Abstract
Catalyzed reporter deposition fluorescence in situ hybridization combined with microautoradiography (MICRO-CARD-FISH) is increasingly being used to obtain qualitative information on substrate uptake by individual members of specific prokaryotic communities. Here we evaluated the potential for using this approach quantitatively by relating the measured silver grain area around cells taking up 3H-labeled leucine to bulk leucine uptake measurements. The increase in the silver grain area over time around leucine-assimilating cells of coastal bacterial assemblages was linear during 4 to 6 h of incubation. By establishing standardized conditions for specific activity levels and concomitantly performing uptake measurements with the bulk community, MICRO-CARD-FISH can be used quantitatively to determine uptake rates on a single-cell level. Therefore, this approach allows comparisons of single-cell activities for bacterial communities obtained from different sites or growing under different ecological conditions.
In situ hybridization with fluorescently labeled oligonucleotide probes (FISH) allows enumeration of specific prokaryotic groups or individual phylotypes with a microscope. Recently, this technique was modified to enhance the detection efficiency, and the new method was called catalyzed reporter deposition FISH (CARD-FISH) (25). CARD-FISH uses oligonucleotide probes labeled with horseradish peroxidase, which results in a sensitivity comparable to that obtained with FISH with polynucleotide probes (26). The CARD-FISH protocol includes a permeabilization step for bacterial cells using lysozyme (25) or for Archaea using proteinase K (28). This CARD-FISH protocol combined with microautoradiography (MICRO-CARD-FISH) was used to determine the number of bacterial and archaeal cells taking up the enantiomeric amino acid aspartic acid in waters of the North Atlantic down to a depth of 4,000 m (29).
Microautoradiography combined with FISH is increasingly being used to obtain qualitative information on the substrate utilization by specific prokaryotic populations in complex natural communities (6, 24). Cottrell and Kirchman (6) demonstrated that the method commonly used to assess the prokaryotic activity of entire communities (leucine and thymidine incorporation) indeed covers the entire prokaryotic community. Using MICRO-FISH, it has been shown that some groups in a bacterial community are more active than expected based on their relative abundance (7, 11, 17-19). Also, it has been shown that Cytophaga utilizes high-molecular-weight material more efficiently (7), while members of the SAR11 cluster and Roseobacter sp. apparently also efficiently utilize dimethylsulfoniopropionate (18, 19).
Generally, however, our knowledge about the phylogenetic composition of prokaryotic communities is increasing much faster than our understanding of the function of these phylotypes in their natural environments. Thus, methods that more closely link phylogenetic information with functional aspects are required, and one of these methods is MICRO-CARD-FISH. Thus far, this method has been used mainly in a qualitative way; however, it also has the potential to quantify substrate uptake under specific conditions (23). The size of the halo area around cells generated by autoradiography indicating uptake of a specific radiolabeled substrate depends on several factors in the processing of the samples (6). If conditions during preparation of samples are kept constant, however, the halo area should be proportional to the uptake rate, at least for a certain period of time.
In this study, we evaluated the potential of MICRO-CARD-FISH for obtaining not only qualitative information but also quantitative information concerning substrate uptake by specific prokaryotic populations. We also addressed the largely unsolved problem of the intra- and interspecific variability of substrate uptake in natural bacterioplankton populations. The range of activity levels within a given bacterial population might be wide or might be narrow, with similar levels of activity in all the active cells. As the activity level likely influences a number of key ecological features, such as viral infection (13, 14) and flagellate grazing (9), this kind of information about specific bacterioplankton populations is essential for a more in-depth understanding of the ecology of specific phylotypes. Thus, to obtain metabolic rate estimates on a single-cell level, we quantified the halo area obtained by MICRO-CARD-FISH using image analysis. The total halo area obtained was then related to concurrently obtained bulk activity measurements.
MATERIALS AND METHODS
Laboratory experiments with natural bacterial assemblages to relate bulk activity and halo area.
