Abstract
The gram-positive bacterium Bacillus subtilis secretes high levels of proteins into its environment. Most of these secretory proteins are exported from the cytoplasm in an unfolded state and have to fold efficiently after membrane translocation. As previously shown for α-amylases of Bacillus species, inefficient posttranslocational protein folding is potentially detrimental and stressful. In B. subtilis, this so-called secretion stress is sensed and combated by the CssRS two-component system. Two known members of the CssRS regulon are the htrA and htrB genes, encoding potential extracytoplasmic chaperone proteases for protein quality control. In the present study, we investigated whether high-level production of a secretory protein with two disulfide bonds, PhoA of Escherichia coli, induces secretion stress in B. subtilis. Our results show that E. coli PhoA production triggers a relatively moderate CssRS-dependent secretion stress response in B. subtilis. The intensity of this response is significantly increased in the absence of BdbC, which is a major determinant for posttranslocational folding of disulfide bond-containing proteins in B. subtilis. Our findings show that BdbC is required to limit the PhoA-induced secretion stress. This conclusion focuses interest on the BdbC-dependent folding pathway for biotechnological production of proteins with disulfide bonds in B. subtilis and related bacilli.
The gram-positive eubacterium Bacillus subtilis is well known for its high capacity to secrete proteins, both in its natural habitat (the soil) and in biotechnological applications. Recent studies have suggested that most secretory proteins of this organism are translocated across the cytoplasmic membrane in an unfolded state via the Sec pathway (14, 38, 39, 42). When the posttranslocational folding of secretory proteins, such as the α-amylase AmyQ from Bacillus amyloliquefaciens, is not efficient enough, extracytoplasmic accumulation of malfolded proteins can harm the cell (12, 33). B. subtilis uses the CssR-CssS two-component system to sense this stress situation and combat its effects (12). In general, two-component systems mediate the relay between specific stimuli and adequate transcriptional behavior (28, 36). The B. subtilis CssRS system consists of a typical sensor kinase, CssS, and its cognate cytoplasmic response regulator, CssR. Most likely, the membrane protein CssS senses stimuli at the membrane-cell wall interface (12). This information is then transmitted to CssR, using a phosphorylation-dephosphorylation relay. Finally, the CssR protein modulates the transcription of certain genes in response to the initial stimuli. So far, the CssRS two-component system is known to respond not only to stress induced by overproduction of α-amylases from different Bacillus species but also to heat (6, 12). Collectively, the conditions that trigger a CssRS-dependent cellular response have been termed secretion stress. At least four genes have been identified as members of the CssRS regulon: htrA, htrB, cssR, and cssS, with cssR and cssS forming an operon (6, 12, 13). In particular, the levels of both htrA transcription and htrB transcription are significantly increased in secretion-stressed cells. Consequently, expression of these genes can be used as a reliable indicator for this physiological state of the cell.
HtrA and HtrB are homologues of the HtrA chaperone protease of Escherichia coli. In the case of secretion stress, both the protease and foldase activities of HtrA-like proteins can play important roles; while the protease activity results in degradation of malfolded proteins that congest the membrane and cell wall, the foldase activity helps fold and rescue these proteins (17, 35). A different class of folding catalysts is represented by the thiol-disulfide oxidoreductases and the cognate quinone oxidoreductases. Together, these enzymes catalyze the formation of disulfide bonds that are required for the stability and activity of certain proteins (25, 30). In B. subtilis, two putative thiol-disulfide oxidoreductases (BdbA and BdbD) and two putative quinone oxidoreductases of the E. coli DsbB type (BdbB and BdbC) have been identified (4, 10, 23). Computer-assisted predictions have suggested that BdbA and BdbD are synthesized with N-terminal membrane anchors (41), whereas BdbB and BdbC are integral membrane proteins with four transmembrane segments (4, 23, 39). It has been proposed that BdbC and BdbD are a functional pair in which BdbD is a thiol-disulfide oxidoreductase that oxidizes a substrate protein and BdbC is a quinone oxidoreductase that reoxidizes BdbD (9, 23). The BdbC and BdbD proteins are essential for development of natural competence, most likely because of their indispensable role in the biogenesis of the pseudopilin ComGC. This pseudopilin is a critical component in the DNA uptake machinery of B. subtilis (23). Furthermore, BdbC and BdbD play an important role in the posttranslocational folding into a protease-resistant conformation of the PhoA alkaline phosphatase of E. coli, when this protein is produced and secreted by B. subtilis (4). Importantly, both ComGC and PhoA contain intramolecular disulfide bonds that are essential for their activity and stability (5, 34). In contrast, the posttranslocational folding and secretion of proteins lacking disulfide bonds, such as the AmyQ α-amylase of B. amyloliquefaciens, are not affected by bdbC or bdbD mutations (4). The available data therefore support the view that BdbC and BdbD are required for the formation of disulfide bonds in ComGC and PhoA, thereby preventing extracytoplasmic degradation of these proteins. In contrast, BdbA and BdbB of B. subtilis, the paralogues of BdbD and BdbC, respectively, are dispensable for ComGC biogenesis and secretion of active PhoA.
The aim of the present study was to answer the question whether high-level production of a secretory protein, which is significantly different from Bacillus α-amylases with respect to folding catalyst requirements, can trigger a secretion stress response in B. subtilis. The E. coli PhoA protein was chosen for this study because it is the only known BdbC- and BdbD-dependent disulfide bond-containing secretory protein that can be secreted at high levels by B. subtilis. Additionally, E. coli PhoA is an attractive model protein for studying the biotechnological production of heterologous secretory proteins with multiple disulfide bonds in B. subtilis, which is generally considered problematic (42). Our results show for the first time that high-level expression of E. coli PhoA can induce a secretion stress response in B. subtilis. In the absence of BdbC, the intensity of this stress response is significantly increased, which implies that unfolded PhoA is a direct or indirect stimulus for the CssRS system. Accordingly, it seems that the BdbC-dependent folding pathway helps limit PhoA-induced secretion stress.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
The plasmids and bacterial strains used are listed in Table 1. B. subtilis was grown with agitation at 37°C in TY medium (1% tryptone, 0.5% yeast extract, 1% NaCl). Antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 5 μg/ml; erythromycin, 1 μg/ml; kanamycin, 10 μg/ml; spectinomycin, 100 μg/ml; and tetracycline, 15 μg/ml. To visualize α-amylase activity (encoded by the amyE gene), TY medium plates were supplemented with 1% starch.
TABLE 1.
