Abstract
The immunoglobulin G (IgG) binding ZZ domain of protein A from Staphylococcus aureus was fused to the N terminus of the polyhydroxyalkanoate (PHA) synthase from Cupriavidus necator. The fusion protein was confirmed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry and mediated formation of ZZ domain-displaying PHA granules in recombinant Escherichia coli. The IgG binding capacity of isolated granules was assessed using enzyme-linked immunosorbent assay and could be enhanced by the overproduction of the ZZ-PHA synthase. ZZ-PHA granules enabled efficient purification of IgG from human serum.
The polyhydroxyalkanoate (PHA) synthase is the key enzyme of PHA biosynthesis and PHA granule formation. PHA granules (biopolyester particles) are formed inside bacterial cells, based on the activity and biochemical properties of the PHA synthases (8, 9). The PHA granule core is composed of high-molecular-weight PHA, which is biodegradable and biocompatible. The surface of the PHA granule is composed of a phospholipid membrane with embedded or attached proteins. Evidence was provided that the PHA synthase is covalently attached to the PHA granule core (5). Recently, the enzyme β-galactosidase and green fluorescent protein (GFP) were immobilized to the PHA granules by use of PHA synthase engineering (5, 6). Phasin proteins have been also subjected to protein engineering in order to enable purification of proteins fused to these proteins, which hydrophobically attach to the preformed PHA granules (1, 2). In this study, we targeted the PHA synthase with respect to display of the immunoglobulin G (IgG) binding domain ZZ of protein A at the PHA granule surface. Only the PHA synthase provides covalent attachment to the PHA granule surface and hence provides a robust particle-based purification system (8, 10).
Construction of plasmids mediating ZZ-PHA granule production in Escherichia coli.
The plasmid pCWE, encoding the PHA synthase from Cupriavidus necator, and plasmid pEZZ18 (GE Healthcare) (providing the ZZ domain- and signal peptide-encoding sequences; GenBank accession no. M74186) were used to generate plasmids encoding the respective PHA synthase fusion proteins (Table 1) (4). The DNA regions encoding the ZZ domain with or without the signal peptide were amplified from vector pEZZ18 by using oligonucleotides introducing NdeI sites at each end of the PCR product (Table 1). Each PCR product was then inserted into the NdeI site of plasmid pCWE, resulting in plasmids pCWE-ZZ(+)phaC and pCWE-ZZ(−)phaC, respectively (Table 1). Each hybrid gene was subcloned into XbaI/BamHI sites of plasmid pBHR69 upstream of the genes phbA and phbB, which mediate provision of the activated precursor for polyhydroxybutyrate synthesis (3). This resulted in plasmids pBHR69-ZZ(+)phaC and pBHR69-ZZ(−)phaC (Table 1). To investigate whether the entire open reading frame encoding the respective fusion protein could be overproduced at the PHA granule surface, the respective hybrid genes were also subcloned into overexpression vector pET14b downstream of the strong T7 promoter (Table 1). The resulting plasmids pET14b-ZZ(+)phaC and pET14b-ZZ(−)phaC, encoding ZZ-PhaC with or without the signal peptide, respectively, were transformed into E. coli BL21(DE3)/pLysS harboring pMCS69 (phbA phbB). The PHA synthase function of the fusion proteins was assessed by analyzing PHA accumulation of respective cells by gas chromatography-mass spectrometry analysis as previously described (6). No major differences in PHA accumulation could be detected compared to cells harboring pCWE or pHAS and pMCS69 as a control (data not shown). These data suggested that the ZZ-PHA synthase fusion protein mediates PHA biosynthesis and PHA granule formation. The presence or absence of the signal peptide did not affect PHA synthase function.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Descriptiona | Source or reference |