Experiments were conducted with water samples collected at the Royal Netherlands Institute for Sea Research jetty at the southern entrance of the North Sea into the Dutch Wadden Sea during high tide so that incoming North Sea water was collected. Two different sets of experiments were conducted. For the first set of experiments, seawater was filtered through 0.8-μm or 3-μm polycarbonate filters, and 200 ml of each filtered seawater sample was supplemented with [3H]leucine (final concentration, 20 nM; specific activity, 160 Ci mmol−1; Amersham) and incubated at the in situ temperature for up to 12 h. Duplicate subsamples (2 to 4 ml for bulk bacterial production and 2.5 to 5 ml for MICRO-CARD-FISH) were taken at regular intervals over a 12-h period. Immediately after the tracer was added to a 200-ml seawater batch culture, subsamples were taken, fixed immediately with formaldehyde (final concentration, 2%), and used as killed controls for leucine incorporation by the bulk bacterial community and for MICRO-CARD-FISH. Bacterial abundance was determined at the beginning and at the end of the experiments by epifluorescence microscopy following 4′,6′-diamidino-2-phenylindole (DAPI) staining (final concentration, 2 μg ml−1). The experiment was carried out twice in July 2004 and once in February 2005.
For the other set of experiments, a dilution culture was established at the beginning of the spring phytoplankton bloom in April 2005. Seawater filtered through 0.2-μm polycarbonate filters (Millipore) was combined with seawater filtered through 0.8-μm filters at a ratio 9:1 and incubated at the in situ temperature. Bacterial abundance and leucine incorporation into the bulk bacterial community were monitored at roughly 12-h intervals for 5 days, and MICRO-CARD-FISH samples were taken. Samples used for determination of bacterial abundance were fixed with formaldehyde (final concentration, 2%), and abundance was measured by flow cytometry (BD FACScalibur) after SYBR green I staining (22). To determine leucine incorporation into the bacterial community, 5-ml subsamples were taken in duplicate along with one formaldehyde-killed control, and 2- to 10-ml subsamples for MICRO-CARD-FISH were supplemented with [3H]leucine (final concentration, 20 nM), incubated at the in situ temperature for 3 h, and then fixed with formaldehyde.
Relating bulk leucine incorporation to the halo area obtained by MICRO-CARD-FISH for bacteria inhabiting the sea surface microlayer.
Samples from the sea surface microlayer (SML) (16) and the underlying water (ULW) (30 cm below the surface) were collected at five stations along a trophic gradient from the Mauritanian upwelling to the North Atlantic subtropical gyre during the BADE-2 cruise of R/V Pelagia in September 2004. The sea surface microlayer was collected with glass plate samplers as described by Agogue et al. (1). Basic physicochemical parameters were determined together with bacterial abundance, bacterial production, and MICRO-CARD-FISH. Bacterial abundance was measured by flow cytometry as described above. For analysis of bacterial production, duplicate 5-ml samples were used along with one formaldehyde-killed control, and for MICRO-CARD-FISH 4-ml samples were supplemented with [3H]leucine (final concentration, 20 nM), incubated at the in situ temperature for 3 h, and then fixed with formaldehyde.
MICRO-CARD-FISH.
After 18 h of fixation with formaldehyde (final concentration, 2%), samples were filtered onto 0.2-μm white polycarbonate filters (Millipore) using a 0.45-μm cellulose nitrate supporting filter (HAWP; Millipore). The method used for processing the filters has been described in detail elsewhere (25, 28). Briefly, the bacterial cell walls were permeabilized by incubating the filters with a lysozyme solution (10 mg ml−1; pH 8) at 37°C for 1 h. The filters were cut into sections for hybridization with the following different oligonucleotide probes: Eub338-I (3), Eub338-II, and Eub338-III (8) to target Bacteria, Non338 (4), CF319a for the Cytophaga-Flavobacter group (21), and SAR86-1249 targeting the SAR86 cluster (10). The horseradish peroxidase-labeled probe was added at a final concentration of 2.5 ng/μl, and hybridization was performed at 35°C for 12 to 15 h. After the washing steps, amplification was carried out by adding H2O2 and tyramide-Alexa488 and incubating the preparation at 37°C for 30 min (28).