Plasmids, bacterial strains, and primers
| Plasmid, strain, or primer | Relevant properties or sequence (5′-3′) | Reference(s) or source |
|---|---|---|
| Plasmids | ||
| pDG1514 | pMTL23 derivative; contains the tetracycline resistance marker from Streptococcus agalactiae; Apr Tcr | 11 |
| pJM1 | pUC18 derivative; contains the gene encoding the preprolipase from S. hyicus under control of the regulatory elements of the lac operon; Apr | 21 |
| pJM1-23 | pJM1 derivative with an SnaBI site at the position corresponding to the junction between the propeptide and mature lipase of S. hyicus; Apr | 22 |
| pKTH10L | pUB110 derivative containing the amyQ gene of B. amyloliquefaciens; Kmr | 12 |
| pPA12 | pJM1-23 derivative containing a precise fusion between the prepro part of the preprolipase of S. hyicus and the coding sequence for mature PhoA; Apr | This study |
| pPS2 | pLipPS1 derivative containing the constitutive promoter of the S. hyicus preprolipase gene (pLip) followed by a multiple cloning site; Cmr | 22 |
| pPSPhoA5 | pPS2 derivative carrying the fusion between the preprolip part of the preprolipase of S. hyicus and the mature PhoA coding sequence from pPA12 (preprolip-PhoA); Cmr | 4; this study |
| pUC19 | Plasmid containing ColE1, φ80dlacZ, lac promoter; Apr | 27 |
| pUC19bdbC | pUC19 derivative containing the bdbC gene; Apr | This study |
| pX | Vector for the integration of genes in the amyE locus; the integrated gene is transcribed from the xylA promoter; carries the xylR gene; Apr Cmr | 15 |
| pXTC | pX derivative containing a tetracycline resistance marker instead of a chloramphenicol resistance marker; Apr Tcr | This study |
| pXTCbdbC | pXTC derivative carrying bdbC under transcriptional control of the xylA promoter; Apr Tcr | This study |
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 endA1 recA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Life Technologies, Inc. |
| JM109 | F′ traD36 lacIqZΔM15 proA+B+/e14− (McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rK− mK+) relA1 supE44 recA1 | 49 |
| MC1061 | F−araD139 Δ(ara-leu)7696 Δ(lac)X74 galU galK hsdR2 mcrA mcrB1 rspL | 47 |
| B. subtilis strains | ||
| 168 | trpC2 | 1 |
| bdbC-Km | 168 bdbC::Km; Kmr | 10 |
| BFA3041 | 168 htrB::pMutin4; Emr | 6 |
| BV2001 | 168 cssS::Sp; Spr | 12 |
| BV2003 | 168 htrA::pMutin2; Emr | 12 |
| BV2015 | 168 cssS::Sp htrB::pMutin4; Emr Spr | 6 |
| BV2031 | 168 htrA::pMutin2 bdbC::Km; Emr Kmr | This study |
| BV2032 | 168 htrB::pMutin4 bdbC::Km; Emr Kmr | This study |
| BV2033 | 168 cssS::Sp htrB::pMutin4 bdbC::Km; Emr Kmr Spr | This study |
| BV2034 | 168 htrB::pMutin4 amyE::XTCbdbC; Emr Tcr | This study |
| BV2035 | 168 htrB::pMutin4 bdbC::Km amyE::XTCbdbC; Emr Kmr Tcr | This study |
| Primersa | ||
| pho4 | GGGATTTAAATGATATCACGTGTTAACCGGGCTGCTCAGGGCGATAT | This study |
| pho5 | TTTAAAGCTTGGATCCTTATTTCAGCCCCAGAGCGGC | This study |
| yvgU1 | GAAATTCTA GAGACAATAGAAAAAGAGCTGAAAGGGAAGTAA C | 10 |
| yvgU2 | GCGAAATGGATCCTTAGTTCAGGTCCTCCTCGCTGATTAATTTTTGTTCAGATTTTTCGCCTTTCAGCAGGCAC | This study |
Underlining indicates restriction sites used for cloning.
DNA techniques.
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of competent E. coli cells were carried out as described by Sambrook et al. (32). Enzymes were obtained from New England Biolabs, Life Technologies, or Roche Molecular Biochemicals. PCR were carried out with the Taq or Pwo DNA polymerase, using chromosomal DNA as the template (44). B. subtilis was transformed as described by Leskela et al. (20) or Kunst and Rapoport (18). The nucleotide sequences of primers used for PCR are listed in Table 1. Constructs were first made in E. coli DH5α or MC1061 and then introduced into B. subtilis.
Plasmid pPSPhoA5, which encodes a precise fusion between the signal peptide plus the pro region of the Staphylococcus hyicus lipase and mature PhoA of E. coli (preprolip-PhoA), was constructed using plasmid pJM1-23. The latter plasmid carries a copy of the S. hyicus lipase gene in which an SnaBI site was introduced at the position that corresponds to the junction between the propeptide and the mature lipase. Next, the phoA gene of E. coli JM109 was PCR amplified with primers pho4 and pho5. The resulting fragment was cleaved with HpaI and HindIII and used to replace the SnaBI-HindIII fragment of pJM1-23, which encodes the mature lipase. This resulted in plasmid pPA12. Finally, pPSPhoA5 was obtained by insertion of a 2.16-kb SacI-HindIII fragment from pPA12, which encoded preprolip-PhoA, into the corresponding sites of pPS2.
To construct pXTC, the chloramphenicol resistance marker of plasmid pX was replaced with the tetracycline resistance marker of plasmid pDG1514, using the flanking BamHI and SphI sites. In the first step in the construction of pXTCbdbC, the bdbC gene of B. subtilis 168 was PCR amplified with primers yvgU1 and yvgU2 and cloned into the XbaI and BamHI sites of plasmid pUC19. The resulting plasmid was designated pUC19bdbC. Next, the bdbC gene was excised from pUC19bdbC with BamHI and XbaI and ligated into the BamHI and SpeI sites of pXTC. This resulted in plasmid pXTCbdbC.
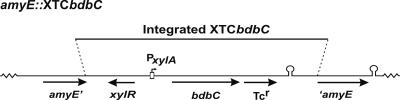
The B. subtilis 168 htrB::pMutin4 amyE::XTCbdbC (BV2034) strain was generated by transformation of B. subtilis 168 htrB::pMutin4 (BFA3041) with plasmid pXTCbdbC, subsequent selection for tetracycline resistance, and screening for an AmyE-negative phenotype on starch-containing plates. The integration of the XTCbdbC cassette into the chromosomal amyE locus is shown schematically in Fig. 1. B. subtilis 168 htrA::pMutin2 bdbC::Km (BV2031), B. subtilis 168 htrB::pMutin4 bdbC::Km (BV2032), B. subtilis 168 cssS::Sp htrB::pMutin4 bdbC::Km (BV2033), and B. subtilis 168 htrB::pMutin4 bdbC::Km amyE::XTCbdbC (BV2035) were generated by transformation of B. subtilis 168 htrA::pMutin2 (BV2003), B. subtilis 168 htrB::pMutin4 (BFA3041), B. subtilis 168 cssS::Sp htrB::pMutin4 (BV2015), and B. subtilis 168 htrB::pMutin4 amyE::XTCbdbC (BV2034), respectively, with chromosomal DNA from B. subtilis 168 bdbC::Km (bdbC-Km) and selection for kanamycin resistance. It should be noted that deletion of bdbC severely affects the development of competence (23). Therefore, introduction of the bdbC mutation was the final step in most strain construction procedures.
FIG. 1.
Construction of mutant strains: schematic diagram of the chromosomal amyE region of B. subtilis strains containing the XTCbdbC cassette. The cassette includes the bdbC gene from B. subtilis 168 controlled by a xylose-inducible promoter (PxylA). The amyE gene was disrupted via a double-crossover recombination event with the XTCbdbC cassette from plasmid pXTCbdbC. Tcr, tetracycline resistance marker; xylR, gene encoding the XylR repressor protein; amyE′, 3′-truncated amyE gene; ′amyE, 5′-truncated amyE gene.