|---|---|---|
| E. colistrains | ||
| BL21(DE3)/pLysS | F−; ompT hsdSB(rB− mB−) gal dcm(DE3); pLysS (Camr) | Invitrogen |
| XL1-Blue | E. coli cloning strain | Stratagene |
| Plasmids | ||
| pEZZ18 | Apr; Lac promoter, encoding the IgG binding domain ZZ from protein A | GE Healthcare |
| pHAS | pET-14b containing NdeI/BamHI-inserted phaCWe gene from C. necator | 11 |
| pET14b | Apr; T7 promoter | Novagen |
| pET14b-ZZ(−)phaC | pET14b containing XbaI/BamHI fragment comprising gene ZZ-phaC without the signal sequence-encoding region | This study |
| pET14b-ZZ(+)phaC | pET14b containing XbaI/BamHI fragment comprising gene ZZ-phaC | This study |
| pCWE | pBluescript SK(−) derivative containing the PHA synthase gene from C. necator | 6 |
| pCWE-ZZ(−)phaC | pCWE derivative containing the ZZ domain-encoding NdeI fragment lacking the signal sequence-encoding region | This study |
| pCWE-ZZ(+)phaC | pCWE derivative containing the ZZ domain-encoding NdeI fragment | This study |
| pBHR69-ZZ(−)phaC | pBHR69 derivative containing the hybrid PHA synthase gene from pCWE-ZZ(−)phaC upstream of genes phaA and phaB of C. necatorcolinear to lacpromoter | This study |
| pBHR69-ZZ(+)phaC | pBHR69 derivative containing the hybrid PHA synthase gene from pCWE-ZZ(−)phaC upstream of genes phaA and phaB of C. necatorcolinear to lacpromoter | This study |
| pBHR69 | pBluescript derivative containing genes phaA and phaB of C. necatorcolinear to lac promoter | 7 |
| pMCS69 | pBBR1MCS derivative containing genes phaA and phaB of C. necatorcolinear to lac promoter | 3 |
| Oligonucleotides | ||
| 5′-ZZ (+)-NdeI | 5′-GCGCGCATATGACTTTACAAATACATACAGGGGGTATTAATTTG-3′ | This study |
| 3′-ZZ-NdeI | 5′-GTAATCATATGGGGTACCGAGCTCGAATTCGCGTCTAC-3′ | This study |
Restriction enzyme recognition sites are underlined.
Production of the ZZ-PhaC fusion proteins.
The ZZ domain of protein A was chosen in this study as an example of a binding domain to be covalently attached to the PHA granule surface. PHA granules, whose formation was mediated by the respective fusion proteins, were isolated and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis as previously described (5). ZZ-PhaC plus the N-terminal signal peptide has a theoretical molecular weight of 83,981 and a protein with an apparent molecular mass of 84 kDa could be detected as the predominant protein (data not shown). Without the signal peptide the fusion protein has a theoretical molecular weight of 79,338, and a protein with an apparent molecular mass of 80 kDa appeared as the predominant protein (data not shown). The identities of these proteins were confirmed by peptide fingerprinting using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Thus, both open reading frames could be efficiently and completely expressed in E. coli. The plasmids pET14b-ZZ(+)phaC and pET14b-ZZ(−)phaC, encoding ZZ-PhaC with and without the signal peptide, respectively, mediated overproduction of ZZ-PhaC at the PHA granule surface. Overall, these findings were consistent with previous studies, which demonstrated that GFP and LacZ could be fused to the N terminus of PHA synthases, enabling production of GFP-labeled PHA granules as well as PHA granules with immobilized LacZ (5, 6).
Display of the ZZ domain at the PHA granule surface and binding capacity of ZZ-PHA granules.
