Abstract
During the brewing of Japanese sake, Saccharomyces cerevisiae cells produce a high concentration of ethanol compared with other ethanol fermentation methods. We analyzed the gene expression profiles of yeast cells during sake brewing using DNA microarray analysis. This analysis revealed some characteristics of yeast gene expression during sake brewing and provided a scaffold for a molecular level understanding of the sake brewing process.
During the process of brewing Japanese sake, rice starch is saccharified by enzymes produced by koji (Aspergillus oryzae), and the resultant glucose is fermented to ethanol by sake yeast (Saccharomyces cerevisiae). This process allows a highly condensed mash to be made without accumulation of high levels of sugars, which inhibit yeast cell growth and ethanol fermentation. Thus, yeast cells in the sake mash can produce ethanol to a concentration of 20% (vol/vol), the highest concentration among alcoholic beverages that are made without distillation (12). During this process, yeast cells are simultaneously exposed to various stresses, including hypoxia, low temperature, low pH, high osmotic pressure, and an increasing concentration of ethanol (12). Global gene expression analysis of yeast cells during the sake brewing process would be helpful for understanding how yeast cells adapt to these multiple stresses. It would also be useful to determine which processes are compromised by stresses and thus should be manipulated to improve the fermentation performance of yeast cells in the sake mash. In the present study, we used DNA microarray analysis to analyze the global gene expression profile of yeast cells during the sake brewing process. The resulting data revealed several characteristic features of gene expression by sake yeast during the brewing process.
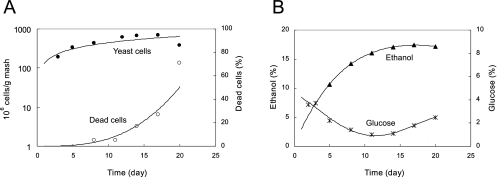
The sake yeast used in the present study was kyokai no. 701 (obtained from the Brewing Society of Japan). Yeast cells were cultured in YPD (1% yeast extract, 1% peptone, and 2% glucose) medium at 15°C for 4 days without shaking. The sake mash was made as shown in Fig. S1 in the supplemental material. Raw materials were added on three separate occasions (SOE, NAKA, and TOME in the Japanese language) at 15°C. The mash was incubated at 15°C for 20 days. Sampling was carried out nine times during the brewing process (S2, N1, T1, T2, T3, T5, T8, T11, and T14; day 2, day 3, day 4, day 5, day 6, day 8, day 11, day 14, and day 17 from the first addition, respectively; see Fig. S1 in the supplemental material). Analyses of the mash were performed after T2, because the mash was very viscous and contained only a small amount of liquid during the early stages of the brewing process (S2, N1, and T1). Various parameters were measured at each sampling. Ethanol concentration was measured with an ethanol analyzer (Riken Keiki, Shizuoka, Japan). Yeast cell number was taken as the number of CFU growing on YPD plates. The percentage of dead cells was measured with a microscope by using the methylene blue staining method (3). Glucose concentration was assayed by using a glucose CII test (Wako, Osaka, Japan). Representative data are shown in Fig. 1.
FIG. 1.
Representative fermentation profiles for the sake mash after the third addition. (A) Total number of yeast cells and percentage of dead yeast cells. (B) Ethanol and glucose concentrations.
For RNA preparation, an aliquot (approximately 200 ml) of mash was scooped up and filtered using stainless steel mesh (2-mm pore size) to remove rice and koji particles. Samples containing yeast cells were centrifuged at 4,620 × g at 15°C for 5 min. After the turbid supernatant was removed by decantation, yeast cells were recovered from the pellets and stored at −80°C. Total RNA was extracted from 3 to 5 g of yeast cells by the hot phenol method (13). An equal volume of 4 M lithium acetate was added to the crude RNA solution, and then the solution was incubated at 4°C overnight. After centrifugation at 4,620 × g at 4°C for 15 min, the pellet was dissolved in water, and ethanol precipitation was performed. The resultant pellet was dissolved in water, and the solution was centrifuged at 23,200 × g at 4°C for 10 min to remove insoluble materials. This sample was further purified using the RNeasy mini kit (QIAGEN, Valencia, CA) and was stored as the total RNA sample at −80°C. The quality of the total RNA samples was checked by using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA).