Proteomics.
Cells of B. subtilis were grown at 37°C with vigorous agitation in 1 liter of TY medium. After 1 h of postexponential growth, cells were separated from the growth medium by centrifugation. The secreted proteins in the growth medium were collected for two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE), gels were stained with silver nitrate or the SYPRO Ruby protein gel stain (Molecular Probes Inc.), and protein spots were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and/or N-terminal sequencing as previously described (3, 14). To visualize possible differences in extracellular protein composition, an image analysis of gels stained with the SYPRO Ruby protein gel stain (Molecular Probes Inc.) was performed using the DECODON Delta 2D software (http://www.decodon.com). Green protein spots were present predominantly in the image of the extracellular proteins of parental strain 168; red protein spots were present predominantly in the image of the extracellular proteins of cells containing plasmid pPSPhoA5; and yellow protein spots were present in both images. All quantifications were relative, and each spot volume was related to the cumulative spot volumes of the entire gel. Using the DECODON Delta 2D software, it is not possible to obtain absolute quantification or to use externally applied protein standards for calibration. All ratios were calculated from the percentage of the specific spot volumes (related to the total spot volumes of the entire gels) of the sample image divided by the percentage for the master image. Each experiment was performed at least twice.
Western blotting.
Sodium dodecyl sulfate (SDS)-PAGE was performed as described by van Dijl et al. (43). Western blot analysis was performed using a semidry Western blot apparatus as described by Kyhse-Andersen (19). After separation by SDS-PAGE, proteins were transferred to a nitrocellulose membrane (Protran; Schleicher & Schuell) and detected with specific rabbit antibodies and horseradish peroxidase-anti-rabbit immunoglobulin G conjugates.
Localization of PhoA.
The subcellular localization of PhoA was determined using the protocol described by Tjalsma et al. (40). Briefly, cells were grown overnight at 37°C in TY broth and separated from the growth medium by centrifugation. Next, the cells were resuspended in protoplast buffer (20 mM potassium phosphate [pH 7.5], 15 mM MgCl2, 20% sucrose) supplemented with 1 mg/ml lysozyme. After 30 min of incubation at 37°C, proteins released from the cells by protoplasting (i.e., the cell wall fraction) were separated from the protoplasts by centrifugation. The protoplasts were resuspended in protoplast buffer and divided into three aliquots, which were supplemented with either phosphate-buffered saline (PBS) containing 1% trypsin, PBS containing 1% trypsin and 1% Triton X-100, or PBS with nothing added (PBS contained [per liter] 8 g NaCl, 2.68 g Na2HPO4, 0.2 g KCl, and 0.24 g KH2PO4, and the pH was 7.4). After 30 min of incubation at 37°C, trypsin was inactivated by adding Complete protease inhibitors from a 10× stock solution to a final concentration of 1× as recommended by the supplier (Roche Molecular Biochemicals). All fractions were used for SDS-PAGE. PhoA and the control protein SipS were visualized by Western blotting and immunodetection with specific polyclonal antibodies.
Alkaline phosphatase assays.
Alkaline phosphatase (PhoA) activity assays were carried out essentially as described by Nicholson and Setlow (26). Strains were grown in TY medium with appropriate antibiotics. Cells were separated from the medium by centrifugation. To determine PhoA activity in medium fractions, 750-μl aliquots of the fractions were mixed with 500 μl of freshly prepared substrate (1 g/liter p-nitrophenylphosphate in 1 M Tris [pH 8.1]). The reaction mixtures were incubated at room temperature for 10 to 30 min, and the reactions were stopped by addition of 500 μl of 2 M NaOH. Fresh TY medium was used as a blank. PhoA activities, expressed in U/ml/unit of optical density at 600 nm (OD600), were determined by measuring changes in the optical density at 405 nm as a function of the time of incubation (in minutes) and the OD600. To do this, the following formula was used: [2/3 × (OD405× 352)]/(t × OD600), where t is the time of incubation. To determine PhoA activities in cellular fractions, cells were washed in 750 μl of 1 M Tris-HCl (pH 8.1) and resuspended in 750 μl of 1 M Tris-HCl to which 10 μl of 0.1% SDS and 20 μl of chloroform were added. After incubation for 5 min at room temperature, 500 μl of substrate was added. The PhoA activities in the resulting mixtures were determined as described above. All experiments were repeated at least three times.
β-Galactosidase activity assays.
To assay β-galactosidase activities, overnight cultures were diluted in fresh TY medium and grown at 37°C. Samples were taken at different times to determine the OD600 and β-galactosidase activity. The β-galactosidase assays and calculation of β-galactosidase units (Miller units; nmol/min/unit of OD600) were performed as described by Hyyryläinen et al. (12). Experiments were repeated at least twice, starting with independently obtained transformants. For all experiments, the relevant control experiments were performed in parallel. Although some differences in the absolute β-galactosidase activities were observed, the ratios of the activities in the various strains tested were largely constant. A ratio of about 1.5 was reproducible.
RESULTS
Proteomics of E. coli PhoA secretion by B. subtilis.