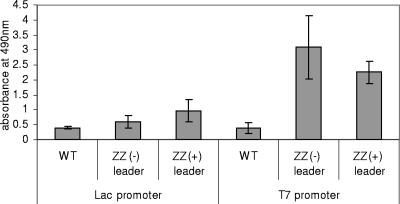
Since the current model of PHA granule formation suggests that the PHA synthase stays covalently attached to the emerging biopolyester granule (5, 8-10), the ZZ domain is presumably exposed at the surface of the PHA granule. To localize the ZZ domain at the PHA granule surface, PHA granules of E. coli harboring plasmid pCWE-ZZ(+)phaC, pCWE-ZZ(−)phaC, pET14b-ZZ(+)phaC, or pET14b-ZZ(−)phaC, as well as PHA granules produced by wild-type PHA synthase (pCWE or pHAS), were isolated and used for enzyme-linked immunosorbent assay (ELISA) as previously described (5). Specific binding of IgG to PHA granules isolated from E. coli harboring any plasmid encoding a ZZ-PHA synthase fusion protein was suggested by at least a twofold increase in absorption at a wavelength of 490 nm compared to the wild-type PHA granules (Fig. 1). These data suggested a functional display of the ZZ domain at the PHA granule surface. The presence or absence of the signal peptide did not affect IgG binding capacity. However, PHA granules whose formation was mediated by overproduction of ZZ-PhaC showed significantly increased binding capacity (Fig. 1).
FIG. 1.
ELISA using various PHA granules and anti-IgG antibodies for the detection of IgG bound to PHA granules. PHA granules were isolated from recombinant E. coli harboring various plasmids. Plasmids contained either the lac promoter or the T7 promoter for gene expression. The following versions of the PHA synthase mediated production of PHA granules: WT, wild-type PHA synthase; ZZ(−), ZZ-PHA synthase without signal peptide; ZZ(+), ZZ-PHA synthase plus signal peptide. Goat polyclonal anti-human IgG-horseradish peroxidase conjugates were used for detection of bound human IgG. Equal amounts of PHA granule protein (0.37 μg), corresponding to 2.6 μg polyhydroxybutyrate, were added to each well. Measurements were conducted in quadruplicate, and the mean value and the standard deviation are indicated.
Purification of IgG from human serum by using ZZ-PHA granules and stability of ZZ-PHA granules.
PHA granules displaying the IgG binding domain ZZ from protein A derived from pET14b-ZZ(−)phaC were used for IgG purification from human serum. For comparative analysis, protein A-Sepharose beads with immobilized, recombinant protein A were also used to purify IgG. IgG purification was conducted according to protein A-Sepharose 4B bead purification protocols (Sigma). SDS-PAGE analysis of eluted proteins showed that the immunoglobulins (a protein representing the heavy chains, with an apparent molecular mass of 50 kDa, and a protein representing the light chains, with an apparent molecular weight of 25 kDa) were purified from human serum by using the ZZ-PHA granules displaying the ZZ domain as part of the PHA synthase on the surfaces of the granules. The immunoglobulins eluted from PHA granules at pH 2.7 and showed a high degree of purity comparable to that of the commercially available protein A-Sepharose beads (Fig. 2). PHA granules formed by wild-type PHA synthase did not show elution of proteins, suggesting that unspecific binding of serum proteins does not interfere with IgG purification and that the ZZ domain mediates IgG purification (Fig. 2). ZZ-PHA granules were subjected to repeated purification cycles, demonstrating consistent purification performance and strong stability (data not shown). Temperature stability was tested by subjecting ZZ-PHA granules to increasing temperatures and then assessing the IgG binding capacity by ELISA. At 60°C, the binding capacity was dropping to 60%, suggesting that the ZZ domain was partially unfolding (data not shown). Control PHA granules containing only wild-type PHA synthase showed only low levels of unspecific binding which were temperature independent.
FIG. 2.
SDS-PAGE analysis of proteins bound in vitro to either ZZ-PHA granules or protein A-Sepharose and released after elution. Lanes: M, molecular weight standard; 1, human serum; 2, proteins eluted from protein A-Sepharose beads; 3, proteins eluted from wild-type PHA granules; 4, proteins eluted from ZZ-PHA granules displaying the ZZ domain without signal sequence. The heavy and light chains of IgG are indicated.
