540971] with no similarities to pNCC293) as described previously (1), except that 0.5 M sucrose was omitted from the culture medium and the agar plates. Aliquots were plated on MRS-cys plates containing 200 μg/ml spectinomycin and incubated for up to 3 days at 37°C under anaerobic conditions.
Transformants were obtained for all plasmids. The enzyme activity was measured as described above, and the results are shown in Table 1. In the cell extracts of recombinants NCC2705 (pGUSA) and NCC2705 (pGUSB) we detected β-glucuronidase activities of 171.7 and 8.6 U for the pr-BL1363 and pr-BL1613 promoters, respectively. While the generation times of NCC2705 (pGUSA) and NCC2705 (pGUSB) were not significantly different from the host strain NCC2705, the standard deviation of the generation time increased, possibly suggesting some toxic effects of β-glucuronidase expression or a problem with plasmid stability or copy number.
The effects of GusA expression on plasmid stability was investigated as follows. Plasmid copy number relative to the bacterial chromosome was determined using quantitative PCR (qPCR) as described previously (16), with the exception that the target chromosomal DNA was not digested by a restriction enzyme. qPCR primers were designed for plasmid pDP870 and the chromosomal gene BL0003 using the program Primer Express (Applied Biosystems, Rotkeutz, Switzerland) and tested on plasmid pDP870-BL0003, where the BL0003 qPCR target region was amplified using primers BL0003-5′ and BL0003-3′, digested with the restriction enzymes HindIII and XbaI, and cloned into HindIII- and SpeI-digested pDP870. Plasmid pDP870-BL0003, ensuring a 1:1 ratio of both qPCR targets for the assessment of amplification efficiency, was used to generate standard curves using 10-fold serial dilutions using SYBR green (Applied Biosystems, Rotkeutz, Switzerland) on an ABI Prism 7000 machine (Applied Biosystems, Rotkeutz, Switzerland) with eight replicates for each dilution. Plasmid copy number was determined by qPCR with primers qPCR-pDP870-5′ plus qPCR-pDP870-3′ and qPCR-BL0003-5′ plus qPCR-BL0003-3′ on approximately 1 pg of total DNA with eight replicates for each sample. The average of the cycle threshold (CT) values was used to determine the difference between the two qPCR targets, ΔCT, and the plasmid copy number calculated using the following equation: plasmid copy number = 2ΔCT. The plasmid copy numbers for all plasmids are shown in Table 1. Plasmid pMDY23 without a promoter in NCC2705 showed an average copy number of 12.7, which was reduced to 10.4 for the highest GusA expression level with plasmid pGUSA. Plasmid segregation stability was determined on 100 individual colonies after 10 generations of nonselective replication. Plasmid pMDY23 in NCC2705 showed 100% segregation stability, while plasmids pGUSA and pGUSB in NCC2705 showed a slightly reduced stability of 98% and 99%, respectively. Plasmid structural stability was investigated by using total DNA extracted from NCC2705 (pGUSA) to transform E. coli strain TG1. Plasmid DNA was extracted from 24 spectinomycin-resistant colonies and digested with the restriction enzyme RsaI, giving seven visible bands on a 1% agarose gel. All 24 plasmid digests showed indistinguishable digestion patterns. These results showed that the E. coli gusA gene was translated into an active and stable enzyme in B. longum NCC2705 and that the highest expression level described here from plasmid pGUSA did not result in a structural instability of the plasmid but may have induced a slight reduction in segregation stability and relative copy number.
Many bacterial genes are expressed in response to changes in environmental conditions, either physical or nutritional, and the corresponding regulated promoters are interesting, as they allow the modulation of the level of heterologous or homologous gene expression. This is especially of interest if the gene product is detrimental to the proliferation of the producing cell. The gene BL1518 encoding the enzyme α-galactosidase is likely a component of the multi-sugar metabolism operon (BL1520 to BL1525) required for the transport and metabolism of various sugars, including raffinose. The promoter of gene BL1518 was identified as being potentially regulated by the sugar raffinose and selected for analysis. The 291-bp pr-BL1518 amplicon (genome coordinates 1692388 to 1692679) was amplified with primers pr-BL1518-5′ and pr-BL1518-3′ and ligated into pMDY23 to give plasmid pGUSC. The plasmid was used to transform B. longum NCC2705. The transformant was cultivated as described above using either 2% glucose, 2% raffinose, or 1% of each sugar as a carbon source, and the β-glucuronidase activity was measured at OD600 of 0.5 to 0.6. β-Glucuronidase activity measured in NCC2705 (pGUSC) with glucose as a carbon source was relatively low at 5.1 U (Table 1). A similar culture with raffinose as a carbon source showed a β-glucuronidase activity of 62 U, a 12× increase in enzyme activity that suggests an equivalent increase in promoter activity. Plasmid pGUSC in NCC2705 showed a 100% segregation stability when cultivated in either glucose or raffinose as a carbon source. This result confirms that the α-galactosidase gene expression is indeed induced when raffinose is supplied as the sole carbon source but that a residual activity is also detectable in the presence of glucose as the sole carbon source. Finally, when NCC2705 (pGUSC) was grown with glucose and raffinose, a β-glucuronidase activity of 10.6 U was measured, a 2× increase in enzyme activity compared to glucose alone (Table 1), which indicates a significant but not complete repression of the raffinose-induced expression by glucose.