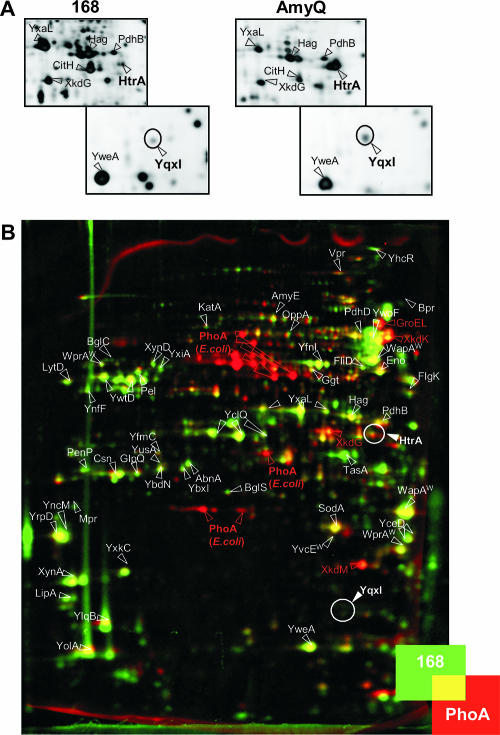
Our previous studies of the secretion stress response in B. subtilis 168 showed that high-level production of the α-amylase AmyQ results in important increases in the extracellular levels of HtrA (2). The increased extracellular HtrA level was readily visualized by proteomics (Fig. 2A). Therefore, proteomics was used as a first approach to investigate whether E. coli PhoA triggers a secretion stress response when it is produced in B. subtilis 168. To address this question, PhoA was fused to the signal peptide (prelip) and pro region (prolip) of an S. hyicus preprolipase. The combined pre and pro regions are known to act as a productive secretion signal for use in gram-positive bacteria (7, 21, 22, 37), and for efficient secretion of E. coli PhoA by B. subtilis, the preregion alone is insufficient (unpublished observations). The plasmid encoding the resulting hybrid preprolip-PhoA precursor was designated pPSPhoA5. Next, the impact of PhoA secretion on the composition of the B. subtilis exoproteome, which includes all extracellular proteins of this organism, was analyzed by 2D PAGE, MALDI-TOF MS, and dual-channel imaging. As shown in Fig. 2B, the exoproteomes of B. subtilis 168 and B. subtilis 168 producing PhoA differed significantly. This was primarily due to the presence of multiple PhoA-specific protein species in the extracellular proteome of the PhoA-producing cells (Fig. 2B, E. coli PhoA red spots). The different PhoA-specific spots correspond to different processing and degradation products of the translocated and secreted prolip-PhoA protein. Notably, detection of such processing and degradation products of prolip-PhoA was anticipated, because the prolip peptide is known to be proteolyzed upon secretion of the corresponding proproteins into the growth medium (7, 31). More unexpectedly, the levels of the prophage PBSX-specific proteins XkdG, XkdK, and XkdM were significantly greater in the growth medium of the PhoA-producing cells (Fig. 2B, red spots). The increased extracellular levels of these three proteins may have been related directly or indirectly to PhoA production (50). In contrast, many other protein spots in the 2D gel obtained with the medium fraction of PhoA-secreting cells appeared to be somewhat less intense than the corresponding spots in the 2D gel obtained with the medium fraction of parental strain 168 (Fig. 2B, yellow-green spots). This reflected a mild “dilution effect” related to the loading of equal amounts of total extracellular protein onto the two gels; due to the relatively high PhoA production level, the absolute amount of homologous extracellular proteins loaded onto a 2D gel was slightly smaller for the medium fraction of PhoA-producing cells than for the medium fraction of parental strain 168. Most important for the present study, both extracellular proteomes contained comparably small amounts of the HtrA protein (Fig. 2B, overlapping HtrA red and green spots). Quantification of the data showed that the ratio of the relative HtrA spot volumes was less than 1.5. This implies that there was not a significant difference between the HtrA levels in the two extracellular proteomes compared, a conclusion that was confirmed by an independent 2D PAGE analysis. Moreover, PhoA production did not result in extracellular appearance of the YqxI protein (in Fig. 2B the position where the YqxI spot would have appeared upon AmyQ-induced secretion stress is circled). This protein has been implicated in sporulation, but its precise role in this process is not known yet (24). As documented in our previous studies (2), detection of increased levels of extracellular YqxI would have been diagnostic for a significant secretion stress response that resulted in synthesis of elevated levels of HtrA, because increased extracellular levels of HtrA were paralleled by increased extracellular levels of YqxI (Fig. 2A).
FIG. 2.
Profile of extracellular HtrA and YqxI levels determined by proteomics. (A) B. subtilis cells producing AmyQ were grown in TY medium at 37°C, and the exoproteome of these cells was compared to the exoproteome of parental strain 168. Plasmid pKTH10L was used for AmyQ production. Extracellular proteins were harvested 1 h after entry of the cells into the stationary phase. The extracellular proteins were analyzed by 2D PAGE, MALDI-TOF MS, or N-terminal sequencing. Gels were stained with silver nitrate. Only the gel sections that are relevant for visualization of HtrA and YqxI production by AmyQ-producing cells and parental control strain 168 are shown. The proteins identified are indicated, and the HtrA and YqxI spots are circled. (B) The exoproteome of B. subtilis cells producing PhoA was compared to the exoproteome of parental strain 168 as described above for panel A. Plasmid pPSPhoA5 was used for PhoA production. After 2D PAGE, the gels were stained with SYPRO Ruby and analyzed by dual-channel imaging. Green protein spots are present predominantly in the master image of the exoproteome of parental strain 168; red protein spots are present predominantly in the image of the exoproteome of the strain producing PhoA; and comparable numbers of yellow spots are present in both exoproteomes. The image was obtained by dual-channel imaging of two representative warped 2D gels. The proteins identified are indicated, and the positions of the HtrA and YqxI spots are circled. Note that HtrB has not been detected in 2D gels so far.
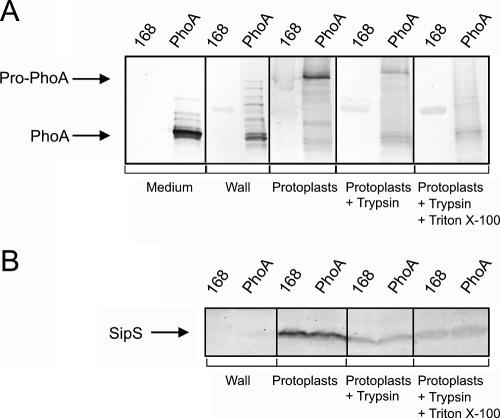
As shown by Western blotting using both growth medium and subcellular fractions (Fig. 3A), the largest amounts of PhoA produced by the cells were secreted into the growth medium. Nevertheless, significant amounts of PhoA were also present in the cells. Specifically, mature-size PhoA and various processing and degradation products of preprolip-PhoA were detected in the protoplast supernatant fraction, which was composed largely of proteins residing in the cell wall at the time of protoplasting. By contrast, the largest precursor form of PhoA was detected exclusively in protoplasts. Most likely, this precursor form represented either preprolip-PhoA or processed prolip-PhoA (designated Pro-PhoA) (Fig. 3A). Some of this Pro-PhoA was accessible to trypsin in intact protoplasts, suggesting that it was translocated across the protoplast membrane. Another fraction of Pro-PhoA was degraded by trypsin only when 1% Triton X-100 was added to lyse the protoplasts, suggesting that it represented nontranslocated prolip-PhoA. The present subcellular fractionation data were supported by the immunodetection of a membrane marker protein that was exposed on the extracytoplasmic side of the membrane, i.e., the SipS signal peptidase (40) (Fig. 3B). As shown in Fig. 3B, no SipS was detected in the wall fraction, indicating that very little lysis occurred during protoplasting. Furthermore, all trypsin-degradable SipS in intact protoplasts was degraded by incubation of the protoplasts with trypsin, whereas this was clearly not the case for PhoA. Complete degradation of the cellular PhoA precursor form required protoplast lysis with 1% Triton X-100 (Fig. 3A). The presence of translocated PhoA in the cell wall fraction is fully consistent with the results of our previously documented fractionation studies of pulse-labeled B. subtilis cells producing PhoA (40).
FIG. 3.
Secretion and subcellular localization of PhoA. Fractionation experiments were performed to localize precursor and mature forms of PhoA produced by B. subtilis 168. Plasmid pPSPhoA5 was used for PhoA production (lanes PhoA). Parental strain 168 was used as a negative control (lanes 168). Cells grown in TY medium were protoplasted, and the protoplasts were separated from the cell wall fraction by centrifugation. Protoplasts were incubated for 30 min in the presence of 1 mg/ml of trypsin with or without 1% Triton X-100, as indicated. Samples were used for SDS-PAGE and Western blotting, and specific antibodies were used to detect the precursor and mature forms of PhoA (A) or the SipS signal peptidase (B). The positions of mature-size PhoA (PhoA) and preprolip-PhoA or prolip-PhoA (Pro-PhoA) are indicated. Note that the medium and wall fractions of cells producing PhoA contained PhoA species that migrated more slowly than mature-size PhoA but faster than pro-PhoA. These PhoA species of intermediate size were most likely degradation and processing products of preprolip-PhoA.