Microautoradiography was performed with the previously hybridized filter sections by transferring them onto slides coated with photographic emulsion (Kodak NTB-2). Subsequently, the slides were placed in a light-tight box with silica gel as a drying agent and exposed at 4°C for 24 h, after which the amount of active cells, as indicated by the attached silver grains, did not increase (28). After this, the slides were developed and fixed using the specifications of the manufacturer. Cells were counterstained with a DAPI mixture (5.5 parts Citifluor, 1 part Vectashield, and 0.5 part phosphate-buffered saline with DAPI at a final concentration of 1 μg ml−1).
The slides were examined with a Zeiss Axioplan 2 microscope equipped with a 100-W Hg lamp and appropriate filter sets for DAPI and Alexa488. The presence of silver grains surrounding cells was determined by using the transmission mode of the microscope (5). In the killed controls, less than 0.5% of the total DAPI-stained cells were associated with two or more silver grains. More than 800 DAPI-stained cells were counted per sample.
Analysis of the image of the silver grain area surrounding active cells.
For each sample, three images of the bacteria were acquired, including one image for bacteria stained with DAPI and one image for the specific stain Alexa488, both in epifluorescence mode. An image of the silver grain areas was acquired by switching to the transmission mode of the microscope. The images were acquired with a digital camera (AxioCam MRc5) mounted on the microscope, and the three images were overlaid to obtain a composite image. Pictures were taken for 10 to 40 fields per FISH probe used. Images of at least 400 to 800 DAPI-stained cells and 20 to 200 probe-positive cells (cells with overlapping signals in the DAPI and Alexa488 images) per filter section were recorded. Overlapping signals in the DAPI images and the transmitted light images (silver grains) indicated cells that had assimilated the radioactive leucine. The analysis of the pictures was conducted with the KS300 3.0 software (Carl Zeiss). This program allowed us to record the number of DAPI cells, the number of probe-positive cells, the area of the DAPI-stained cells, and the silver grain area.
The average silver grain area for the assimilating cells was calculated from the data for all DAPI-stained cells with associated silver grains. Also, the total silver grain area for the whole community was calculated and expressed as the silver grain area per liter. The total silver grain area was then related to the bulk leucine incorporation of the bacterial community.
Additionally, the equivalent spherical diameter (ESD) was calculated for each cell from the area of the DAPI-stained cell.
Bulk leucine incorporation.
The bulk bacterial production was measured by determining the incorporation of [3H]leucine (final concentration, 20 nM; specific activity, 160 Ci mmol−1; Amersham) into bacterial cells. Fixed samples were filtered onto 0.2-μm polycarbonate filters and rinsed three times with 10 ml of 5% ice-cold trichloroacetic acid. After this, the filters were transferred into scintillation vials and dried at room temperature. Subsequently, 8 ml of scintillation cocktail (FilterCount; Canberra Packard) was added to the vials and counted with a liquid scintillation counter (model 1212; LKB Wallac) after 18 h. The dpm values obtained were converted to leucine incorporation rates.
RESULTS AND DISCUSSION
Relationship between the silver grain area and bulk leucine incorporation.
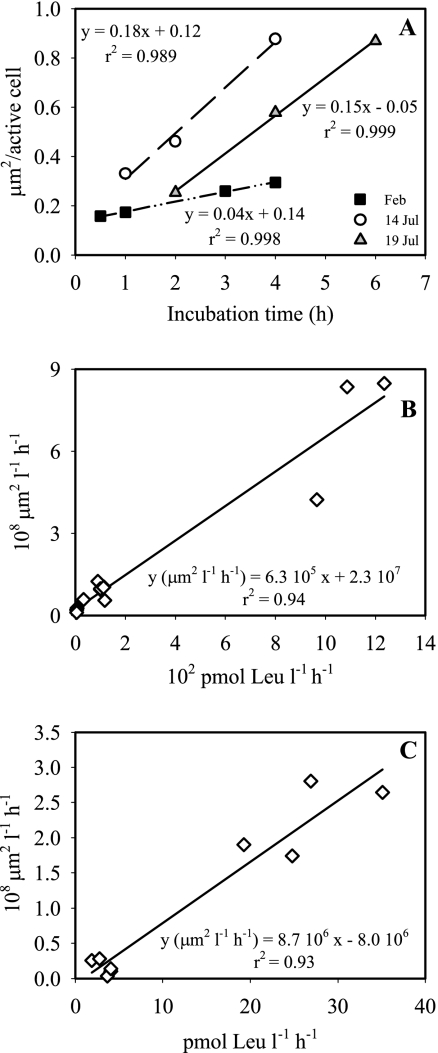
The average silver grain area per active cell increased linearly with incubation time for the initial 4 to 6 h both in the summer, when leucine incorporation was high, and in the winter, when bulk activity was low (Fig. 1A). Thus, for further experiments and for freshly collected samples, a 3-h incubation period was used to ensure linearity of the increase in the silver grain area.