The GeneChip yeast genome S98 array (Affymetrix, Santa Clara, CA) was used as the microarray in this study. Total RNA was labeled using the GeneChip one-cycle target labeling and control reagents (Affymetrix). Labeling of total RNA, hybridization to the DNA microarray, and scanning with the GeneChip scanner 2500 (Affymetrix) were performed in accordance with the manufacturer's instructions. Microarray analysis of the samples from each time point was repeated three times using total RNA samples obtained from three independent sake brewing experiments, except for sample S2, for which there were two replicates, rather than three. Microarray data were analyzed using GeneSpring 7 software (Agilent Technologies). To compare the relative changes in gene expression levels, the signal intensity of gene A in sample X was divided by the median of every measurement taken for gene A throughout the experiment (per gene normalization). The averages of raw and normalized values for each gene at each sampling time are shown in Table S1 in the supplemental material. All standard S. cerevisiae open reading frames in the Saccharomyces Genome Database (http://www.yeastgenome.org/) were used for cluster analysis by using GeneSpring 7 software. K-mean cluster analysis was performed using the expression profiles. Standard correlation was used for similarity measures.
Before expression profile analysis, the correlation coefficients for each set of gene expression data (three replicates from the same sampling time) were calculated to evaluate the reproducibility of the experiments. A high correlation coefficient (greater than 0.95) was obtained for comparisons between all possible pairs of samples for each sampling time (see Table S2 in the supplemental material), indicating good reproducibility.
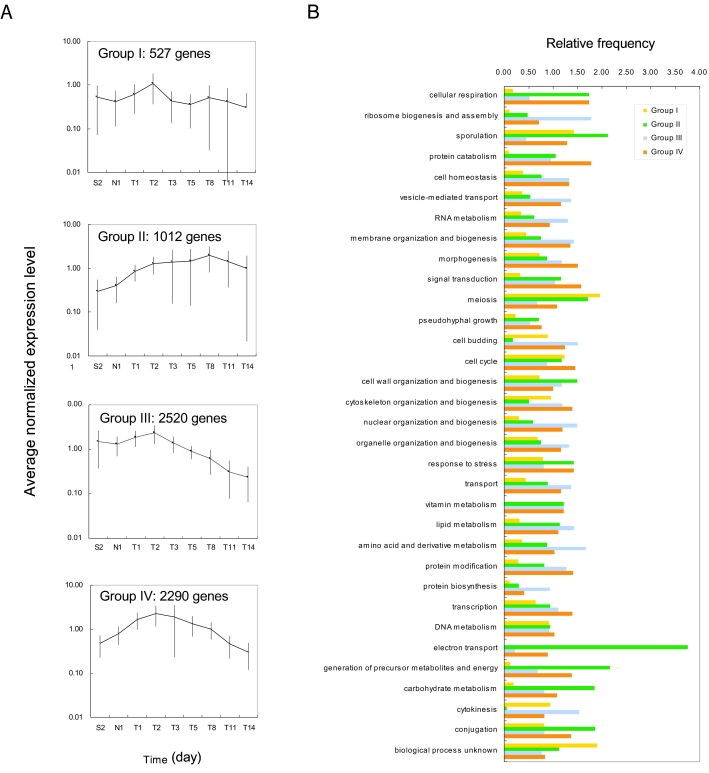
Genes were classified into four groups by K-mean cluster analysis of the expression profiles (Fig. 2A), and the genes were categorized in accordance with the biological process they were involved in (Fig. 2B) by using the Saccharomyces Genome Database Gene Ontology Slim Mapper (http://db.yeastgenome.org/cgi-bin/GO/goTermMapper). Group I contained genes whose expression levels were almost constant during the brewing process. This group included 527 genes, representing 8.3% of the total genes. Group II contained genes whose expression levels increased during the brewing process. This group included 1,012 genes, representing 15.9% of the total genes. This group contained many genes that are involved in response to stress, cell wall organization and biogenesis, and electron transport. Because these genes were expressed after ethanol accumulation, they may be involved in ethanol tolerance. Group III contained genes whose expression levels were high during the early growth phase but decreased during the late stages. This group comprised 2,520 genes, representing 39.7% of the total genes. This group contained genes whose biological roles are related to cell growth, including genes involved in RNA metabolism, amino acid and derivative metabolism, ribosome biogenesis and assembly, protein biosynthesis, nuclear organization and biogenesis, cytokinesis, and cell budding. This is consistent with the fact that cell growth occurs only during the early stages of the brewing process. Group IV contained genes whose expression levels increased during the early stages, reached maximum levels during the middle stages, and decreased during the late stages. This group included 2,290 genes, representing 36.1% of the total genes. This group contained many genes involved in signal transduction, response to stress, and protein catabolism. In the following text, we discuss characteristic gene expression patterns relevant to the sake brewing process.
