Abstract
Wolbachia pipientis is an obligate intracellular bacterium found in a wide range of invertebrate taxa. While over ecological timescales Wolbachia infections are maintained by strict maternal inheritance, horizontal transfer events are common over evolutionary time. To be horizontally transferred between organisms, Wolbachia bacteria must pass through and survive an extracellular phase. We used BacLight live-dead staining, PCR, and fluorescence in situ hybridization to assess the ability for purified Wolbachia bacteria to survive in cell-free media. We found that purified Wolbachia bacteria were able to survive extracellularly for up to 1 week with no decrease in viability. While no replication was observed in the extracellular phase, purified Wolbachia bacteria were able to reinvade cells and establish stable infections at all time points. The ability of Wolbachia bacteria to survive outside host cells may increase the probability of successful horizontal transfer and the exploitation of new ecological niches. Our development of methods to purify and maintain viable Wolbachia bacteria from cultured cells will be useful for other researchers studying Wolbachia biology.
Wolbachia pipientis is an obligate intracellular bacterium found in a wide range of invertebrate hosts, including arthropods and filarial nematodes (18). Wolbachia bacteria cause alterations in host reproductive phenotypes, including cytoplasmic incompatibility, feminization, parthenogenesis, male killing, fitness alterations, and obligate symbiosis (5, 18). There has been a great amount of recent interest in Wolbachia as an agent to manipulate pest and vector populations (13, 14, 15, 16, 18, 22, 27) and control diseases caused by filarial nematodes (19, 21) and as a system to address questions related to the ecology and evolution of symbiosis (3, 4, 23) and speciation events (2, 20).
Over ecological timescales, Wolbachia infections are maintained by strict maternal inheritance (18). However, comparative phylogenetics indicate that over evolutionary timescales, Wolbachia horizontal transfer events are common, resulting in closely related infections in very distantly related host taxa (18, 28). Potential nonexclusive mechanistic hypotheses for Wolbachia transfer between nonrelated hosts include parasitoids, cofeeding on the same host plant, or predation events (11, 17, 24). In all cases, to be horizontally transferred between hosts, Wolbachia bacteria must spend some time outside the intracellular environment. In transferring infection from embryos to cell culture, Dobson and colleagues incidentally showed that Wolbachia bacteria must remain viable outside a host cell for a brief period of time (6). Recently, Frydman and colleagues (7) demonstrated that when injected into the abdomen of Drosophila melanogaster, Wolbachia remained viable in the extracellular hemolymph environment long enough to establish intracellular infection of the ovarian stem cells. These results suggest that the ability of Wolbachia bacteria to survive outside a host cell may greatly increase the probability of successful horizontal transfer and the exploitation of new niches for the bacterium. In this research, we assessed the ability of Wolbachia pipientis to remain viable for up to 1 week when suspended in cell-free medium. Our observations suggest that Wolbachia is capable of surviving for extended periods in the absence of a host cell, an ability that may facilitate successful horizontal transmission of the bacteria.
MATERIALS AND METHODS
Cell maintenance.
Experiments were performed using cell line Aa23, which was originally derived from Aedes albopictus embryos that were infected with 2 Wolbachia strains (wAlbA and wAlbB). The derived cell line is constitutively infected with only one Wolbachia strain (wAlbB) (12). A split from this cell line was cured of Wolbachia infection by treatment with tetracycline (Aa23T) (6). Cells were grown without antibiotics in 25-cm2 plastic culture flasks at room temperature in Schneider medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and passaged approximately once per week.
Wolbachia purification.
While sonication and detergent lysis have been previously shown to be useful methods for purifying Wolbachia from infected cells or embryos (9, 10), we were concerned that these methods would not preserve bacterial viability. We therefore isolated Wolbachia from infected cells using a protocol modified from one developed to purify Rickettsia from cultured tick cells (1).
For purifications, Aa23 cells were grown in 50-cm2 culture flasks to ∼90% confluence. Originally, cells were washed with phosphate-buffered saline and trypsinized to create a suspension; subsequent work showed that trypsin was unnecessary and that sufficient suspensions could be created by vigorously shaking flasks to dislodge cells. Cells were pelleted by centrifugation at 2,500 × g and 4°C for 10 min, the supernatant was removed, and cells were resuspended in 10 ml media in a 50-ml conical tube. Cells were vortexed for 5 min with approximately 100 sterile 3-mm borosilicate glass beads to lyse cells. The lysate was centrifuged at 2,500 × g and 4°C for 10 min to pellet large cellular debris. The supernatant was decanted, passed through a 5-μm Millex syringe filter (Millipore, Billerica, MA), and centrifuged at 18,400 × g and 4°C for 5 min on a 250 mM sucrose cushion to pellet Wolbachia. The Wolbachia pellet was resuspended in 1 ml Schneider's media with 10% FBS and passed through a 2.7-μm syringe filter (Whatman, Florham Park, NJ) to remove residual cellular debris.
