Abstract
Malaria parasites are sexually reproducing protozoa, although the extent of effective meiotic recombination in natural populations has been debated. If meiotic recombination occurs frequently, compared with point mutation and mitotic rearrangement, linkage disequilibrium between polymorphic sites is expected to decline with increasing distance along a chromosome. The rate of this decline should be proportional to the effective meiotic recombination rate in the population. Multiple polymorphic sites covering a 5-kb region of chromosome 9 (the msp1 gene) have been typed in 547 isolates from six populations in Africa to test for such a decline and estimate its rate in populations of Plasmodium falciparum. The magnitude of two-site linkage disequilibrium declines markedly with increasing molecular map distance between the sites, reaching nonsignificant levels within a map range of 0.3–1.0 kb in five of the populations and over a larger map distance in the population with lowest malaria endemicity. The rate of decline in linkage disequilibrium over molecular map distance is at least as rapid as that observed in most chromosomal regions of other sexually reproducing eukaryotes, such as humans and Drosophila. These results are consistent with the effective recombination rate expected in natural populations of P. falciparum, predicted on the basis of the underlying molecular rate of meiotic crossover and the coefficient of inbreeding caused by self-fertilization events. This is conclusive evidence to reject any hypothesis of clonality or low rate of meiotic recombination in P. falciparum populations. Moreover, the data have major implications for the design and interpretation of population genetic studies of selection on P. falciparum genes.
The malaria parasite Plasmodium falciparum is the most important eukaryotic pathogen of humans, being responsible for most of the cases of malaria illness and mortality. Malaria parasites are protozoa with a haploid genome of approximately 3 × 107 bp organized in 14 chromosomes (1). The parasites replicate asexually during blood stage infections of humans and differentiate into male and female gametocyte forms that may be acquired by a feeding mosquito. During infection of the mosquito host, fertilization between male and female gametes produces a diploid zygote in which meiosis proceeds, allowing recombination between homologous chromosomes and assortment of recombinant chromosomes in haploid progeny. Experimental crosses between laboratory clones of P. falciparum show independent segregation of genes on different chromosomes (2, 3). Recombination along each chromosome occurs such that a 1-centiMorgan genetic map distance corresponds to a molecular map distance of approximately 1.5–3.0 × 104 bp on average (4). Analysis of meiotic progeny allows fine-scale mapping of major trait loci (5, 6) and subsequent identification of candidate gene loci (6).
The effective recombination rate between heteroallelic chromosomes in nature is likely to be somewhat lower than its maximum in the laboratory. Cross-fertilization between parasite gametes of different genotypes can occur in the mosquito only when more than one genotype is present in the blood meal. Self-fertilization between identical gametes can also occur in this case, and self-fertilization must exclusively occur when the human blood contains only one parasite genotype. The inbreeding coefficient F gives the extent to which the effective recombination rate is likely to be lowered in a particular local population because of self-fertilization (7). If the meiotic crossover rate (number of Morgans per kb) = c, the effective recombination rate, c′ = c (1 − F). The value of F is probably in the range of 0.4–0.9 in many populations (8, 9).
The level of inbreeding existing in P. falciparum populations is not sufficient to generate linkage disequilibrium (i.e., gametic phase disequilibrium) between alleles of genes located on different chromosomes. Several studies, involving genotypic analysis of hundreds of isolates in defined populations, have all shown no disequilibria between unlinked loci (8–12), even when multiallelic data are analyzed by statistically powerful permutation testing (12). Thus, reassortment of chromosomes at meiosis occurs frequently enough for their combinations to be effectively randomized. In contrast to some other parasitic protozoa (13) and many prokaryotic pathogens (14), P. falciparum clearly does not have a “clonal” population genetic structure. Further inferences about recombination rate cannot be derived from these data.
Patterns of linkage disequilibria between polymorphic loci within a small region of a chromosome may be more sensitive for estimation of effective recombination rate in populations. Analysis of polymorphic sites within the csp gene, in a sample of 25 published sequences, revealed no decline in linkage disequilibrium with increasing nucleotide distance between pairs of sites over a distance of approximately 1.0 kb, leading to the suggestion that meiotic recombination might be rare in natural populations of P. falciparum (15). To study linkage disequilibrium in a statistically appropriate manner, larger population samples are necessary (16). Previous evidence for interallelic recombination in the msp1 (merozoite surface protein 1) antigen gene (17–20) provided a basis for prospectively designing a study of linkage disequilibrium and molecular map distance. Here, 547 P. falciparum isolates from six populations in Africa (the continent with 90% of all P. falciparum infections) have been studied, with multiple polymorphic sites genotyped throughout the msp1 gene, covering a nucleotide distance of approximately 5 kb in a nontelomeric region of chromosome 9 (1). A very strong and highly significant negative relationship between linkage disequilibrium and molecular map distance between the sites is demonstrated. The rates of decline in linkage disequilibrium with molecular map distance in each of the populations are compared with those expected from the underlying molecular recombination rate and likely range of inbreeding coefficients.