FIG. 1.
(A) Increase in the average silver grain area per active cell with incubation time, determined for bacterioplankton of the coastal North Sea in winter and summer. (B and C) Relationship between the total average silver grain area and bulk leucine incorporation by bacterioplankton of the coastal North Sea (B) and the subtropical North Atlantic (C).
The average silver grain area of the cells in a bacterial community taking up leucine was linearly correlated with the bulk leucine incorporation rate of the same community, as exemplified by the bacterial communities collected from the coastal North Sea (Fig. 1B) and from the open North Atlantic Ocean (Fig. 1C). Plotting the data on a log scale also revealed highly significant relationships, indicating that the relationships obtained were not simply caused by some high values.
These results indicate that keeping the incubation time (this study), fixation time (23), and exposure time (6) for MICRO-CARD-FISH constant allows estimation of single-cell uptake rates by relating the silver grain area to bulk substrate uptake. The appropriate incubation time and exposure time have to be checked before analysis for each system and each substrate used (2, 11). For reliability of the single-cell estimates it is essential to determine the bulk activity of the water sample at the same time under identical conditions (Fig. 1B and C) and to use the same fixation protocol (23).
For routine use with specific samples and conditions, the linear relationship between the bulk leucine uptake and the silver grain area needs to be established. Subsequently, for repeated measurements for the same site or in a specific experiment, the silver grain area of each cell can be converted to cell-specific uptake rates by using a factor calculated by dividing the measured bulk uptake for a specific sample by the silver grain area obtained for the same bacterial community.
Applicability of quantitative MICRO-CARD-FISH to natural bacterial communities: intra- and interspecific variability of leucine uptake.
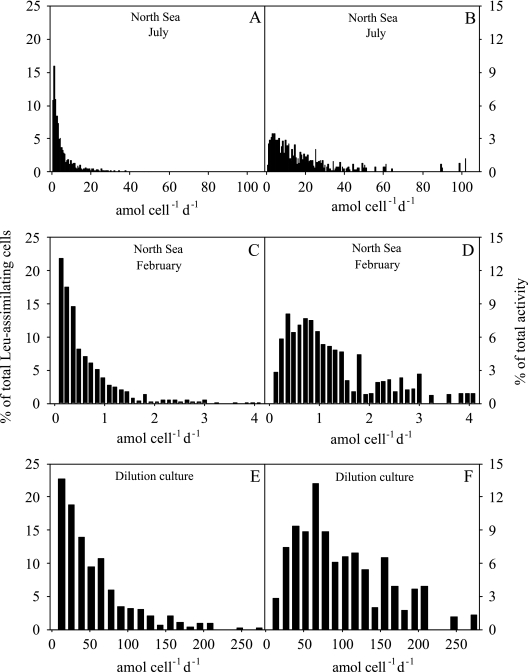
The distribution of leucine uptake on a single-cell level was determined for natural bacterial assemblages collected in the coastal North Sea during the summer and winter by relating the bulk leucine uptake to the silver grain area. About 70% of all the leucine-assimilating cells took up leucine at rates that were less than 5.4 amol cell−1 day−1 in the summer (Fig. 2A), accounting for only 25% of the total activity (Fig. 2B). About 30% of the cells took up leucine at rates between 5.4 and 30 amol cell−1 day−1, accounting for about 60% of the total leucine uptake by the community, and 2% of the cells exhibited uptake rates greater than 30 amol cell−1 day−1, accounting for 16% of the uptake by the community (Fig. 2A and B). In the winter, the cell-specific leucine uptake rates were generally about 1 order of magnitude lower than those in the summer; however, the general distribution pattern for single-cell activity was similar, with the majority of the cells exhibiting activity levels at the lower end of the spectrum (Fig. 2C). About 70% of the Leu-positive cells took up leucine at rates that were less than 0.6 amol cell−1 day−1, accounting for 30% of the total activity of the bacterial community (Fig. 2D).