To our surprise, the engineered ZZ-PHA granules performed equally to commercial protein A-Sepharose beads with respect to IgG purification (Fig. 2). This result in combination with the strong stability of the ZZ-PHA granules outside the bacterial cell opens up a new and interesting field of biotechnological applications for these biopolyester particles.
This study demonstrated that protein engineering of the PHA synthase provides a platform technology for efficient covalent enzyme/protein immobilization (5). Commercial protein A beads require the in vitro production of polymer beads and subsequently the chemical cross-linking of purified protein A. PHA granule-based beads with covalently attached protein function are produced in one step by recombinant bacteria, suggesting a commercially viable biotechnological production process (5). The PHA synthase contains all the inherent properties required for PHA synthesis as well as PHA granule formation and can be produced in a variety of organisms (9). These unique properties and covalent binding to the PHA granule make these enzymes an ideal tool for functionalization of PHA granules (10).
Acknowledgments
This study was supported by research grants from Massey University and PolyBatics Ltd. V.P. received a Ph.D. scholarship from Massey University.
Proteomic analysis was performed by S. König (Integrated Functional Genomics, Interdisciplinary Center for Clinical Research, University of Münster, Germany). We thank J. Sommer-Knudsen (Innovation Purification Technologies, Sydney, Australia) for scientific discussion.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Banki, M. R., T. U. Gerngross, and D. W. Wood. 2005. Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association. Protein Sci. 14:1387-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, G. C., J. D. McCool, D. W. Wood, and T. U. Gerngross. 2005. Integrated recombinant protein expression and purification platform based on Ralstonia eutropha. Appl. Environ. Microbiol. 71:5735-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann, N., A. A. Amara, B. B. Beermann, Q. Qi, H. J. Hinz, and B. H. A. Rehm. 2002. Biochemical characterization of the Pseudomonas putida 3-hydroxyacyl ACP:CoA transacylase, which diverts intermediates of fatty acid de novo biosynthesis. J. Biol. Chem. 277:42926-42936. [DOI] [PubMed] [Google Scholar]
- 4.Lowenadler, B., B. Jansson, S. Paleus, E. Holmgren, B. Nilsson, T. Moks, G. Palm, S. Josephson, L. Philipson, and M. Uhlen. 1987. A gene fusion system for generating antibodies against short peptides. Gene 58:87-97. [DOI] [PubMed] [Google Scholar]
- 5.Peters, V., and B. H. A. Rehm. 2006. In vivo enzyme immobilization by use of engineered polyhydroxyalkanoate synthase. Appl. Environ. Microbiol. 72:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters, V., and B. H. A. Rehm. 2005. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol. Lett. 248:93-100. [DOI] [PubMed] [Google Scholar]
- 7.Qi, Q., and B. H. A. Rehm. 2001. Polyhydroxybutyrate biosynthesis in Caulobacter crescentus: molecular characterization of the polyhydroxybutyrate synthase. Microbiology 147:3353-3358. [DOI] [PubMed] [Google Scholar]
- 8.Rehm, B. H. A. 2006. Genetics and biochemistry of polyhydroxyalkanoate granule self-assembly: the key role of polyester synthases. Biotechnol. Lett. 28:207-213. [DOI] [PubMed] [Google Scholar]
- 9.Rehm, B. H. A. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehm, B. H. A. 2006. Biopolyester particles produced by microbes or using polyester synthases: self-assembly and potential applications, p. 1-34. In B. H. A. Rehm (ed.), Microbial bionanotechnology: biological self-assembly systems and biopolymer-based nanostructures. Horizon Bioscience, Wymondham, United Kingdom.
- 11.Yuan, W., Y. Jia, J. Tian, K. D. Snell, U. Muh, A. J. Sinskey, R. H. Lambalot, C. T. Walsh, and J. Stubbe. 2001. Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch. Biochem. Biophys. 394:87-98. [DOI] [PubMed] [Google Scholar]