MATERIALS AND METHODS
P. falciparum Isolates.
Blood samples (n = 547) were obtained from P. falciparum-infected individuals in six geographical locations in Africa. Blood was collected from consenting volunteers as 1-ml samples in EDTA or as 20-μl samples spotted and dried on filter paper, and nucleic acids were prepared from these samples by proteinase K digestion followed by phenol:chloroform extraction and ethanol precipitation. The sources of isolates were as follows: 91 isolates from Basse, Upper River Division, The Gambia collected in 1994–1995; 107 isolates from Ibadan, south western Nigeria collected in 1996; 124 isolates from Lambarene district, Gabon, collected in 1996; 66 isolates from Daraweesh village, north eastern Sudan collected in 1992–1995; 86 isolates from Muheza district, north eastern Tanzania collected in 1995; and 73 isolates from KwaZulu/Natal, South Africa collected in 1996. All samples were obtained with informed consent from patients and guardians in studies of malaria, which were in ethical compliance with the relevant ethical committees of the collaborating institutes and governments.
Analysis of Polymorphic Sites in the msp1 Gene.
Four pairs of oligonucleotide primers were used in separate PCR reactions to amplify four regions of the msp1 gene from genomic DNA of each of the 547 isolates. The codon numbers given here refer to the sequence alignment of Miller et al. (21), and the block numbers refer to the scheme of Tanabe et al. (17). The four regions and primer pairs were (i) codons 34–234 (blocks 1–3), primers BK1F: 5′-gctttagaagatgcagtattgaca, BK3R: 5′-gtaatcttccatttttcctacatta; (ii) codons 198–359 (blocks 3–5), primers BK3F: 5′-ttcaatcttaaaattcgtgca, BK5R: 5′-aaatttaatagttttggcaatttcttt; (iii) codons 1235–1638 (blocks 12–16), primers BK12F: 5′-aaaaattatacaggtaattctccaag, BK16R: 5′-tacgcattggtgttgtgaaatgtt; and (iv) codons 1630–1735 (block 17), BK17F: 5′-aacatttcacaacaccaatgcgta, BF17R: 5′-tattaataagaatgatattcctaagaa. PCR amplifications were performed in 20-μl volumes in 96-well plates, by using 100 nM primers, with initial template denaturation for 4 min at 94°C and 45 cycles for 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. After amplification, products were denatured for 2 min at 94°C and cooled rapidly to 4°C, and 1.5-μl aliquots were dotted on replicate nylon membranes (MagnaGraph; Microcon Separations) in 96-dot arrays.
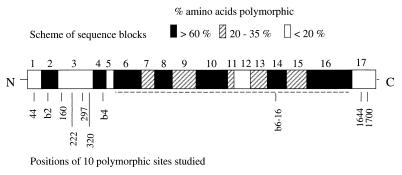
Replicate membranes were probed with digoxigenin-labeled sequence-specific oligonucleotides representing allelic sequences at 10 polymorphic positions throughout the msp1 gene (Table 1; Fig. 1). The names of the probes correspond to allelic amino acids at particular codon positions (21), or to allelic types at particular sequence blocks (17), to which they hybridize. Allelic probes were labeled in separate tubes simultaneously, under identical conditions by using the Boehringer Mannheim 3′-end-labeling kit, and labeled probes were used for hybridization at a final concentration of 2 nM in hybridization buffer (3 M tetramethylammonium chloride/50 mM Tris, pH 8.0/2 mM EDTA, pH 8.0/0.1% SDS). After the membranes were blocked for 30 min in 1% milk powder, hybridization was performed at 53°C for 90 min and the membranes were washed at low stringency twice for 10 min at room temperature [in 2× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA)/0.1% SDS] and at high stringency twice for 10 min at 56°C in hybridization buffer. Detection of hybridized digoxigenin-labeled probes on the membranes was performed by probing with anti-digoxigenin Fab fragment conjugated with alkaline phosphatase (Boehringer Mannheim) followed by detection with CSPD substrate (Boehringer Mannheim) and exposure on Hyperfilm-ECL (Amersham), according to Boehringer Mannheim recommendations. These conditions allowed clear and accurate discrimination between alleles, including those differing at a single nucleotide position, as confirmed by the inclusion of allele-specific controls to be typed in each assay.
Table 1.
Allele-specific oligonucleotide probes used for typing 10 intragenic polymorphic sites (loci) in the P. falciparum msp1 gene
| Locus* | Allele | Oligonucleotide sequence | |||||
|---|---|---|---|---|---|---|---|
| aa 44 | S | aca | ggt | tat | agt | tta | ttt |
| G | aca | ggt | tat | ggt | tta | ttt | |
| block 2 | MAD-like | aca | agt | gga | aca | gct | gtt |
| K1-like | gca | tca | gct | gga | ggg | ctt | |
| RO33-like | gtt | gtt | gca | aag | cct | gca | |
| aa 160 | Q | cat | aga | gtt | caa | aat | tac |
| R | cac | aga | gta | cga | aat | tac | |
| aa 222 | A | gca | tgt | gcc | aat | agt | tat |
| V | gta | tgt | gct | aat | gat | tat | |
| aa 297-8 | TK | gca | act | aaa | gaa | gaa | gaa |
| DN | gca | gat | aat | gaa | gaa | gga | |
| aa 320 | E | aaa | caa | tta | gaa | gaa | gca |
| Q | aaa | caa | tta | caa | gaa | gca | |
| block 4 b | Kl-like | ctt | gat | aag | aac | aaa | aaa |
| MAD-like | cct | gag | aat | aag | aaa | aaa | |
| block 6-16 | Kl-like | agt | tca | gga | tcc | aca | aaa |
| MAD-like | tcc | gac | aca | tta | gaa | caa | |
| aa 1644 | E | caa | tgt | cca | gaa | aat | tct |
| Q | caa | tgt | cca | caa | aat | tct | |
| aa 1700-1 | SR | ggt | agc | agc | aga | aag | aaa |
| NG | ggt | agc | aac | gga | aag | aaa | |
Loci are given either as polymorphic amino acid position(s) in the deduced protein sequence [numbered as in the alignment of Miller et al. (21)] or as polymorphic blocks of sequence (following the scheme of Tanabe et al. (17)] Bases that differ between allelic probes are in bold and underlined. Previously described very rare alleles at block 4b (RO33-like) and aa 1700-01 (SG) were absent (in the case of the former) or extremely rare (in the case of the latter) in a large subsample of more than half the total number of isolates, and hybridization patterns for all of the other intragenic loci did not suggest the existence of any previously undescribed additional alleles of these sequences.