FIG. 2.
Cluster analysis using the K-mean method. (A) Expression profiles of the four groups of genes. The number of genes in each group is shown. (B) Genes in each group were classified in accordance with the major biological process they are involved in by using the Saccharomyces Genome Database Gene Ontology Slim Mapper. The relative frequency of each biological process category was calculated as follows:
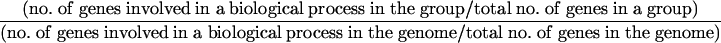 |
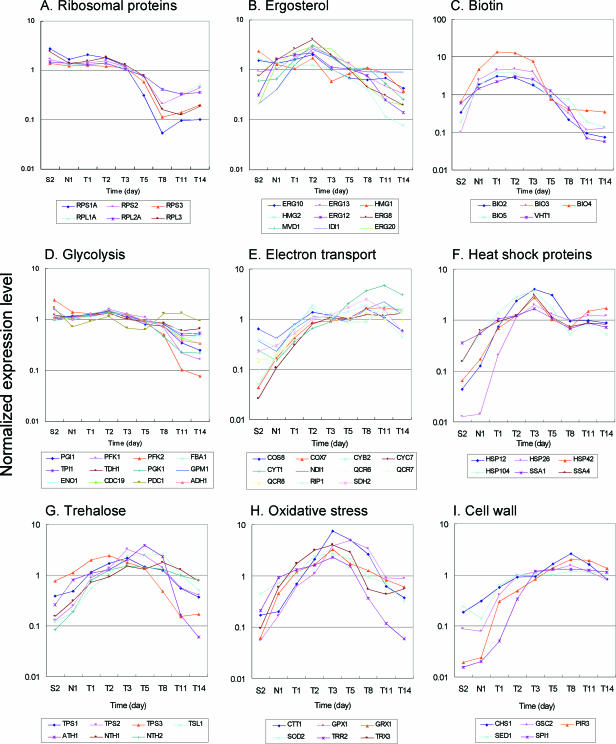
Cell growth in the sake mash was limited during the early stages of sake brewing, as shown in Fig. 1. Consistent with this, the expression levels of many genes involved in cellular growth were high during the cell growth phase and decreased during the late stages of fermentation. These genes included genes related to cytosolic ribosomal proteins, amino acid biosynthesis and transport, nucleotide biosynthesis, and fatty acid biosynthesis (data not shown). Typical expression patterns were found for genes encoding cytosolic ribosomal proteins as shown in Fig. 3A, suggesting that these genes are coordinately regulated.
FIG. 3.
Characteristic gene expression profiles in sake brewing.
Ergosterol is an important constituent of the plasma membrane, and ergosterol content is important for ethanol tolerance (8). Because its biosynthesis requires molecular oxygen (9, 18), yeast cells under anaerobic fermentation conditions require oxygen during the cell growth phase. In fact, aeration to supply oxygen is frequently performed in wine making (22). Sake brewing does not require the addition of oxygen because ergosterol is provided by koji, which is cultured in aerobic conditions and contains ergosterol. As shown in Fig. 3B, however, expression levels of many genes related to ergosterol biosynthesis increased during the cell growth phase, and their signal intensities were relatively high throughout the sake brewing process (data not shown). It has been reported that expression of some ergosterol synthesis genes are induced in anaerobic conditions (19). In microarray analysis of wine fermentation, sufficient ergosterol was added to facilitate yeast cell growth, and increased expression of the ergosterol synthesis genes was not observed (22). Taken together, these data seem to show that ergosterol supply is not sufficient during the cell growth phase in sake mash and that expression of ergosterol biosynthetic genes is therefore induced.