Wolbachia viability assays.
The viability of purified Wolbachia suspended in Schneider's media with 10% FBS was assessed 0 h, 2 h, 24 h, and 1 week postpurification by BacLight live-dead staining and by reinfection of purified Wolbachia into uninfected cells. All assays were performed in triplicate at each time point.
(i) BacLight live-dead assay.
BacLight live-dead staining (Molecular Probes, Carlsbad, CA) was first used to examine survival of purified Wolbachia. The BacLight assay is a differential stain that uses cell membrane integrity to assess bacterial viability, in which the SYTO 9 (green) stain penetrates all cells while the propidium iodide (red) stain only penetrates cells with damaged cell membranes. Thus, intact (“live”) cells appear green while nonintact (“dead”) cells appear red. Fifty microliters of purified Wolbachia suspension was stained according to the manufacturer's suggested protocol and viewed on an Olympus BX-41 compound microscope fitted with epifluorescent optics. Wolbachia viability was estimated by dividing the number of green Wolbachia bacteria visible in the field by the total number visible (green plus red). As a control, the assay was repeated using Wolbachia bacteria that had been killed by heating at 95°C for 10 min. To confirm that what we observed by BacLight was really Wolbachia, purified bacteria were placed on an eight-well chamber slide (Nalge Nunc International, Rochester, NY), allowed to air dry, fixed for 15 min with 4% formalin, and viewed using Wolbachia-specific fluorescence in situ hybridization (FISH) as described below.
(ii) Experimental infection assay.
To confirm live-dead stain results, purified Wolbachia bacteria were reintroduced into uninfected Aa23T cells using the modified shell-vial technique (6). In initial infection assays, we did not attempt to quantify the number of Wolbachia bacteria used for experiments and thus qualitatively assessed Wolbachia viability by the presence of one or more visibly infected cells. In later experiments (below), we standardized both the amount of Wolbachia bacteria used for infections and the number of recipient cells. For all infections, cells were passaged twice prior to visualization.
(a) Experiment one (qualitative).
Purified Wolbachia bacteria were held in suspension after purification in Schneider's media with 10% FBS at room temperature for 0 h, 2 h, 24 h, or 1 week and then reinfected into Aa23T cells. Aa23T monolayers were grown in the wells of 48-well plates to ∼90% confluence. Wolbachia suspension (500 μl) was layered onto cells, and the plate was centrifuged at 4,000 × g at 15°C for 1 h. Cells were left undisturbed overnight (approximately 18 h) and then transferred into a six-well plate containing 1 ml media plus 10% FBS. After approximately 5 days, cells were transferred to a 25-cm2 flask and cultured as described. Successful transfections were assessed by PCR and by direct visualization of infected cells using FISH (see below). As a control, purified Wolbachia bacteria were killed by heating at 95°C for 10 min prior to reinfection into uninfected cells.
(b) Experiment two (quantitative).
Purified Wolbachia bacteria were held as described above for 0 h, 24 h, or 1 week at room temperature. Cells were grown to 80% confluence in the wells of a six-well culture plate (approximately 75,000 cells per well). For each time point, an aliquot of purified Wolbachia was live-dead stained and the bacterial concentration was determined using a hemocytometer as described previously (1). Wolbachia bacteria were adjusted to a final concentration of 2 × 108 live bacteria per ml. For each time point, 1 ml of purified Wolbachia bacteria was placed into each well, for a final inoculum load of approximately 2,600 bacteria per cell, and infections were carried out as described above.
Wolbachia-specific fluorescence in situ hybridization.
Wolbachia bacteria were visualized with 2 probes 5′ end labeled with rhodamine: W1, 5′-AATCCGGCCGARCCGACCC-3′; W2, 5′-CTTCTGTGAGTACCGTCATTATC-3′ (8). Cell monolayers were grown on eight-well chamber slides to ∼50% confluence and then fixed for 10 min with 4% formalin. A well with Aa23T cells was included as a negative control. One hundred nanograms of each probe was added to 20 ml hybridization buffer [50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 200 g/liter dextran sulfate, 250 mg/liter poly(A), 250 mg/liter salmon sperm DNA, 250 mg/liter tRNA, 0.1 M dithiothreitol (DTT), 0.5× Denhardt's solution], and slides were incubated at 37°C overnight (approximately 18 h). The next day, slides were washed twice in 1× SSC, 10 mM DTT and twice in 0.5× SSC, 10 mM DTT at 55°C. Slides were rinsed in deionized water, mounted with glycerol, and viewed on an Olympus BX-41 compound microscope fitted with epifluorescent optics.