Figure 1.
Scheme of the P. falciparum msp1 gene with positions of the 10 polymorphic sites studied. The coding sequence of the gene covers 5.1 kb, and there are no introns. 5′- and 3′ ends of the gene are marked by the corresponding N and C termini of the protein. The 17 block scheme of Tanabe et al. (17) is shown above, indicating the amount of sequence polymorphism in different parts of the gene. The positions of the 10 sequence sites typed in this study are marked at the bottom (sequences of allelic oligonucleotide probes are given in Table 1). Dashed lines under blocks 6–16 indicate that this whole region exists as either one of two highly divergent dimorphic sequence types, between which no recombinants have been detected (18, 19); therefore, typing of a sequence at only one representative dimorphic site (in block 14) was performed.
Scoring of Alleles at Each Site and Scoring of Two-Site Haplotypes.
Alleles at each of the 10 sites in the msp1 gene were scored in each of the 547 isolates. Many isolates contained more than one allele at a particular site (because of the presence of more than one P. falciparum genotype in the infection). For the analyses of linkage disequilibrium here, two-site haplotypes were scored from only isolates that had a single allele at each of the two sites under consideration; therefore, there would be no possibility of a false haplotype being scored because of the presence of mixed genotypes in some infections.
Statistical Analyses of Linkage Disequilibrium Between Polymorphic Sites in the msp1 Gene.
Linkage disequilibria were calculated in pairwise analyses of all sites studied in the msp1 gene. Two indicators of linkage disequilibrium were calculated: the D′ index of Lewontin (22) and the r2 (square of correlation coefficient) index of Hill and Robertson (23). The D′ index has a potential range from 0 to 1.0 (or −1.0). The test of disequilibrium considers the most common allele at each site; therefore, the positive or negative sign of the index merely reflects which alleles these were, whereas the magnitude of disequilibrium is indicated by its departure from 0 in either direction. When one of the potential haplotypes is missing from a population sample, D′ takes a value of 1.0 or −1.0, even though this may be nonsignificant and merely because of low expected frequency of the missing type. The r2 index has a potential range from 0 to 1.0 and is more conservative in that its value is not automatically skewed by a single missing haplotype. The statistical significance of each pairwise test of linkage disequilibrium on these haploid data was tested by Fisher’s exact test.
For analysis of the relationship between linkage disequilibrium and molecular map distance between sites, the correlation coefficient may be presented to indicate the trend but is not an appropriate basis for statistical testing, because not all pairwise analyses between sites are fully independent because of the linkage disequilibrium itself (24). Instead, the proportion of significant (P < 0.05) pairwise tests was compared for pairs of sites separated by different molecular map distance ranges: <0.3, 0.3–1.0, and >1.0 kb. The comparisons of proportions of significant tests, over different distance ranges, were tested by χ2. For each population, an exponential decline in linkage disequilibrium over increasing molecular map distances was fitted to the data points by using glim software.
RESULTS
Scoring of msp1 Alleles and Haplotypes in Each Population Sample.
At the majority of the 10 intragenic loci typed, there were significant differences in allele frequencies among the six populations (P < 0.001); therefore, analyses of linkage disequilibrium were performed separately for each population. Polymorphic loci with frequencies of the most common allele <0.90 were used for analyses of linkage disequilibrium, because these were highly statistically informative. This included almost all of the sites in each of the populations, although the common allele at block 6–16 in the gene (MAD-like) had a frequency of >0.90 in all populations except Sudan, and this was therefore included only in analysis of linkage disequilibrium in Sudan. After isolates containing a mixture of alleles at the relevant two loci were excluded, two-locus haplotypes were scored from an average of 78 isolates in The Gambia, 95 in Nigeria, 107 in Gabon, 76 in Tanzania, 56 in Sudan, and 65 in South Africa.
Linkage Disequilibrium Between Sites in the msp1 Gene.
For each of the six populations, linkage disequilibrium indices, D′ and r2, and statistical significance values were calculated for all pairwise combinations of informative loci. Table 2 shows results for The Gambia, with the two-locus combinations ranked by increasing nucleotide distance between the loci. A decline in linkage disequilibrium with increasing nucleotide distance is apparent, with some large and significant values of linkage disequilibrium between closely situated sites (within a few hundred nucleotides) but not between more distantly separated sites.