The production and secretion of active PhoA were assessed by determining alkaline phosphatase activities in growth medium and cellular fractions. Consistent with the results of the Western blot analysis, the highest levels of PhoA activity were detected in the growth medium (17.2 ± 2.4 U/ml/unit of OD600), and much lower levels of PhoA activity were detected in the corresponding cells (1.8 ± 0.1 U/ml/unit of OD600). Notably, these activities were significantly greater than the background levels of alkaline phosphatase activity determined using the growth medium of parental strain 168 (2.1 ± 0.5 U/ml/unit of OD600) and the corresponding cells (0.3 ± 0.04 U/ml/unit of OD600).
Induction of secretion stress by PhoA.
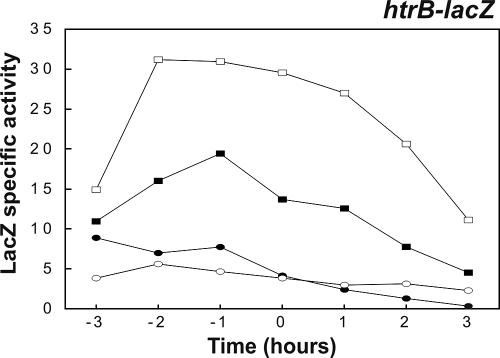
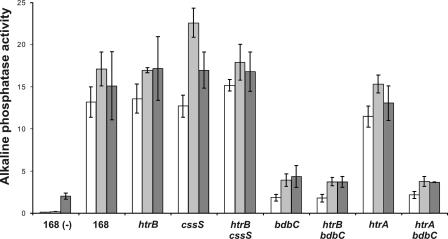
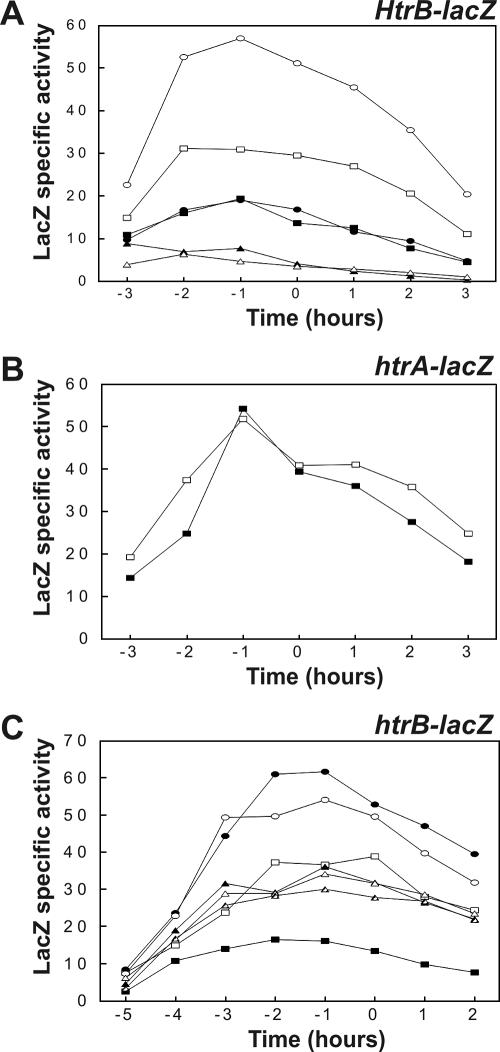
To study whether production of PhoA triggers a secretion stress response despite the lack of a detectable effect on extracellular HtrA and YqxI levels (Fig. 2B), the PhoA-encoding plasmid pPSPhoA5 was introduced into a strain (BV2003) that contains a transcriptional htrA-lacZ gene fusion (note that the htrA gene in strain BV2003 is disrupted). Next, the expression of htrA-lacZ in PhoA-producing cells grown in TY broth was monitored as a function of time. Consistent with the proteomics data, the production of PhoA did not detectably affect the expression of htrA-lacZ (data not shown). To further investigate a potential PhoA-induced secretion stress response, PhoA was also expressed in a B. subtilis strain (BFA3041) that contained a transcriptional htrB-lacZ gene fusion (note that the htrB gene of strain BFA3041 is disrupted). The rationale for studying potential effects of PhoA expression on htrB transcription was that previous studies had shown that htrB promoter activity is more sensitive to overexpression of α-amylases than htrA promoter activity is (6, 12). As judged by plating and culturing in broth, the production of PhoA affected neither the growth nor the viability of B. subtilis htrB-lacZ cells in a detectable way. The transcription of htrB was analyzed by determining β-galactosidase activity as a function of time. As shown in Fig. 4, the expression of htrB-lacZ was increased about twofold in the strain producing PhoA compared to the expression in the control strain. To verify that the increase represented a genuine secretion stress response, PhoA was produced in a strain (BV2015) that contained a cssS::Sp mutation in addition to the htrB-lacZ fusion. In this cssS mutant strain producing PhoA, htrB-lacZ was expressed at approximately the same low level that it was expressed in the cssS mutant control strain that did not produce PhoA. Interestingly, the htrB (BFA3041) and htrB cssS (BV2015) mutant strains, containing the htrB-lacZ reporter fusion and producing PhoA, secreted amounts of active PhoA into the growth medium similar to the amounts secreted by parental strain 168 producing PhoA (Fig. 5). This result was confirmed by Western blotting using PhoA-specific antibodies (not shown). Together, these observations show that disruption of htrB and/or cssS did not have a significant impact on the extracellular accumulation of active PhoA, despite the fact that PhoA production triggered a secretion stress response which resulted in an approximately twofold increase in the level of htrB expression.
FIG. 4.
Production of PhoA triggers a secretion stress response. The effect of PhoA production on htrB-lacZ transcription was analyzed in the presence and absence of an intact cssS gene. Transcriptional htrB-lacZ gene fusions were used to determine the time courses of htrB expression in cells grown in TY medium at 37°C. Plasmid pPSPhoA5 was used for PhoA production. Samples used for determination of β-galactosidase activities (in nmol/min/unit of OD600) were removed at the times indicated. Zero time was the transition point between the exponential and postexponential growth phases. The relevant genotypes and properties of the B. subtilis 168 strains used for the analyses are indicated as follows: solid rectangles, htrB-lacZ; open rectangles, htrB-lacZ and PhoA production; solid ovals, cssS htrB-lacZ; open ovals, cssS htrB-lacZ and PhoA production (see Table 1 for details). B. subtilis parental strain 168 was used as a negative control (data not shown).
FIG. 5.
Secretion of PhoA by cssS, htrA, htrB, and bdbC mutant strains. Alkaline phosphatase (PhoA) activities in the growth media of htrA-lacZ, htrB-lacZ, cssS, and bdbC single- and double-mutant strains were determined. Plasmid pPSPhoA5 was used for PhoA production. Cells were grown overnight in TY medium at 37°C, and samples were removed to determine the PhoA activities in the growth media (dark gray bars). After dilution of the overnight culture in fresh TY medium to obtain an OD600 of 0.025 and continued incubation at 37°C, samples for determination of PhoA activity were removed after 3 h (open bars) and 8 h (light gray bars). PhoA activities are expressed in U/ml/unit of OD600. The error bars indicate standard deviations. The strains used are indicated as follows: 168 (−), parental strain 168 not containing pPSPhoA5; 168, strain 168 producing PhoA; htrA, strain containing the htrA-lacZ reporter and a disrupted htrA gene; htrB, strain containing the htrB-lacZ reporter and a disrupted htrB gene; cssS, strain containing the cssS gene disrupted with an Spr marker; bdbC, strain containing the bdbC gene disrupted with a Kmr marker. All strains except parental strain 168 not containing pPSPhoA5 contained pPSPhoA5 for PhoA production (see Table 1 for details).