Biotin (vitamin H) is a cofactor of many enzymes related to carboxylation reactions. Typical S. cerevisiae strains cannot synthesize biotin and must incorporate it from an extracellular medium (17). Thus, elevated expression of biotin biosynthetic and transport genes has been observed during the cell growth phase of wine (22) and beer (10) fermentation. In contrast, sake yeast cells possess genes encoding the complete biotin biosynthetic pathway and so do not require biotin for their growth (26). However, in the present study, expression of the genes involved in biotin synthesis and incorporation (BIO2, BIO3, BIO4, BIO5, and VHT1) coordinately increased during the cell growth phase and decreased during the late phases of sake brewing as shown in Fig. 3C. These results suggest an unexpected shortage of biotin in the growth phase cells. The expression patterns of genes involved in inositol and thiamine biosyntheses were similar (data not shown). It is likely that large quantities of these vitamins are required for yeast cell growth under sake brewing conditions.
Production of high concentrations of ethanol in the sake mash suggests that genes involved in ethanol fermentation (glycolysis) and stress responses have characteristic expression patterns. The expression profiles of genes related to glycolysis are summarized in Fig. 3D. Many genes were expressed at high levels from the beginning of sake brewing (data not shown [see Table S1 in the supplemental material]). However, their expression levels gradually decreased during the late stages. In particular, the expression levels of PFK1 and PFK2, which encode proteins involved in a rate-limiting step of glycolysis (15), decreased to less than 1/10 their initial values. In contrast, such decreases in the expression levels of glycolytic genes have not been observed in wine making (22).
In anaerobic sake brewing conditions, it is unlikely that gene products related to respiration and electron transport are functional. However, the expression levels of many genes related to this process increased during the middle to late stages of sake brewing as shown in Fig. 3E. Although a similar observation has been reported for wine making (25), the significance of the elevated expression of these genes is unknown.
During the sake brewing process, yeast cells are subjected to various environmental stresses, including ethanol exposure. Heat shock proteins protect proteins from the deleterious effects of stresses by acting as chaperones or facilitating the degradation of proteins denatured by stress (20). The expression patterns of some heat shock proteins (HSP12, HSP26, HSP42, HSP104, SSA1, and SSA4) are shown in Fig. 3F. Their expression levels increased during the early stages of fermentation and were maintained at this level until the late stages, suggesting that their expression reflects accumulation of ethanol in the sake mash. In fact, these genes are also known to be induced by ethanol (21). It appears that they play important roles in ethanol tolerance in sake brewing.
Trehalose is also known to directly protect yeast cells from various stresses, such as dehydration, heat, oxygen, and ethanol, by stabilizing proteins (6). As shown in Fig. 3G, the expression levels of genes involved in trehalose synthesis and degradation increased during the middle stages but decreased during the late stages. In wine making, decreases in the expression levels of the trehalose metabolism genes have not been found, even during the late stages of fermentation (22).
Because sake brewing is carried out under anaerobic conditions, it is likely that the concentration of intracellular reactive oxygen is low and oxidative stress is weak. However, as shown in Fig. 3H, the expression levels of many genes related to oxidative stress increased during the middle stages of sake brewing. A similar phenomenon has also been reported for brewery yeast cells under ethanol fermentation conditions (10). It is known that ethanol exacerbates oxidative stress in aerobic conditions (4). Even in anaerobic conditions, yeast cells may produce oxidative stress substances via an unknown pathway in the presence of ethanol.
Because cell walls are essential components of yeast cells, many genes involved in cell wall biosynthesis showed elevated expression levels during the cell growth phase (data not shown). However, it is known that cell wall integrity is an important factor of response to stress, including ethanol tolerance (16, 24). As shown in Fig. 3I, the expression levels of some genes (CHS1, GSC2, PIR3, SED1, and SPI1) related to cell wall biosynthesis increased during the late stages of sake brewing. GSC2 (FKS2) encodes a 1,3-β-glucan synthase that is similar to Fks1p (7), and its expression is induced in various stress conditions via the calcineurin and Rho1p-Pkc1p-Mpk1p pathways (14, 27). SED1 and SPI1 encode glycosylphosphatidylinositol-anchored serine/threonine-rich cell wall proteins and are involved in cell wall integrity in stationary-phase cells (23) and ethanol tolerance (16), respectively. The increased expression of these genes suggests their positive roles in the ethanol tolerance of sake yeast cells during the late stages of sake brewing.