Table 2.
Linkage disequilibrium between alleles at pairs of sites in the msp1 gene in the P. falciparum population sample from The Gambia
| Two-site combination | Nucleotide distance, bp | Haplotype frequency
|
Linkage disequilibrium
|
||
|---|---|---|---|---|---|
| Expected | Observed | D′ | r2 | ||
| aa44s × bk2k1 | 30 | 0.47 | 0.42 | −0.767 | 0.069* |
| bk2k1 × aa160q | 48 | 0.17 | 0.02 | −0.861 | 0.421*** |
| aa320e × bk4k1 | 141 | 0.58 | 0.65 | 0.376 | 0.122** |
| aa1644e × aa1700s | 168 | 0.05 | 0.05 | 0.134 | 0.001 |
| aa160q × aa222a | 186 | 0.03 | 0.11 | 0.853 | 0.334*** |
| bk2k1 × aa222a | 238 | 0.14 | 0.04 | −0.732 | 0.221*** |
| aa44s × aa160q | 258 | 0.23 | 0.26 | 1.0 | 0.046 |
| aa222a × aa320e | 294 | 0.13 | 0.10 | −0.24 | 0.037 |
| aa222a × bk4k1 | 435 | 0.14 | 0.14 | 0.085 | 0.0005 |
| aa44s × aa222a | 444 | 0.17 | 0.20 | 1.0 | 0.029 |
| aa160q × aa320e | 480 | 0.19 | 0.14 | 0.294 | 0.08* |
| bk2k1 × aa320e | 528 | 0.41 | 0.41 | 0.008 | 0.00001 |
| aa160q × bk4k1 | 621 | 0.21 | 0.19 | −0.139 | 0.011 |
| bk2k1 × bk4k1 | 669 | 0.41 | 0.42 | 0.108 | 0.0037 |
| aa44s × aa320e | 738 | 0.69 | 0.69 | −0.032 | 0.00004 |
| aa44s × bk4k1 | 879 | 0.69 | 0.69 | −0.032 | 0.00004 |
| bk4k1 × aa1644e | 3798 | 0.09 | 0.09 | −0.006 | 0.00002 |
| bk4k1 × aa1700s | 3966 | 0.10 | 0.08 | −0.204 | 0.019 |
| aa320e × aa1644e | 3972 | 0.34 | 0.39 | 0.472 | 0.05 |
| aa320e × aa1700s | 4140 | 0.07 | 0.08 | 0.349 | 0.003 |
| aa222a × aa1644e | 4266 | 0.09 | 0.10 | 0.07 | 0.002 |
| aa222a × aa1700s | 4434 | 0.01 | 0.01 | −0.103 | 0.0002 |
| aa160q × aa1644e | 4452 | 0.12 | 0.14 | 0.122 | 0.006 |
| bk2k1 × aa1644e | 4500 | 0.25 | 0.26 | 0.042 | 0.001 |
| aa160q × aa1700s | 4620 | 0.02 | 0.03 | 0.16 | 0.007 |
| aa44s × aa1644e | 4620 | 0.39 | 0.37 | −0.218 | 0.007 |
| bk2k1 × aa1700s | 4668 | 0.04 | 0.04 | −0.023 | 0.00005 |
| aa44s × aa1700s | 4788 | 0.10 | 0.12 | 1.0 | 0.022 |
Statistical significance (Fisher’s exact test): ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
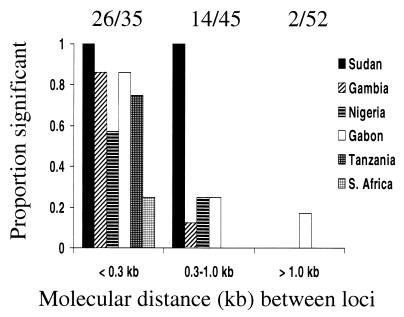
A negative correlation between linkage disequilibrium and nucleotide distance is also seen in each of the other populations (r range from −0.26 in Sudan to −0.67 in Tanzania). Fig. 2 shows the proportion of statistically significant linkage disequilibria according to nucleotide distance between loci. The three nucleotide distance categories of <0.3, 0.3–1.0, and 1.0–5.0 kb were chosen because they contain approximately equal numbers of the two-site combinations analyzed. Over all the populations, there was a much higher proportion of significant linkage disequilibria over distances of <0.3 kb (74%, 26/35 tests) than over distances of 0.3–1.0 kb (31%, 14/45 tests; χ2, P = 0.00013). Moreover, the latter proportion was much higher than the proportion of significant tests over 1.0–5.0 kb (4%, 2/52 tests; P = 0.0003). For the comparison of proportions of significant tests over the smallest (<0.3 kb) and largest (1.0–5.0 kb) nucleotide distance ranges, the P value is <10−7.
Figure 2.
Proportions of significant linkage disequilibrium tests between pairs of loci within the msp1 gene in each of six countries, according to molecular distance between the loci. Overall proportions are shown above the histogram, illustrating a highly significant decline with increasing molecular distance (P < 10−7, see text).