FIG. 2.
Distribution of cell-specific leucine uptake by bacterioplankton of the coastal North Sea expressed as percentages of the abundance of leucine-assimilating DAPI-stained cells (left panels) and as percentages of the total leucine uptake (right panels) in July (A and B) and February (C and D) and in a dilution culture during exponential growth (E and F) (see Fig. 3). The activity classes are those used by Freedman and Diaconis (12).
During exponential growth of bacteria in a seawater dilution culture (Fig. 2E and F), cells with leucine uptake rates higher than about 78 amol cell−1 day−1 accounted for about 60% of the bulk leucine uptake by the community; however, these cells were only 24% of the community (Fig. 2E).
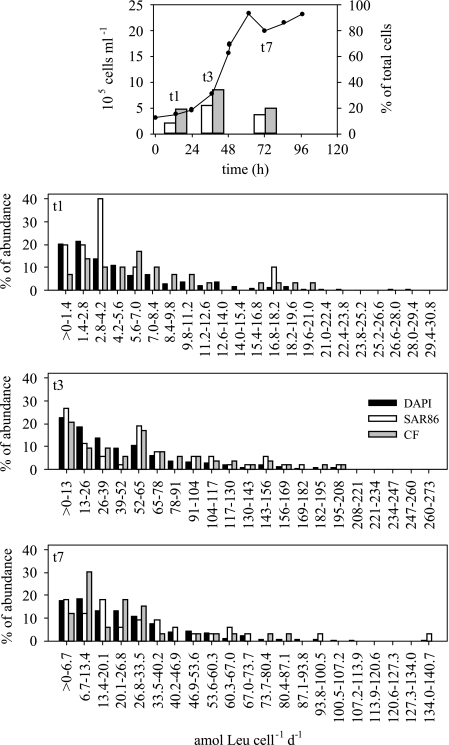
The distribution of single-cell activities for the Cytophaga-Flavobacter group and the SAR86 cluster during the initial, mid-exponential, and stationary growth phases in a dilution culture is shown in Fig. 3, together with the distribution for the DAPI-stained cells. The SAR86 cluster accounted for between 8 and 22% of the bacterial community, and the Cytophaga-Flavobacter group accounted for between 19 and 34% of the bacterial community (Fig. 3, upper panel). Both groups contributed most to the total bacterial abundance during the exponential phase. During the initial growth phase in the seawater dilution culture, the majority (70%) of DAPI-stained cells actively taking up leucine exhibited uptake rates that were less than 5.6 amol cell−1 day−1. Forty percent of the cells affiliated with the SAR86 cluster took up leucine at rates between 2.8 and 4.2 amol cell−1 day−1. About 60% of the Cytophaga-Flavobacter cells took up leucine at rates between 2.8 and 8.4 amol cell−1 day−1 (Fig. 3). In the mid-exponential growth phase (Fig. 3), the cell-specific leucine uptake rate was generally about 1 order of magnitude higher than that in the early exponential phase. Despite this, the distribution pattern for DAPI-stained cells taking up leucine was similar to that in the early exponential phase. About 75% of the cells exhibited leucine uptake rates that were less than 65 amol cell−1 day−1. Leucine-assimilating cells of members of the SAR86 and Cytophaga-Flavobacter clusters showed an uptake distribution pattern different from that of DAPI-stained cells (Fig. 3), with large contributions at the lower end of the cell-specific uptake rates (<13 amol cell−1 day−1) and at intermediate rates (52 to 65 amol cell−1 day−1). During the stationary phase (Fig. 3), the distribution of the single-cell activity for the whole community (DAPI-stained cells) showed that there were higher percentages of cells at the low end of the uptake spectrum and that the percentages gradually decreased toward the high end of the uptake spectrum. SAR86 cells with leucine uptake rates of <20.1 amol cell−1 day−1 comprised more than 50% of the leucine-assimilating SAR86 cells. Cytophaga-Flavobacter leucine-assimilating cells showed two peaks of activity, one at uptake rates between 6.7 and 13.4 amol cell−1 day−1 and the other at uptake rates between 20.1 and 26.8 amol cell−1 day−1. Generally, the cell-specific activity in the stationary phase was about one-half of that detected during the mid-exponential phase (Fig. 3).