Sake brewing is carried out under anaerobic growth conditions, in which the composition of cell wall proteins is very different from that under aerobic conditions (11). In the present study, the four TIR family genes (TIR1, TIR2, TIR3, and TIR4) and DAN1 were expressed at high levels from the beginning of sake brewing (data not shown). Expression of these genes is induced by cold shock and anaerobic conditions (1), both of which occur in sake brewing. However, induction by a low temperature is usually temporary and not persistent (5). It thus appears that the high expression levels of these genes are due to the anaerobic conditions of the sake mash. In fact, a typical anaerobic gene (ANB1) (2) was expressed at high levels from the beginning of sake brewing (data not shown).
In the present study, we investigated the gene expression profiles of a sake yeast strain (K701) during the entire sake brewing process. Genes involved in some important metabolic pathways or functional categories of ethanol fermentation, such as glycolysis, biosynthesis and transport of ergosterol, and fatty acid biosynthesis, were expressed at high levels throughout the sake brewing process. These expression profiles have also been observed in fermentations for other alcoholic beverages, such as in wine making and beer brewing (10, 22), suggesting aspects in common for alcoholic beverage fermentations involving yeast cells, for example ethanol fermentation in anaerobic conditions and increasing ethanol concentration during the fermentation process.
However, differences in gene expression have also been observed for each kind of fermentation. These differences reflect specific conditions in fermentations for different alcoholic beverages. In wine making, nitrogen exhaustion causes yeast cells to stop growing and to enter the stationary phase, which affects the expression of hundreds of genes involved in various cellular processes, such as amino acid biosynthesis and transport, and the general stress response (22). For sake brewing, however, nutrition including sugars and amino acids is supplied at sufficient levels by rice and koji throughout the brewing process. Thus, the high concentration of ethanol (about 10% at T5) may be the reason sake yeast cells stop growing and enter the stationary phase. Transcriptional reprogramming mainly occurred in cellular growth-related genes, such as genes involved in protein and amino acid synthesis.
This study comprises the first transcriptome analysis of yeast cells during the sake brewing process. We found various transcription patterns among yeast genes in this brewing process. These different expression patterns can be used to express homologous and heterologous genes during fermentation. By selecting an appropriate promoter region, we can easily create any expression pattern for any gene. If a gene should be expressed at a high level throughout the brewing process, it can be placed under the control of the promoter region of a glycolysis gene. If elevated expression of a gene inhibits yeast cell growth, it can be controlled by the promoter of a gene induced after the middle stages of sake brewing. In the present report, we analyzed only sake mash made with sake yeast. Comparisons of the expression profiles presented here with those of laboratory strains or other industrial strains will be helpful for better understanding the characteristics of sake yeasts that make them more suitable for sake brewing than other yeast strains.
Supplementary Material
Acknowledgments
This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
Footnotes
Published ahead of print on 22 September 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abramova, N. E., B. D. Cohen, O. Sertil, R. Kapoor, K. J. Davies, and C. V. Lowry. 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157:1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian, B., C. V. Lowry, and R. S. Zitomer. 1993. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol. Cell Biol. 13:6071-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borzani, W., and M. L. Vairo. 1958. Quantitative adsorption of methylene blue by dead yeast cells. J. Bacteriol. 763:251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa, V., M. A. Amorim, E. Reis, A. Quintanilha, and P. Moradas-Ferreira. 1997. Mitochondrial superoxide dismutase is essential for ethanol tolerance of Saccharomyces cerevisiae in the post-diauxic phase. Microbiology 143:1649-1656. [DOI] [PubMed] [Google Scholar]