Rate of Decline of Linkage Disequilibrium over Nucleotide Distance in Different Populations.
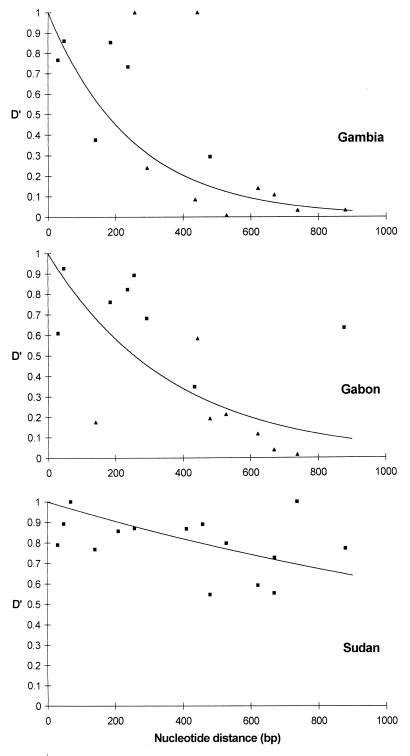
The rate of decline of linkage disequilibrium with increasing nucleotide distance was not identical in all populations. As indicated in Fig. 2, in Sudan all linkage disequilibria were significant over distances of <1.0 kb, whereas in the other populations the majority of tests were significant only over distances of <0.3 kb. To examine these patterns more fully in each country, the magnitude of linkage disequilibrium (D′ index) was plotted according to nucleotide distance between pairs of sites in the most intensively surveyed 1.0-kb 5′ region of the gene, and a negative exponential was fitted to the data points. Scatter plots of linkage disequilibrium vs. nucleotide distance between pairs of polymorphic sites in msp1 in The Gambia, Gabon, and Sudan are shown in Fig. 3. The rate of decline of linkage disequilibrium with increasing nucleotide distance was highest in The Gambia (fitted exponential = 3.97) and lowest in Sudan (exponential = 0.50). The rate of decline in Gabon (exponential = 2.80) was almost as high as in The Gambia and was similar to the rates in Nigeria (2.88), Tanzania (2.58), and South Africa (2.43).
Figure 3.
Decline in linkage disequilibrium (D′) with increasing nucleotide distance between pairs of sites in the msp1 gene, in three countries. The magnitude of D′ is considered, irrespective of sign (positive or negative, which is reversible if an alternative allele is considered). Squares indicate the points that are significantly different from 0, and triangles indicate those not significantly different from 0. Only the distance range of <1.0 kb is plotted, because almost all tests for linkage disequilibrium over a larger nucleotide distance were nonsignificant (Fig. 2). The exponential decay curves (1 − e−xn) were fitted by using glim statistical software, with n = nucleotide distance in kilobases between sites (exponential function x = 3.97 in The Gambia, 2.80 in Gabon, and 0.50 in Sudan). Note the slower rate of decline in Sudan compared with the other countries. Rates of decline in Nigeria, Tanzania, and South Africa were similar to that in Gabon (see text).
The much lower rate of decline in linkage disequilibrium with increasing nucleotide distance in Sudan, compared with the other five populations, is consistent with the much lower endemicity of malaria in the area of Sudan from which the parasite isolates were collected (25). This lower endemicity is expected to cause a lower proportion of multiple genotype infections in humans and, hence, a lower rate of outcrossing between gametes of the parasite and a lower effective recombination rate (26, 27). The proportion of mixed isolates (having more than one detectable msp1 allele at any of the polymorphic sites typed) was indeed much lower in Sudan (24%) than in the other populations (The Gambia, 66%; Nigeria, 45%; Gabon, 61%; Tanzania, 62%; and South Africa, 49%). This difference is significant (P < 0.01 for comparison of Sudan with each of the other populations).
DISCUSSION
The meiotic recombination rate in natural populations of P. falciparum is high. Threefold evidence for this is presented here. First, in all six African populations studied, there is a decline in linkage disequilibrium with increasing molecular map distance between polymorphic sites within the P. falciparum msp1 gene. This is highly statistically significant (P < 10−7) and indicates that multiple sites throughout this gene have been involved in chiasma formation and interallelic recombination and that at each meiosis the likelihood of interallelic recombination is proportional to the distance between loci. Second, in most populations, the decline in linkage disequilibrium with molecular map distance is very steep; therefore, there is little linkage disequilibrium over distances of >1 kb. This is a more rapid decline in linkage disequilibrium than is seen in humans (28) and Drosophila melanogaster (29, 30) and indicates that P. falciparum has a higher effective recombination rate than these species. Third, one population studied had a less steep decline in linkage disequilibrium than the others, and this was the population in which the infection was least endemic and in which most blood stage infections contained only a single detectable msp1 haplotype. This indicates that inbreeding caused by self-fertilization between parasites modifies the effective recombination rate, as expected (7).