Wolbachia-specific PCR.
Cells were pelleted by centrifugation, and media were removed. Total DNA from cells was extracted using DNEasy kits (QIAGEN, Valencia, CA) according to the manufacturer's suggested protocol. Wolbachia infection was confirmed by diagnostic PCR amplification of a 440-bp fragment of the Wolbachia 16S rRNA gene using primers WspecF (5′-CATACCTATTCGAAGGGATAG-3′) and WspecR (5′-AGCTTCGAGTGAAACCAATTC-3′) as previously described (25). Reaction mixtures containing DNA from known infected (Aa23) and uninfected (Aa23T) cells were included in all assays as positive and negative controls, respectively. PCR amplicons were separated by 1% agarose gel electrophoresis, stained with ethidium bromide, and visualized with UV light.
RESULTS AND DISCUSSION
Wolbachia purification.
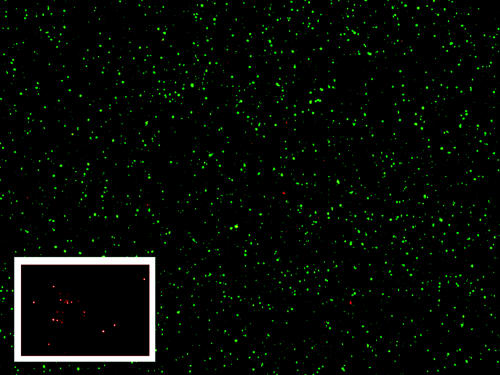
Our purification protocol resulted in numerous bacteria indicated by punctate staining using the BacLight assay (Fig. 1). No bacteria were visible when the purification protocol was attempted using uninfected cells. Because the BacLight assay is nonspecific and will stain any bacteria, we fixed purified bacteria to a slide and specifically stained them using Wolbachia-specific FISH probes (8). Bacteria were again indicated by punctuate staining (Fig. 1, inset) that was lacking when uninfected cells were used as donor material for purification. We therefore conclude that the bacteria visible by BacLight are Wolbachia bacteria.
FIG. 1.
Purified Wolbachia organisms visualized by the BacLight live-dead assay. Live Wolbachia organisms are stained green; dead Wolbachia organisms are stained red. Inset: purified Wolbachia stained by FISH (magnification, ×1,000).
Viability time series.
The BacLight stain was used to assess the viability of purified Wolbachia bacteria when held in suspension in culture media at room temperature. Wolbachia bacteria that had been killed by heat were stained uniformly red.
Non-heat-treated Wolbachia bacteria remained viable (>95% green) for up to 1 week, with no apparent reduction in viability over time. Since the BacLight assay relies on cell membrane integrity to assess bacterial viability, we considered the hypothesis that we were observing dead Wolbachia bacteria that nevertheless had intact cell membranes and that our results were an artifact of the staining mechanism.
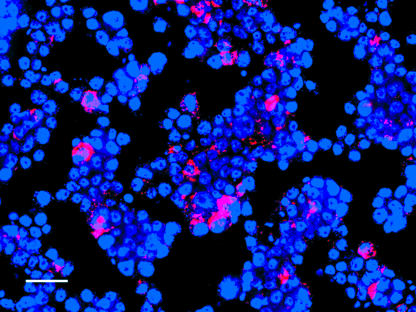
To address this issue, we reinoculated purified Wolbachia bacteria into uninfected Aa23T cells. Successful Wolbachia infection of cells was assessed by FISH and PCR for 5 passages postinfection, starting on passage 2. Cells were positive by both specific staining using FISH (Fig. 2) and by amplification of a diagnostic 440-bp PCR amplicon at all time points. Wolbachia bacteria were not detectable in cells by FISH or PCR when heat killed prior to infection attempts.
FIG. 2.
Establishment of Wolbachia-infected cells by inoculating purified Wolbachia organisms into uninfected Aa23 cells and visualization by fluorescence in situ hybridization. Results are typical of all examined time points. Red, Wolbachia; blue, cell nuclei stained with 4′,6′-diamidino-2-phenylindole (DAPI). Bar, 20 μm.