BdbC helps limit PhoA-induced secretion stress.
The BdbC protein, which is a homologue of the E. coli DsbB quinone oxidoreductase, has an important role in the folding of E. coli PhoA into an active and protease-resistant conformation during its export by B. subtilis (4). Figure 5 clearly supports this view and shows that a bdbC::Km mutation (referred to as the bdbC mutation) resulted in significantly reduced extracellular levels of active PhoA protein. Therefore, the effects of this bdbC mutation on htrB-lacZ expression (strain BV2032) were analyzed as a function of time under PhoA production conditions. As shown in Fig. 6A, the expression of htrB-lacZ was not affected by the bdbC mutation in strains not producing PhoA. In contrast, the production of PhoA resulted in approximately twofold induction of htrB-lacZ expression in cells containing an intact bdbC gene, similar to the results shown in Fig. 4, and fourfold induction in bdbC mutant cells. As expected, the level of active PhoA in the growth medium protein decreased significantly upon disruption of bdbC in the htrB-lacZ reporter strain (referred to as htrB bdbC) (Fig. 5). The high levels of htrB-lacZ expression in cells lacking BdbC and producing PhoA depended entirely on CssS; when PhoA was produced in an htrB-lacZ bdbC cssS mutant strain (BV2033), the levels of htrB-lacZ expression were comparable to those observed in the htrB-lacZ cssS control strain (BV2015) (Fig. 6A). Furthermore, bdbC expression was influenced neither by production of PhoA nor by disruption of cssS, as demonstrated using strains containing a transcriptional bdbC-lacZ fusion (data not shown). In contrast to htrB-lacZ expression, htrA-lacZ expression was not affected by PhoA production even in a bdbC mutant strain (BV2031) (Fig. 6B), despite the fact that introduction of the bdbC mutation into this strain resulted in significantly reduced levels of active PhoA secretion (referred to as htrA bdbC) (Fig. 5).
FIG. 6.
Effects of bdbC and cssS mutations on the PhoA-induced secretion stress response. Time courses of htrB-lacZ and htrA-lacZ expression were determined for cells grown in TY medium at 37°C. Samples for determination of β-galactosidase activities (in nmol/min/unit of OD600) were removed at the times indicated. Zero time was the transition point between the exponential and postexponential growth phases. In all experiments, B. subtilis parental strain 168 was used as a negative control (data not shown). (A) Effects of a bdbC mutation on the expression of an htrB-lacZ transcriptional fusion were analyzed in strains with and without an intact cssS gene. Plasmid pPSPhoA5 was used for production of PhoA. The relevant genotypes and properties of the B. subtilis 168 strains used for the analyses are indicated as follows: solid rectangles, htrB-lacZ; open rectangles, htrB-lacZ and PhoA production; solid ovals, htrB-lacZ bdbC; open ovals, htrB-lacZ bdbC and PhoA production; solid triangles, htrB-lacZ cssS; open triangles, htrB-lacZ cssS bdbC and PhoA production. (B) Analysis of the possible effects of a bdbC mutation on expression of an htrA-lacZ transcriptional fusion. The relevant genotypes and properties of the B. subtilis 168 strains used for the analyses are indicated as follows: solid rectangles, htrA-lacZ; open rectangles, htrA-lacZ bdbC and PhoA production. (C) The observed effects of the bdbC mutation on htrB-lacZ expression in cells producing PhoA were verified by ectopic expression of bdbC from a chromosomally integrated, xylose-inducible XTCbdbC cassette. The relevant genotypes and properties of the B. subtilis 168 strains used for the analyses are indicated as follows: solid rectangles, htrB-lacZ; open rectangles, htrB-lacZ and PhoA production; solid ovals, htrB-lacZ bdbC and PhoA production; open ovals, htrB-lacZ bdbC XTCbdbC and PhoA production in the absence of xylose; solid triangles, htrB-lacZ bdbC XTCbdbC and PhoA production in the presence of 0.5% xylose; open triangles, htrB-lacZ bdbC XTCbdbC and PhoA production in the presence of 1% xylose; striped triangles, htrB-lacZ bdbC XTCbdbC and PhoA production in the presence of 2% xylose (see Table 1 for details concerning the strains).
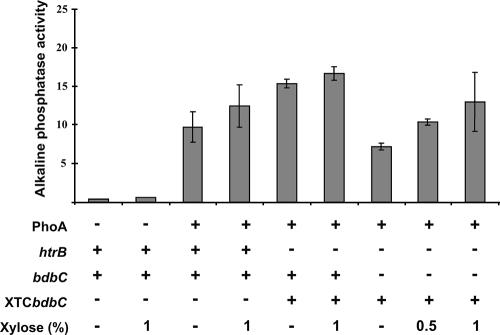
To verify that the effects of a bdbC mutation on PhoA production and the concomitant secretion stress response could be reversed by ectopic bdbC expression, we investigated the expression of htrB-lacZ in a bdbC mutant strain that contained a xylose-inducible bdbC gene cassette (BV2035). This so-called XTCbdbC cassette was integrated into the amyE locus of this strain (Fig. 1). When organisms were grown in TY broth without xylose, the expression of htrB-lacZ in bdbC XTCbdbC cells producing PhoA (Fig. 6C) was similar to that in bdbC mutant cells lacking the XTCbdbC cassette. When increasing amounts of xylose (0.5%, 1%, and 2%) were added to cultures of PhoA-producing bdbC mutant cells containing the XTCbdbC cassette, an approximately twofold decrease in the level of expression of htrB-lacZ was observed (Fig. 6C). Thus, the levels of htrB-lacZ expression in these xylose-induced cells were the same as those in the PhoA-producing control strain with a wild-type bdbC gene. As shown by determination of PhoA activity, the xylose-induced expression of the XTCbdbC cassette restored secretion of active PhoA in a bdbC mutant strain (Fig. 7). This result was confirmed by Western blotting (not shown). Notably, the htrB-lacZ bdbC XTCbdbC strain grown in the absence of xylose secreted a relatively large amount of active PhoA (Fig. 5 and 7). This was probably due to incomplete repression of the xylA promoter. Taken together, these observations show that BdbC helps the B. subtilis cells limit the PhoA-specific generation of stimuli that induce a CssS-dependent secretion stress response.
FIG. 7.
Restoration of PhoA secretion through ectopic bdbC complementation. The activities of PhoA secreted by htrB bdbC mutant strains carrying the XTCbdbC cassette for ectopic xylose-inducible bdbC expression were determined. Plasmid pPSPhoA5 was used for PhoA production. Alkaline phosphatase assays (activity is expressed in U/ml/unit of OD600) were performed with growth medium fractions after overnight growth of the strains in TY medium at 37°C. The standard deviations are indicated by error bars. The presence and absence of pPSPhoA5 for PhoA production, intact htrB and bdbC genes, and the XTCbdbC cassette are indicated by plus and minus signs, respectively (see Table 1 for details). The amounts of xylose added to the growth media are also indicated.