- 5.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hincha, D. K., E. Zuther, E. M. Hellwege, and A. G. Heyer. 2002. Specific effects of fructo- and gluco-oligosaccharides in the preservation of liposomes during drying. Glycobiology 12:103-110. [DOI] [PubMed] [Google Scholar]
- 7.Inoue, S. B., N. Takewaki, T. Takasuka, T. Mio, M. Adachi, Y. Fujii, C. Miyamoto, M. Arisawa, Y. Furuichi, and T. Watanabe. 1995. Characterization and gene cloning of 1,3-beta-D-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 231:845-854. [DOI] [PubMed] [Google Scholar]
- 8.Inoue, T., H. Iefuji, T. Fujii, H. Soga, and K. Satoh. 2000. Cloning and characterization of a gene complementing the mutation of an ethanol-sensitive mutant of sake yeast. Biosci. Biotechnol. Biochem. 64:229-236. [DOI] [PubMed] [Google Scholar]
- 9.Jahnke, L., and H. P. Klein. 1983. Oxygen requirements for formation and activity of the squalene epoxidase in Saccharomyces cerevisiae. J. Bacteriol. 155:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James, T. C., S. Campbell, D. Donnelly, and U. Bond. 2003. Transcription profile of brewery yeast under fermentation conditions. J. Appl. Microbiol. 94:432-448. [DOI] [PubMed] [Google Scholar]
- 11.Kitagaki, H., H. Shimoi, and K. Itoh. 1997. Identification and analysis of a static culture-specific cell wall protein, Tir1p/Srp1p in Saccharomyces cerevisiae. Eur. J. Biochem. 249:343-349. [DOI] [PubMed] [Google Scholar]
- 12.Kodama, K. 1993. Sake-brewing yeast, p. 129-168. In A. H. Rose and J. S. Harrison (ed.), The yeasts, vol. 3. Academic Press, London, United Kingdom. [Google Scholar]
- 13.Köhrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 14.Mazur, P., and W. Baginsky. 1996. In vitro activity of 1,3-beta-D-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 271:14604-14609. [DOI] [PubMed] [Google Scholar]
- 15.Moore, P. A., F. A. Sagliocco, R. M. Wood, and A. J. Brown. 1991. Yeast glycolytic mRNAs are differentially regulated. Mol. Cell. Biol. 11:5330-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa, Y., A. Nitta, H. Uchiyama, T. Imamura, H. Shimoi, and K. Ito. 2000. Tolerance mechanism of the ethanol-tolerant mutant of sake yeast. J. Biosci. Bioeng. 903:313-320. [DOI] [PubMed] [Google Scholar]
- 17.Ohsugi, M., and Y. Imanishi. 1985. Microbiological activity of biotin vitamers. J. Nutr. Sci. Vitaminol. 3:563-572. [DOI] [PubMed] [Google Scholar]
- 18.Osumi, T., S. Taketani, H. Katsuki, T. Kuhara, and I. Matsumoto. 1978. Ergosterol biosynthesis in yeast. Pathways in the late stages and their variation under various conditions. J. Biochem. 83:681-691. [DOI] [PubMed] [Google Scholar]
- 19.Paltauf, F., S. Kohlwein, and S. A. Henry. 1992. Regulation and compartmentalization of lipid synthesis in yeast, p. 415-500. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Piper, P. 1997. The yeast heat shock response, p. 75-99. In S. Hohmann and W. H. Mager (ed.), Yeast stress responses, Chapman & Hall, New York, N.Y.
- 21.Piper, P. W., K. Talreja, B. Panaretou, P. Moradas-Ferreira, K. Byrne, U. M. Praekelt, P. Meacock, M. Recnacq, and H. Boucherie. 1994. Induction of major heat-shock proteins of Saccharomyces cerevisiae, including plasma membrane Hsp30, by ethanol levels above a critical threshold. Microbiology 140:3031-3038. [DOI] [PubMed] [Google Scholar]
- 22.Rossignol, T., L. Dulau, A. Julien, and B. Blondin. 2003. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20:1369-1385. [DOI] [PubMed] [Google Scholar]
- 23.Shimoi, H., H. Kitagaki, H. Ohmori, Y. Iimura, and K. Ito. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180:3381-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi, T., H. Shimoi, and K. Ito. 2001. Identification of genes required for growth under ethanol stress using transposon mutagenesis in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:1112-1119. [DOI] [PubMed] [Google Scholar]
- 25.Varela, C., J. Cardenas, F. Melo, and E. Agosin. 2005. Quantitative analysis of wine yeast gene expression profiles under winemaking conditions. Yeast 22:369-383. [DOI] [PubMed] [Google Scholar]
- 26.Wu, H., H. Shimoi, and K. Itoh. 2005. Identification and characterization of a novel biotin biosynthesis gene in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:6845-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao, C., U. S. Jung, P. Garrett-Engele, T. Roe, M. S. Cyert, and D. E. Levin. 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.