Cell infection rates ranged from approximately 5% to >90% of cells infected, as determined by FISH, with no relationship between the proportion of cells infected and the age of purified Wolbachia bacteria. We did not explicitly quantify the amount of Wolbachia bacteria used for infections in this experiment but noticed qualitatively that the proportion of infected cells was not dependent on the length of time that Wolbachia bacteria were held prior to reinfection but rather on the Wolbachia titer in the donor material used to establish the infection. The more Wolbachia bacteria that were initially added to cells, the more cells became infected. This is similar to previous experiments showing that the probability of establishing in vitro infections from infected Drosophila embryos was proportional to the amount of donor material used (6). We investigated this further by quantifying the number of Wolbachia bacteria and using a standard Wolbachia/cell ratio for infections. When Wolbachia bacteria were inoculated at approximately 2,600 bacteria per cell, infection was established in approximately 60% of cells at all time points, indicating no apparent decrease in Wolbachia viability during the experiment.
Within artificially infected cell lines, Wolbachia titers in individual cells exhibited a range of infection levels, from some cells with only a few visible symbionts to very heavy infections where the entire cytoplasm was filled with Wolbachia bacteria (Fig. 2). In some cases, infection titers were so high that there appeared to be a pathogenic effect on cells, with abnormal cell and nucleus morphology.
The time period for extracellular viability experiments did not extend past 7 days. We have extended live-dead staining for up to 1 month with no increase in red-stained bacteria but have not validated this outer time point by cell reinfection. Consequently, the outer time boundary for extracellular Wolbachia survival is unknown. Because we observed no apparent decrease in extracellular viability during the experiment, we hypothesize that Wolbachia extracellular survival may be significantly longer than 1 week. Storage of purified Wolbachia at cooler temperatures may further extend this time frame. We are currently conducting experiments to determine the environmental limits for extracellular Wolbachia survival.
Our results prompt us to question how an intracellular symbiont can survive for an extended period of time outside a host cell. It is apparent from comparisons between Wolbachia and host phylogenies that horizontal transmission is a common phenomenon over evolutionary time (18, 28). If in natural systems Wolbachia survival in insect hemolymph is similar to results in culture media, extended extracellular survival could be a potential adaptation to increase the probability of successful horizontal transfer. This hypothesis is further supported by in vivo studies (7) suggesting that extracellular Wolbachia survival can facilitate successful establishment of artificial transfections.
Because Wolbachia bacteria cannot be grown in axenic culture and must be grown in eukaryotic host cells, experimental Wolbachia manipulation requires methods to purify live bacteria from cells without contaminating host material. Protocols have been developed to isolate purified Wolbachia bacteria from infected embryos (9) and from cell culture systems (10), but these methods did not preserve the viability of purified bacteria. Our development of methods to purify and maintain viable Wolbachia bacteria from cultured cells will be useful for numerous applications, such as development of Wolbachia transformation protocols, supplying a source of enriched mRNA for microarray experiments and facilitating studies on the mechanisms of Wolbachia cell invasion and infection establishment.
One of the most significant hurdles to experimental investigation of Wolbachia biology is the reliance on a eukaryotic host cell for bacterial propagation. In this study, we have shown that eukaryotic host cells are not required for Wolbachia survival. In our experiments, Wolbachia bacteria were held in medium with high amino acid content (due to the added FBS). Wolbachia bacteria are hypothesized to obtain the majority of their energy from amino acid metabolism (26), so it is possible that the long survival times observed were in part due to the ability of Wolbachia bacteria to subsist on the amino acids in the medium during storage. We did not observe replication of purified Wolbachia, presumably due to lack of the necessary factors in our cell-free medium. However, if the specific cellular factors required for Wolbachia replication can be identified, it may be possible to establish Wolbachia culture in a host cell-free system.
Acknowledgments
This work was supported by a Johns Hopkins Malaria Research Institute Pilot Grant to J.L.R.