FIG. 3.
Development of bacterial abundance in a dilution culture with water from the coastal North Sea and relative contributions of the SAR86 and Cytophaga-Flavobacter (CF) clusters during the different growth phases of the culture (upper panel) and distribution of cell-specific leucine uptake during the early exponential (t1), mid-exponential (t3), and stationary (t7) growth phases expressed as percentages of leucine-positive cells stained by DAPI (solid bars), of SAR86 cells (open bars), and of Cytophaga-Flavobacter cells (gray bars) (lower panels). The activity classes are those used by Freedman and Diaconis (12).
The average cell-specific uptake rates determined for the different experiments, as calculated from the silver grain area, varied from 0.53 amol cell−1 day−1 (during the winter) to 47.4 amol cell−1 day−1 (during exponential growth in the spring). This range is slightly greater than the range of cell-specific uptake rates (0.15 to 26.15 amol cell−1 day−1) obtained by Reinthaler et al. (27) for the open North Sea (April to December). However, Reinthaler et al. (27) calculated the cell-specific uptake rates for the total prokaryotic population and not just for the active cells, as was done in this study. The cell-specific leucine uptake rates obtained in the winter are in the lower range of the rates obtained by Lebaron et al. (15) for different size classes of flow cytometry-sorted prokaryotic communities in the coastal Mediterranean Sea in October (1.5 to 12 amol cell−1 day−1).
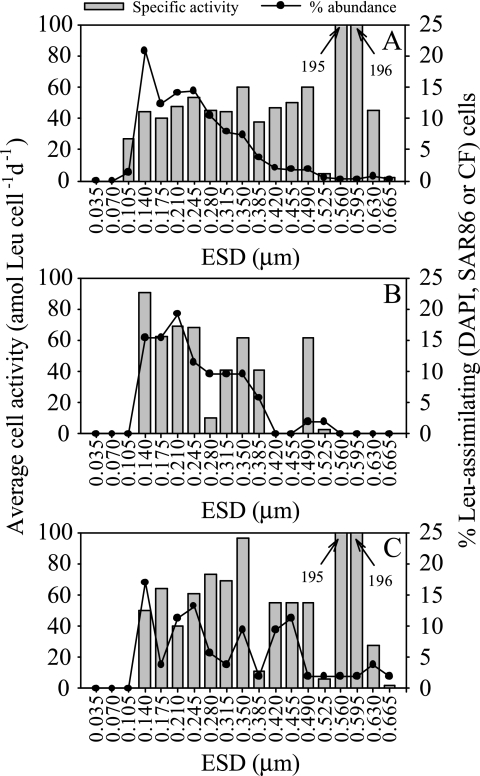
Cell-specific assimilation rates for different cell size classes can also be obtained with this method. For example, the distribution of cell-specific leucine uptake during the exponential growth phase in April is shown in Fig. 4 for the DAPI-stained SAR86 and Cytophaga-Flavobacter leucine-positive cells. No clear relationship was found between the cell size and the average specific uptake rate during the exponential and stationary phases. Only during the early exponential phase was a tendency for increasing cell activity with increasing cell size apparent for the DAPI-stained cells (data not shown), as reported in other studies (15). The smaller cells dominated the leucine-positive prokaryotic community (Fig. 4A), with the number of leucine-assimilating cells decreasing steadily with cell size, while the cell-specific uptake rates increased slightly up to an ESD of 0.245 μm and exhibited distinct peaks for high-ESD classes. The level of SAR86 cells peaked for cell sizes slightly larger than the sizes of the total DAPI-stained bacteria, and SAR86 cells had two main peaks for cell-specific uptake (Fig. 4B). The Cytophaga-Flavobacter cluster seemed to be very heterogeneous, with up to four peaks for abundance classes and cell-specific uptake rates (Fig. 4C).