DISCUSSION
The aim of the present study was to characterize a possible secretion stress response in B. subtilis cells producing a heterologous disulfide bond-containing secretory protein. The PhoA alkaline phosphatase of E. coli was the reporter protein of choice, because it contains two disulfide bonds and relatively high levels of it can be secreted by B. subtilis when it is fused to the preprolip region of a lipase from S. hyicus (4). Accordingly, E. coli PhoA is an appropriate model for studies of potential limitations in the biotechnological production of disulfide bond-containing proteins in Bacillus species. The present results show that the production of PhoA in B. subtilis does induce a secretion stress response. This response is, however, relatively mild compared to the previously documented secretion stress responses that were triggered by high-level production of α-amylases from different Bacillus species in B. subtilis (6, 12). Furthermore, the PhoA-induced response is paralleled by the presence of translocated PhoA molecules in the cell wall. This finding is consistent with the previous observation that the increased accumulation of wall-associated forms of AmyQ is paralleled by an increase in the secretion stress response (12). Notably, these parallel events suggest, but do not prove, that there is a causative connection between the appearance of cell wall-associated forms of PhoA or AmyQ and secretion stress.
E. coli PhoA is known to require the BdbC-dependent pathway for efficient folding during secretion by B. subtilis (4). The present study showed that the intensity of the PhoA-induced secretion stress response depends on the presence of the BdbC protein. In cells containing an intact bdbC gene, the secretion stress response induced by PhoA production resulted in about twofold-higher levels of htrB transcription. This relatively mild stress response could not be counteracted by the xylose- induced ectopic overexpression of bdbC from the XTCbdbC cassette. Together, these observations suggest that under the experimental conditions tested, BdbC does not represent a major bottleneck for the folding of translocated E. coli PhoA in cells of B. subtilis parental strain 168. Thus, it seems that another, unidentified extracytoplasmic folding factor is limiting upon PhoA production.
When PhoA was produced in bdbC mutant cells, the cellular secretion stress response was significantly increased. Most likely, this response was triggered by malfolded translocated PhoA, which was directly or indirectly sensed by CssS. Importantly, this increased stress response could be completely reversed by ectopic expression of BdbC from the XTCbdbC cassette. Taken together, the present observations suggest that the stimuli that are sensed by the CssRS two-component system upon production of PhoA can be derived from at least two stressful events: PhoA accumulation in the cell wall due to an unidentified limiting factor and a block in the BdbCD-dependent pathway for posttranslocational PhoA folding. Both events result in stimulation of htrB transcription. In turn, this should lead to synthesis of elevated amounts of HtrB, which can counteract potentially harmful accumulation of malfolded proteins. Although the present study suggested that inefficient disulfide bonding in PhoA is the primary reason for the increased secretion stress response in cells with impaired BdbCD function, this idea should be verified carefully, for example, by employing a Cys-less variant of preprolip-PhoA. Such studies should also address the question whether a PhoA-induced secretion stress response is also detectable in B. subtilis cells containing an intact htrB gene in order to evaluate whether PhoA molecules with and without Cys residues are effectively degraded by HtrB.
Irrespective of the presence of BdbC, the transcription of htrA was not detectably induced by production of PhoA in B. subtilis. Consistent with this observation, PhoA production did not result in increased extracellular levels of HtrA and YqxI. These observations probably are related to the fact that PhoA induces only a mild secretion stress response that is detectable as a twofold increase in htrB transcription. The finding that PhoA triggers such a mild secretion stress response that does not result in detectable stimulation of htrA transcription is remarkable, because significantly increased htrA transcription is detectable upon AmyQ production at levels that are about threefold lower than the current level of PhoA production (2; unpublished observations). This suggests that the threshold level for induction of htrA expression by AmyQ is significantly lower than the threshold level for induction of htrA expression by PhoA. If this is true, it might indicate that compared to the levels of PhoA, higher levels of malfolded AmyQ are present at critical locations in the B. subtilis cell envelope, at least under the conditions tested. Alternatively, the CssRS system might be more sensitive to malfolded AmyQ than to malfolded PhoA. If expression of PhoA results in a slight increase in htrA transcription or HtrA production, this increase is too small to be clearly detected even in the absence of BdbC. Our previous studies have shown that the intensity of the AmyQ-induced secretion stress response depends on the level of AmyQ production (48). Accordingly, it is conceivable that the current production levels of PhoA are simply too low to trigger a secretion stress response that detectably affects htrA transcription. If this is true, production of significantly higher levels of PhoA should elicit a much stronger secretion stress response, especially if the BdbC-dependent folding pathway becomes limiting for PhoA folding.
Finally, the present observations corroborate the view that B. subtilis employs both CssRS-dependent expressed genes and CssRS-independent genes to combat the detrimental effects of secretion stress. The first class of genes consists of genuine members of the CssRS regulon, such as htrA and htrB (6, 12). The second class consists of CssRS-independent expressed genes that specify posttranslocational protein folding activities. The best-characterized representative of the second class is the prsA gene, which encodes a protein folding catalyst with peptidyl-prolyl cis/trans isomerase activity (45). Importantly, PrsA is indispensable for proper folding of a variety of secretory proteins, including α-amylases (12, 16, 46). In the present study, we found that the bdbC gene, which encodes a major determinant for folding of disulfide bond-containing exported proteins in B. subtilis, also belongs to the class of CssRS-independent expressed genes required to prevent or limit secretion stress. Notably, the CpxAR system of E. coli, which is related to the CssRS system of B. subtilis (12), triggers increased production of periplasmic protein folding catalysts in response to accumulation of malfolded proteins at the inner membrane (8, 29). In addition to the HtrA chaperone protease, these folding catalysts include E. coli proteins involved in disulfide bond handling (DsbA and DsbC) and peptidyl-prolyl cis/trans isomerization (PpiA and PpiD). Therefore, it seems that the CssRS system of B. subtilis and the CpxAR system of E. coli have different impacts on the regulation of extracytoplasmic protein refolding. This leads to the more general conclusion that, even though the fundamental principles for extracytoplasmic protein folding are conserved in both organisms, the compositions and regulation of the extracytoplasmic protein folding machinery are very different. Accordingly, different strategies are needed for construction of E. coli and B. subtilis strains with improved properties for posttranslocational protein folding. The present results focus attention on the BdbC-dependent folding pathway for biotechnological production of proteins with disulfide bonds in B. subtilis and related bacilli.
Acknowledgments
We thank H. Tjalsma, H. Westers, G. E. Zanen, D. Noone, K. Devine, and other members of the Groningen and European Bacillus Secretion Groups for stimulating discussions and Decodon GmbH for cooperation and access to new software tools.
E.D. was supported by the Ubbo Emmius Foundation of the University of Groningen. R.D., J.M., H.A., J.-Y.F.D., R.F., O.P.K., W.J.Q., M.H., S.B., and J.M.V.D. were supported in part by European Union grants BIO4-CT98-0250, QLK3-CT-1999-00413/00917, LSHC-CT-2004-503468, and LSHG-CT-2004-005257. H.A. and M.H. were supported by the “Deutsche Forschungsgemeinschaft,” the “Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie,” and the “Fonds der Chemischen Industrie.”