In this work, we described the construction of a versatile plasmid, pMDY23, that can function as a reporter plasmid for the determination of promoter strength and the identification of promoters containing DNA fragments from a random genome library. On the basis of microarray expression data, we identified candidate promoter regions that were cloned before the gusA gene of pMDY23 to demonstrate its utility as a reporter plasmid by the quantification of one low and one strong active promoter. Finally, a promoter whose activity is regulated by the sugar raffinose was identified from the genome annotation and confirmed using pMDY23. Plasmid pMDY23 therefore provides a new tool for the analysis and understanding of the B. longum genome sequence.
Nucleotide sequence accession number.
The DNA sequence of pDP870 has been submitted to GenBank with the accession number DQ834380.
Acknowledgments
Mary O'Connell-Motherway and Ivana Jankovic are acknowledged for helpful discussions.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Argnani, A., R. J. Leer, N. van Luijk, and P. H. Pouwels. 1996. A convenient and reproducible method to genetically transform bacteria of the genus Bifidobacterium. Microbiology 142:109-114. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Germond, J. E., O. Mamin, and B. Mollet. 2002. Species specific identification of nine human Bifidobacterium spp. in feces. Syst. Appl. Microbiol. 25:536-543. [DOI] [PubMed] [Google Scholar]
- 4.Ivanov, D., C. Emonet, F. Foata, M. Affolter, M. Delley, M. Fisseha, S. Blum-Sperisen, S. Kochhar, and F. Arigoni. 2006. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 281:17246-17252. [DOI] [PubMed] [Google Scholar]
- 5.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. Beta-glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jefferson, R. A., M. Klass, N. Wolf, and D. Hirsh. 1987. Expression of chimeric genes in Caenorhabditis elegans. J. Mol. Biol. 193:41-46. [DOI] [PubMed] [Google Scholar]
- 8.Klijn, A., A. Mercenier, and F. Arigoni. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29:491-509. [DOI] [PubMed] [Google Scholar]
- 9.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, J. H., and D. J. O'Sullivan. 2006. Sequence analysis of two cryptic plasmids from Bifidobacterium longum DJO10A and construction of a shuttle cloning vector. Appl. Environ. Microbiol. 72:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumura, H., A. Takeuchi, and Y. Kano. 1997. Construction of Escherichia coli-Bifidobacterium longum shuttle vector transforming B. longum 105-A and 108-A. Biosci. Biotechnol. Biochem. 61:1211-1212. [DOI] [PubMed] [Google Scholar]
- 12.Matteuzzi, D., P. Brigidi, M. Rossi, and D. Di. 1990. Characterization and molecular cloning of Bifidobacterium longum cryptic plasmid pMB1. Lett. Appl. Microbiol. 11:220-223. [DOI] [PubMed] [Google Scholar]
- 13.Missich, R., B. Sgorbati, and D. J. LeBlanc. 1994. Transformation of Bifidobacterium longum with pRM2, a constructed Escherichia coli-B. longum shuttle vector. Plasmid 32:208-211. [DOI] [PubMed] [Google Scholar]
- 14.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 1999. Sequence analysis of plasmid pKJ50 from Bifidobacterium longum. Microbiology 145:585-592. [DOI] [PubMed] [Google Scholar]
- 15.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Providenti, M. A., J. M. O'Brien, R. J. Ewing, E. S. Paterson, and M. L. Smith. 2006. The copy-number of plasmids and other genetic elements can be determined by SYBR-Green-based quantitative real-time PCR. J. Microbiol. Methods 65:476-487. [DOI] [PubMed] [Google Scholar]
- 17.Rossi, M., P. Brigidi, V. Gonzalez, and D. Matteuzzi. 1996. Characterization of the plasmid pMB1 from Bifidobacterium longum and its use for shuttle vector construction. Res. Microbiol. 147:133-143. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz, U. K., D. M. Lonsdale, and R. A. Jefferson. 1990. Application of the beta-glucuronidase gene fusion system to Saccharomyces cerevisiae. Curr. Genet. 17:261-264. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka, K., K. Samura, and Y. Kano. 2005. Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 69:422-425. [DOI] [PubMed] [Google Scholar]
- 22.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 23.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]