FIG. 4.
Distribution of the abundance of cells taking up leucine and cell-specific uptake rates for DAPI-stained (A), SAR86 (B), and Cytophaga-Flavobacter (C) cells in different size classes during the exponential growth phase in seawater cultures. The size classes were determined by using the equivalent spherical diameter, as described by Freedman and Diaconis (12).
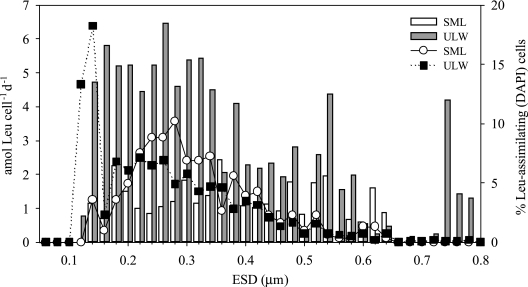
Another example of the ecological information that can be obtained with this method is that different bacterial consortia can be compared, both at a community level and for specific groups or taxa, depending on the probes used. In Fig. 5, the average cell-specific leucine incorporation rate and the percentage of Leu-positive DAPI-stained cells are plotted versus the equivalent spherical diameter for the SML and the ULW collected at a station in the North Atlantic gyre. Generally, the cell-specific activity was significantly higher in the ULW than in the SML (P < 0.001, as determined by the Wilcoxon test), which might reflect the specific conditions in the SML, such as high levels of UV radiation. In the ULW, however, most of the Leu-positive cells were in the smaller size classes (0.10 to 0.14 μm). Several peaks of cell-specific activity were detected in both communities for different bacterial size classes; the data for SML bacteria showed higher cell-specific activity for ESD ranging from 0.16 to 0.18 μm, from 0.28 to 0.30 μm, from 0.34 to 0.36 μm, and from 0.44 to 0.48 μm, while the ULW bacteria exhibited activity peaks for ESD ranging from 0.14 to 0.16 μm and from 0.24 to 0.26 μm, as well as two other peaks toward the higher end of the range (Fig. 5).
FIG. 5.
Distribution of the average cell-specific Leu uptake for different size classes (bars) and percentages of the DAPI-stained leucine-assimilating cells (lines) for the SML and ULW collected in the subtropical North Atlantic gyre. The size classes were determined by using the equivalent spherical diameter, as described by Freedman and Diaconis (12).
Only recently, quantitative MICRO-CARD-FISH has been developed for filamentous bacteria in activated sludge systems by using internal standards (23). Cottrell and Kirchman (6) and their collaborators (11, 17-20) used the silver grain area as an indicator of the cell activity of planktonic heterotrophic bacteria and Synechococcus. However, no attempts have been made yet to calibrate the silver grain area with bulk uptake rates determined for the total bacterial community. Here we demonstrated that, when standardized conditions for MICRO-CARD-FISH are maintained, the silver grain area around leucine-positive cells is directly related to the bulk bacterial activity. This indicates that the uptake rates of individual cells can be derived from quantifying the silver grain area around active cells of a population or community using image analysis and relating the total silver grain area of a specific sample to the conventionally measured bulk uptake rate. The information obtained with this approach has a wide range of potential applications and allows workers to answer questions related to the activity patterns of different populations within a specific community and to the distribution of activity levels within specific populations. Moreover, normalizing the silver grain area to actual uptake rates allows comparisons of single-cell activities for bacterial communities originating from different environments, which is not possible when only the silver grain area of the cells is taken into account.
Acknowledgments
We thank the captain and crew of the R/V Pelagia for their help during work at sea and Karen Stoderegger for her help with the image analysis.
This research was supported by a European Community Marie Curie Fellowship to E.S., by grants from the NWO-ALW (project NWO-ALW 812.03.001) to G.J.H., and by the Commission of the EU (BASICS Project).