The results presented here are consistent with expectations derived from many previous studies. A genetic map distance of 1 centiMorgan was estimated to correspond to a 15- to 30-kb molecular map distance in the genome of P. falciparum (4). This gave a crude recombination rate (c = Morgans/kb) in the order of c ≅ 4 × 10−4, approximately 20 times higher than the average in D. melanogaster (2 × 10−5) (29, 31), 40 times higher than in humans (10−5) (28, 31), and 80 times higher than in mice (32). Inbreeding will lower the frequency of pairing of heteroallelic chromosomes; therefore, the effective recombination rate (c′) will be correspondingly lower: c′ = c (1 − F), where F equals the inbreeding coefficient (7). Even if a fairly high value of F = 0.9 is considered for some populations of P. falciparum (9), the effective recombination rate (c′ ≅ 4 × 10−5) would remain higher than the average in the other eukaryotes cited.
In five of the six populations studied here, the decline in linkage disequilibrium with molecular map distance was very rapid, disequilibrium reaching half its maximal value over a distance of >0.3 kb. The data did not indicate any single “hot spot” of recombination in the msp1 gene, and disequilibrium was not particularly lower between sites separated by a highly polymorphic repeat sequence (within block 2 of the gene). Other studies have indicated that there is a “cold spot” for recombination in a 3.7-kb region of the gene, between blocks 6 and 16 in the scheme of Tanabe et al. (17), in which there are two highly divergent dimorphic sequences and recombination appears to have occurred between haplotypes of the same, but not different, dimorphic types (18, 19). In the present study, one of the block 6–16 dimorphic types was almost at fixation in most populations (MAD-like allele frequency >0.90), and recombination across this region of the gene was not apparently restricted (only 2/52 tests for linkage disequilibrium were significant over distances of >1.0 kb, most of which spanned this block 6–16 sequence).
A rapid decline in linkage disequilibrium with molecular map distance appears to be typical of P. falciparum genes in most African populations. From a population in Mali, a sample of 242 haplotypes of the trap (thrombospondin-related adhesive protein) gene (located on chromosome 13; ref. 1) was typed at several sequence sites, of which five were highly informative (with common allele frequencies of <0.9; four restriction fragment length polymorphism sites and one polymorphic repeat) covering approximately 1.0 kb (33). With these data (full tabulation of haplotypes kindly provided by K. J. H. Robson, Univ. of Oxford), analysis of linkage disequilibrium in 10 pairwise tests among these five sites shows declining linkage disequilibrium with map distance (r = −0.64; map distance = <0.2 kb, 3/3 tests significant; map distance = >0.2 kb, 2/7 tests significant). In contrast, data for the msp1 gene in South American and southeast Asian populations suggest that linkage disequilibrium can extend over molecular map distances of several kilobases (19, 34, 35). This is consistent with a lower effective recombination rate caused by a lower transmission rate and, thus, higher inbreeding of P. falciparum in these regions, compared with Africa. It is not surprising that an analysis of linkage disequilibrium over a distance of <1.0 kb, by using a very small sample size of mainly Thai sequences, yielded uninformative results (15), because a distance effect would be unlikely to be statistically apparent in such an analysis (16, 36).
The high recombination rate has important implications for strategies to identify polymorphic loci under natural selection, including loci encoding targets of chemotherapy and acquired immunity. Different sites within genes segregate as independent meiotic units; therefore, selection on a particular sequence site can generate a discordant pattern of allele frequency distribution at that site compared with other sites within the gene (37, 38). Fine mapping of sites under selection within candidate genes of P. falciparum should be possible with molecular population genetic analyses of multiple sites within the genes (e.g., by analysis of FST indices for each site separately) (39). Molecular evolutionary analyses, which consider each gene or part of a gene as an independent unit for study, are also appropriate for studies of selection on P. falciparum genes (40, 41).
On the other hand, a chromosome-wide or genome-wide search for a locus by using a molecular population genetic approach will require a very dense set of molecular markers, to give a reasonable likelihood that allele frequencies at one of these markers will be “hitch-hiked” by close linkage with the locus under selection (42). Many polymorphic simple sequence repeat microsatellite loci have been identified and developed for use in P. falciparum (43). Currently, >700 of these loci are available (X.-Z. Su & T. E. Wellems, National Institutes of Health, personal communication), equivalent to one on average every ≈40 kb of the genome (i.e., every ≈2–3 centimorgan). Although this density is adequate for segregational mapping of loci by using meiotic progeny of an experimental cross, it would not be sufficient for mapping of loci by using a recently advocated population genetic approach (42). Single nucleotide polymorphisms could potentially give a denser map, e.g., one on average every 1 kb (i.e., every ≈0.05 centimorgan) would involve 30,000 single nucleotide polymorphisms, which could be genotyped by using oligonucleotides on a silicon chip for high throughput hybridization analysis. The single nucleotide polymorphisms would first have to be identified by sequencing the genome of a second P. falciparum clone, after the genome sequence of the 3D7 clone is completed (44, 45).