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 4.Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl. 1999. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274:24531-24538. [DOI] [PubMed] [Google Scholar]
- 5.Chung, Y. S., F. Breidt, and D. Dubnau. 1998. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol. Microbiol. 29:905-913. [DOI] [PubMed] [Google Scholar]
- 6.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demleitner, G., and F. Götz. 1994. Evidence for importance of the Staphylococcus hyicus lipase pro-peptide in lipase secretion, stability and activity. FEMS Microbiol. Lett. 121:189-197. [DOI] [PubMed] [Google Scholar]
- 8.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorenbos, R., J. M. van Dijl, and W. J. Quax. 2005. Thiol-disulfide oxidoreductases in bacteria, p. 237-269. In S. G. Pandalai (ed.), Recent research developments in microbiology, vol. 9. Research Signpost, Kerala, India. [Google Scholar]
- 10.Dorenbos, R., T. Stein, J. Kabel, C. Bruand, A. Bolhuis, S. Bron, W. J. Quax, and J. M. van Dijl. 2002. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 277:16682-16688. [DOI] [PubMed] [Google Scholar]
- 11.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 12.Hyyryläinen, H. K., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Prágai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system of Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 13.Hyyryläinen, H. K., M. Sarvas, and V. Kontinen. 2005. Transcriptome analysis of the secretion stress response of Bacillus subtilis. Appl. Microbiol. Biotechnol. 67:389-396. [DOI] [PubMed] [Google Scholar]
- 14.Jongbloed, J. D. H., H. Antelmann, M. Hecker, R. Nijland, S. Bron, U. Airaksinen, F. Pries, W. J. Quax, J. M. van Dijl, and P. G. Braun. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068-44078. [DOI] [PubMed] [Google Scholar]
- 15.Kim, L., A. Mogk, and W. Schumann. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71-76. [DOI] [PubMed] [Google Scholar]
- 16.Kontinen, V. P., and M. Sarvas. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8:727-737. [DOI] [PubMed] [Google Scholar]
- 17.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 18.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyhse-Andersen, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 20.Leskela, S., V. P. Kontinen, and M. Sarvas. 1996. Molecular analysis of an operon in Bacillus subtilis encoding a novel ABC transporter with a role in exoprotein production, sporulation and competence. Microbiology 142:71-77. [DOI] [PubMed] [Google Scholar]
- 21.Meens, J., E. Frings, M. Klose, and R. Freudl. 1993. An outer membrane protein (OmpA) of Escherichia coli can be translocated across the cytoplasmic membrane of Bacillus subtilis. Mol. Microbiol. 9:847-855. [DOI] [PubMed] [Google Scholar]
- 22.Meens, J., M. Herbort, M. Klein, and R. Freudl. 1997. Use of the pre-pro part of Staphylococcus hyicus lipase as a carrier for secretion of Escherichia coli outer membrane protein A (OmpA) prevents proteolytic degradation of OmpA by cell-associated protease(s) in two different gram-positive bacteria. Appl. Environ. Microbiol. 63:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meima, R., C. Eschevins, S. Fillinger, A. Bolhuis, L. W. Hamoen, R. Dorenbos, W. J. Quax, J. M. van Dijl, R. Provvedi, I. Chen, D. Dubnau, and S. Bron. 2002. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J. Biol. Chem. 277:6994-7001. [DOI] [PubMed] [Google Scholar]
- 24.Molle, V., M. Fujita, S. T. Jensen, P. Eichenberger, J. E. Gonzalez-Pastor, J. S. Liu, and R. Losick. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683-1701. [DOI] [PubMed] [Google Scholar]
- 25.Nakamoto, H., and J. C. Bardwell. 2004. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta 1694:111-119. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom.
- 27.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 28.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 29.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 30.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55:21-48. [DOI] [PubMed] [Google Scholar]
- 31.Rollof, J., and S. Normark. 1992. In vivo processing of Staphylococcus aureus lipase. J. Bacteriol. 174:1844-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Sarvas, M., C. R. Harwood, S. Bron, and J. M. van Dijl. 2004. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1694:311-327. [DOI] [PubMed] [Google Scholar]
- 34.Sone, M., S. Kishigami, T. Yoshihisa, and K. Ito. 1997. Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272:6174-6178. [DOI] [PubMed] [Google Scholar]
- 35.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 36.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturmfels, A., F. Götz, and A. Peschel. 2001. Secretion of human growth hormone by the food-grade bacterium Staphylococcus carnosus requires a propeptide irrespective of the signal peptide. used. Arch. Microbiol. 175:295-300. [DOI] [PubMed] [Google Scholar]
- 38.Tjalsma, H., H. Antelmann, J. D. H. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjalsma, H., S. Bron, and J. M. van Dijl. 2003. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278:15622-15632. [DOI] [PubMed] [Google Scholar]
- 41.Tjalsma, H., and J. M. van Dijl. 2005. Proteomics-based consensus prediction of protein retention in a bacterial membrane. Proteomics 5:4472-4482. [DOI] [PubMed] [Google Scholar]
- 42.van Dijl, J. M., A. Bolhuis, H. Tjalsma, J. D. H. Jongbloed, A. de Jong, and S. Bron. 2001. Protein transport pathways in Bacillus subtilis: a genome-based road map, p. 337-355. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 43.van Dijl, J. M., A. de Jong, H. Smith, S. Bron, and G. Venema. 1991. Non-functional expression of Escherichia coli signal peptidase I in Bacillus subtilis. J. Gen. Microbiol. 137:2073-2083. [DOI] [PubMed] [Google Scholar]
- 44.van Dijl, J. M., A. de Jong, G. Venema, and S. Bron. 1995. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. Structural and functional similarities with LexA-like proteases. J. Biol. Chem. 270:3611-3618. [DOI] [PubMed] [Google Scholar]
- 45.Vitikainen, M., I. Lappalainen, R. Seppala, H. Antelmann, H. Boer, S. Taira, H. Savilahti, M. Hecker, M. Vihinen, M. Sarvas, and V. P. Kontinen. 2004. Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J. Biol. Chem. 279:19302-19314. [DOI] [PubMed] [Google Scholar]
- 46.Vitikainen, M., T. Pummi, U. Airaksinen, E. Wahlstrom, H. Wu, M. Sarvas, and V. P. Kontinen. 2001. Quantitation of the capacity of the secretion apparatus and requirement for PrsA in growth and secretion of alpha-amylase in Bacillus subtilis. J. Bacteriol. 183:1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 48.Westers, H., E. Darmon, G. Zanen, J. W. Veening, O. P. Kuipers, S. Bron, W. J. Quax, and J. M. van Dijl. 2004. The Bacillus secretion stress response is an indicator for alpha-amylase production levels. Lett. Appl. Microbiol. 39:65-73. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 50.Zanen, G., H. Antelmann, H. Westers, M. Hecker, J. M. van Dijl, and W. J. Quax. 2004. FlhF, the third signal recognition particle-GTPase of Bacillus subtilis, is dispensable for protein secretion. J. Bacteriol. 186:5956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]