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Agogue, H., E. O. Casamayor, F. Joux, I. Obernosterer, C. Dupuy, F. Lantoine, P. Catala, M. G. Weinbauer, T. Reinthaler, G. J. Herndl, and P. Lebaron. 2004. Comparison of samplers for the biological characterization of the sea surface microlayer. Limnol. Oceanogr. Methods 2:213-225. [Google Scholar]
- 2.Alonso, C., and J. Pernthaler. 2006. Concentration-dependent patterns of leucine incorporation by coastal picoplankton. Appl. Environ. Microbiol. 72:2141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carman, K. R. 1993. Microautoradiographic detection of microbial activity, p. 397. In B. F. S. P. F. Kemp, E. B. Sherr, and J. F. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 6.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 9.delGiorgio, P. A., J. M. Gasol, D. Vaque, P. Mura, S. Agusti, and C. M. Duarte. 1996. Bacterioplankton community structure: protists control net production and the proportion of active bacteria in a coastal marine community. Limnol. Oceanogr. 41:1169-1179. [Google Scholar]
- 10.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elifantz, H., R. R. Malmstrom, M. T. Cottrell, and D. L. Kirchman. 2005. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware Estuary. Appl. Environ. Microbiol. 71:7799-7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman, D., and P. Diaconis. 1981. On this histogram as a density estimator: L2 theory. Z. Wahrscheinlichkeitstheorie Verwandte Geb. 57:453-476. [Google Scholar]
- 13.Hwang, C. Y., and B. C. Cho. 2002. Uneven growth and different susceptibility to viruses among bacteria increase estimates of virus production in the East Sea based on TEM observation. Aquat. Microb. Ecol. 27:211-218. [Google Scholar]
- 14.Kadavy, D. R., J. J. Shaffer, S. E. Lott, T. A. Wolf, C. E. Bolton, W. H. Gallimore, E. L. Martin, K. W. Nickerson, and T. A. Kokjohn. 2000. Influence of infected cell growth state on bacteriophage reactivation levels. Appl. Environ. Microbiol. 66:5206-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebaron, P., P. Servais, A. C. Baudoux, M. Bourrain, C. Courties, and N. Parthuisot. 2002. Variations of bacterial specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat. Microb. Ecol. 28:131-140. [Google Scholar]
- 16.Liss, P. S., and R. A. Duce. 1997. The sea surface and global change. Cambridge University Press, Cambridge, United Kingdom.
- 17.Malmstrom, R. R., M. T. Cottrell, H. Elifantz, and D. L. Kirchman. 2005. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the northwest Atlantic Ocean. Appl. Environ. Microbiol. 71:2979-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malmstrom, R. R., R. P. Kiene, M. T. Cottrell, and D. L. Kirchman. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 70:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malmstrom, R. R., R. P. Kiene, and D. L. Kirchman. 2004. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 20.Malmstrom, R. R., R. P. Kiene, M. Vila, and D. L. Kirchman. 2005. Dimethylsulfoniopropionate (DMSP) assimilation by Synechococcus in the Gulf of Mexico and northwest Atlantic Ocean. Limnol. Oceanogr. 50:1924-1931. [Google Scholar]
- 21.Manz, W., R. I. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 22.Marie, D., F. Partensky, S. Jacquet, and D. Vaulot. 1997. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen, J. L., D. Christensen, M. Kloppenborg, and P. H. Nielsen. 2003. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ. Microbiol. 5:202-211. [DOI] [PubMed] [Google Scholar]
- 24.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic Archaea take up amino acids. Appl. Environ. Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinthaler, T., C. Winter, and G. J. Herndl. 2005. Relationship between bacterioplankton richness, respiration, and production in the southern North Sea. Appl. Environ. Microbiol. 71:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teira, E., T. Reinthaler, A. Pernthaler, J. Pernthaler, and G. J. Herndl. 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol. 70:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teira, E., H. van Aken, C. Veth, and G. J. Herndl. 2006. Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol. Oceanogr. 51:60-69. [Google Scholar]