Epistatic selection operating on combinations of alleles at two or more loci could potentially be detected by the presence of stronger linkage disequilibrium than that expected between the loci on the basis of their map distance alone. One mathematical model suggests that immune selection on multiple polymorphic antigen loci may allow survival only of particular multilocus genotypes that have nonoverlapping sets of alleles (46), and this could theoretically generate complete linkage disequilibrium between these loci. The data presented here suggest that, if complete linkage disequilibrium was actually observed between two candidate loci situated more than several kilobases apart, within highly endemic African populations, this would indeed be suggestive of epistatic selection. Epistatic selection may be operating in a small region of chromosome 7, to which a major determinant of chloroquine resistance has been mapped (6). In a sequence extending over a distance of >10 kb, incorporating the cg1 and cg2 genes, most chloroquine-resistant parasites in Africa and Asia have an identical haplotype. Chloroquine-sensitive parasites contain many different sequence combinations with no apparent disequilibrium over this region (6). Further studies of haplotype frequencies over the cg1–cg2 gene region could reveal whether linkage disequilibrium has been generated by hitch-hiking of neutral sites flanking a single site under selection (in which case the extended haplotype associated with resistance is likely to break down) or whether epistasis involving more than one sequence site in this region is maintaining the extended haplotype.
Given the high rate of meiotic recombination in natural populations of P. falciparum, well designed molecular population genetic studies will have discriminatory power to identify particular sequence sites under natural selection. In the genomic and postgenomic eras of research on P. falciparum, such studies should informatively interact with other approaches to identifying important loci, including analyses of experimental genetic crosses (6) and functional analysis of alleles (47) and their products (48) in vitro.
Acknowledgments
We thank the following for their help and cooperation with sample collection: Dr. Olumide Ogundahunsi and the field staff of the Postgraduate Institute of Medical Research and Training, University of Ibadan, Nigeria; Drs. Thor Theander and Ibrahim Elhassan and the villagers of Daraweesh, Sudan; Jane Trigg, her team, and the villagers in the Pangani Falls Redevelopment area, Tanga Region, Tanzania; field and laboratory staff at Medical Research Council Laboratories, Basse, The Gambia; staff and patients at the Albert Schweitzer Hospital, Lambarene, Gabon; Janet Freese, Andrew Robinson, Tollie Rossouw, and colleagues at the Medical Research Council Laboratories in Durban, South Africa. We are grateful to many colleagues at the London School of Hygiene and Tropical Medicine, in particular Orin Courtenay and Clive Davies, for helpful advice and support in the use of glim statistical software, and David Warhurst for providing laboratory facilities in the early phase of the work. We thank Professor Kazuyuki Tanabe for helpful discussion and comments concerning the manuscript. This research was supported by the Wellcome Trust (Project Grant 047191/Z).
ABBREVIATION
- msp1
merozoite surface protein 1 gene
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Triglia T, Wellems T E, Kemp D J. Parasitol Today. 1992;8:225–229. doi: 10.1016/0169-4758(92)90118-l. [DOI] [PubMed] [Google Scholar]
- 2.Walliker D, Quakyi I A, Wellems T E, McCutchan T F, Szarfman A, London W T, Corcoran L M, Burkot T R, Carter R. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 3.Wellems T E, Walliker D, Smith C L, do Rosario V E, Maloy W L, Howard R J, Carter R, McCutchan T F. Cell. 1987;49:633–642. doi: 10.1016/0092-8674(87)90539-3. [DOI] [PubMed] [Google Scholar]
- 4.Walker-Jonah A, Dolan S A, Gwadz R W, Panton L J, Wellems T E. Mol Biochem Parasitol. 1992;51:313–320. doi: 10.1016/0166-6851(92)90081-t. [DOI] [PubMed] [Google Scholar]
- 5.Guinet F, Wellems T E. Mol Biochem Parasitol. 1997;90:343–346. doi: 10.1016/s0166-6851(97)00144-8. [DOI] [PubMed] [Google Scholar]
- 6.Su X-Z, Kirkman L A, Fujioka H, Wellems T E. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- 7.Dye C, Williams B G. Proc R Soc London Ser B. 1997;264:61–67. doi: 10.1098/rspb.1997.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babiker H A, Ranford-Cartwright L C, Currie D, Charlwood J D, Billingsley P, Teuscher T, Walliker D. Parasitology. 1994;109:413–421. doi: 10.1017/s0031182000080665. [DOI] [PubMed] [Google Scholar]
- 9.Paul R E L, Packer M J, Walmsley M, Lagog M, Ranford-Cartwright L C, Paru R, Day K P. Science. 1995;269:1709–1711. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- 10.Carter R, McGregor I A. Trans R Soc Trop Med Hyg. 1973;67:830–837. doi: 10.1016/0035-9203(73)90011-4. [DOI] [PubMed] [Google Scholar]