We thank Stephen Dobson and Cynthia Khoo for providing cell lines and Mary Claire Hauer for laboratory assistance.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Baldridge, G. D., N. Burkhardt, M. J. Herron, T. J. Kurtti, and U. G. Munderloh. 2005. Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl. Environ. Microbiol. 71:2095-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bordenstein, S. R., F. P. O'Hara, and J. H. Werren. 2001. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature 409:707-710. [DOI] [PubMed] [Google Scholar]
- 3.Brownlie, J. C., and S. L. O'Neill. 2005. Wolbachia genomes: insights into an intracellular lifestyle. Curr. Biol. 15:R507-R509. [DOI] [PubMed] [Google Scholar]
- 4.Casiraghi, M., S. R. Bordenstein, L. Baldo, N. Lo, T. Beninati, J. J. Wernegreen, J. H. Werren, and C. Bandi. 2005. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology 151:4015-4022. [DOI] [PubMed] [Google Scholar]
- 5.Dobson, S. L., E. J. Marsland, and W. Rattanadechakul. 2002. Mutualistic Wolbachia infection in Aedes albopictus: accelerating cytoplasmic drive. Genetics 160:1087-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobson, S. L., E. J. Marsland, Z. Veneti, K. Bourtzis, and S. L. O'Neill. 2002. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl. Environ. Microbiol. 68:656-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frydman, H. M., J. M. Li, ZD. N. Robson, and E. Wieschaus. 2006. Somatic stem cell niche tropism in Wolbachia. Nature 441:509-512. [DOI] [PubMed] [Google Scholar]
- 8.Heddi, A., A. M. Grenier, C. Khatchadourian, H. Charles, and P. Nardon. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. USA 96:6814-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kose, H., and T. L. Karr. 1995. Organization of Wolbachia pipientis in the Drosophila fertilized egg and embryo revealed by an anti-Wolbachia monoclonal antibody. Mech. Dev. 51:275-288. [DOI] [PubMed] [Google Scholar]
- 10.Mavingui, P., V. T. Van, E. Labeyrie, E. Rances, F. Vavre, and P. Simonet. 2005. Efficient procedure for purification of obligate intracellular Wolbachia pipientis and representative amplification of its genome by multiple-displacement amplification. Appl. Environ. Microbiol. 71:6910-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsuhashi, W., T. Saiki, W. Wei, H. Kawakita, and M. Sato. 2002. Two novel strains of Wolbachia coexisting in both species of mulberry leafhoppers. Insect Mol. Biol. 11:577-584. [DOI] [PubMed] [Google Scholar]
- 12.O'Neill, S. L., M. M. Pettigrew, S. P. Sinkins, H. R. Braig, T. G. Andreadis, and R. B. Tesh. 1997. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 6:33-39. [DOI] [PubMed] [Google Scholar]
- 13.Rasgon, J. L., L. M. Styer, and T. W. Scott. 2003. Wolbachia-induced mortality as a mechanism to modulate pathogen transmission by vector arthropods. J. Med. Entomol. 40:125-132. [DOI] [PubMed] [Google Scholar]
- 14.Rasgon, J. L., and T. W. Scott. 2003. Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics 165:2029-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasgon, J. L., and T. W. Scott. 2004. Impact of population age structure on Wolbachia transgene driver efficacy: ecologically complex factors and release of genetically modified mosquitoes. Insect Biochem. Mol. Biol. 34:707-713. [DOI] [PubMed] [Google Scholar]
- 16.Sinkins, S. P. 2004. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem. Mol. Biol. 34:723-729. [DOI] [PubMed] [Google Scholar]
- 17.Sintupachee, S., J. R. Milne, S. Poonchaisri, V. Baimai, and P. Kittayapong. 2006. Closely related Wolbachia strains within the pumpkin arthropod community and the potential for horizontal transmission via the plant. Microb. Ecol. 51:294-301. [DOI] [PubMed] [Google Scholar]
- 18.Stouthamer, R., J. A. J. Breeuwer, and G. D. D. Hurst. 1999. Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53:71-102. [DOI] [PubMed] [Google Scholar]
- 19.Taylor, M. J. 2000. Wolbachia bacteria of filarial nematodes in the pathogenesis of disease and as a target for control. Trans. R. Soc. Trop. Med. Hyg. 94:596-598. [DOI] [PubMed] [Google Scholar]
- 20.Telschow, A., P. Hammerstein, and J. H. Werren. 2005. The effect of Wolbachia versus genetic incompatibilities on reinforcement and speciation. Evol. Int. J. Org. Evol. 59:1607-1619. [PubMed] [Google Scholar]
- 21.Townson, H. 2002. Wolbachia as a potential tool for suppressing filarial transmission. Ann. Trop. Med. Parasitol. 96(Suppl. 2):S117-S127. [DOI] [PubMed] [Google Scholar]
- 22.Turelli, M., and A. A. Hoffmann. 1999. Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol. Biol. 8:243-255. [DOI] [PubMed] [Google Scholar]
- 23.van der Giezen, M. 2005. Endosymbiosis: past and present. Heredity 95:335-336. [DOI] [PubMed] [Google Scholar]
- 24.Vavre, F., F. Fleury, D. Lepetit, P. Fouillet, and M. Bouletreau. 1999. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16:1711-1723. [DOI] [PubMed] [Google Scholar]
- 25.Werren, J. H., and D. M. Windsor. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. Biol. Sci. Lond. Ser. B 267:1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLOS Biol. 2:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xi, Z., C. C. Khoo, and S. L. Dobson. 2005. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310:326-328. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, W., F. Rousset, and S. L. O'Neill. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. B 265:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]