- 11.Carter R, Voller A. Trans R Soc Trop Med Hyg. 1975;69:371–376. doi: 10.1016/0035-9203(75)90191-1. [DOI] [PubMed] [Google Scholar]
- 12.Conway D J, McBride J S. Parasitology. 1991;103:7–16. doi: 10.1017/s0031182000059229. [DOI] [PubMed] [Google Scholar]
- 13.Tibayrenc M, Kjellberg F, Ayala F J. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rich S M, Hudson R R, Ayala F J. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown A H D. Theor Popul Biol. 1975;8:184–201. doi: 10.1016/0040-5809(75)90031-3. [DOI] [PubMed] [Google Scholar]
- 17.Tanabe K, Mackay M, Goman M, Scaife J G. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 18.Tanabe K, Murakami K, Doi S. Exp Parasitol. 1989;68:470–473. doi: 10.1016/0014-4894(89)90132-x. [DOI] [PubMed] [Google Scholar]
- 19.Conway D J, Rosario V, Oduola A M J, Salako L A, Greenwood B M, McBride J S. Exp Parasitol. 1991;73:469–480. doi: 10.1016/0014-4894(91)90071-4. [DOI] [PubMed] [Google Scholar]
- 20.Kerr P J, Ranford-Cartwright L, Walliker D. Mol Biochem Parasitol. 1994;66:241–248. doi: 10.1016/0166-6851(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 21.Miller L H, Roberts T, Shahabuddin M, McCutchan T F. Mol Biochem Parasitol. 1993;59:1–14. doi: 10.1016/0166-6851(93)90002-f. [DOI] [PubMed] [Google Scholar]
- 22.Lewontin R C. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill W G, Robertson A. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 24.Weir B S, Hill W G. Am J Hum Genet. 1986;38:776–778. [PMC free article] [PubMed] [Google Scholar]
- 25.Roper C, Elhassan I M, Hviid L, Giha H, Richardson W, Babiker H, Satti G M H, Theander T G, Arnot D E. Am J Trop Med Hyg. 1996;54:325–331. doi: 10.4269/ajtmh.1996.54.325. [DOI] [PubMed] [Google Scholar]
- 26.Hill W G, Babiker H A, Ranford-Cartwright L C, Walliker D. Genet Res. 1995;65:53–61. doi: 10.1017/s0016672300033000. [DOI] [PubMed] [Google Scholar]
- 27.Arnot D E. Trans R Soc Trop Med Hyg. 1998;92:580–585. doi: 10.1016/s0035-9203(98)90773-8. [DOI] [PubMed] [Google Scholar]
- 28.Chakravarti A, Buetow K H, Antonarakis S E, Waber P G, Boehm C D, Kazazian H H. Am J Hum Genet. 1984;36:1239–1258. [PMC free article] [PubMed] [Google Scholar]
- 29.Miyashita N, Langley C H. Genetics. 1988;120:199–212. doi: 10.1093/genetics/120.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguade M, Miyashita N, Langley C H. Genetics. 1989;122:607–615. doi: 10.1093/genetics/122.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewin B. Genes VI. Oxford, U.K.: Oxford Univ. Press; 1997. [Google Scholar]
- 32.Nachman M W, Churchill G A. Genetics. 1996;142:537–548. doi: 10.1093/genetics/142.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robson K J H, Dolo A, Hackford I R, Doumbo O, Richards M B, Keita M M, Sidibe T, Bosman A, Modiano D, Crisanti A. Am J Trop Med Hyg. 1998;58:81–89. doi: 10.4269/ajtmh.1998.58.81. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira M U, Liu Q, Zhou M, Kimura M, Kaneko O, Van Thien H, Isomura S, Tanabe K, Kawamoto F. J Eukaryotic Microbiol. 1998;45:131–136. doi: 10.1111/j.1550-7408.1998.tb05080.x. [DOI] [PubMed] [Google Scholar]
- 35.Sakihama, N., Kimura, M., Hirayama, K., Kanda, T., Na-Bangchang, K., Jongwutiwes, S., Conway, D. & Tanabe, K. (1999) Gene, in press. [DOI] [PubMed]
- 36.Schaeffer S W, Miller E L. Genetics. 1993;135:541–552. doi: 10.1093/genetics/135.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berry A, Kreitman M. Genetics. 1993;134:869–893. doi: 10.1093/genetics/134.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald J H. In: Non-Neutral Evolution: Theories and Molecular Data. Golding B, editor. New York: Chapman & Hall; 1994. pp. 88–100. [Google Scholar]
- 39.Conway D J. Parasitol Today. 1997;13:26–29. doi: 10.1016/s0169-4758(96)10077-6. [DOI] [PubMed] [Google Scholar]
- 40.Hughes M K, Hughes A L. Mol Biochem Parasitol. 1995;71:99–113. doi: 10.1016/0166-6851(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 41.Escalante A A, Lal A A, Ayala F J. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hastings I M, Wedgewood-Oppenheim B. Parasitol Today. 1997;13:375–383. doi: 10.1016/s0169-4758(97)01110-1. [DOI] [PubMed] [Google Scholar]
- 43.Su X-Z, Wellems T E. Genomics. 1996;33:430–444. doi: 10.1006/geno.1996.0218. [DOI] [PubMed] [Google Scholar]
- 44.Dame J B, Arnot D E, Bourke P F, Chakrabarti D, Christodoulou Z, Coppel R L, Cowman A F, Craig A G, Fischer K, Foster J, et al. Mol Biochem Parasitol. 1996;79:1–12. doi: 10.1016/0166-6851(96)02641-2. [DOI] [PubMed] [Google Scholar]
- 45.Gardner M J, Tettelin H, Carucci D J, Cummings L M, Aravind L, Koonin E V, Shallom S, Mason T, Yu K, Fujii C, et al. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 46.Gupta S, Maiden M C J, Feavers I M, Nee S, May R M, Anderson R M. Nat Med. 1996;2:437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- 47.Triglia T, Wang P, Sims P F G, Hyde J E, Cowman A F. EMBO J. 1998;17:3807–3815. doi: 10.1093/emboj/17.14.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triglia T, Menting J G, Wilson C, Cowman A F. Proc Natl Acad Sci USA. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]