Abstract
We present the first detailed phylogenetic analysis of Buggy Creek virus (BCRV), a poorly known alphavirus with transmission cycles involving a cimicid swallow bug (Oeciacus vicarius) vector and cliff swallows (Petrochelidon pyrrhonota) and house sparrows (Passer domesticus) as the principal avian hosts. Nucleotide sequences of a 2,075-bp viral envelope glycoprotein-coding region, covering the entire PE2 gene, were determined for 33 BCRV isolates taken from swallow bugs at cliff swallow colonies in Nebraska and Colorado in the summer of 2001 and were compared with the corresponding region of BCRV isolates collected from Oklahoma in the 1980s. We also analyzed isolates of the closely related Fort Morgan virus (FMV) collected from Colorado in the 1970s. Phylogenetic analysis indicated that BCRV falls into the western equine encephalomyelitis complex of alphaviruses, in agreement with antigenic results and a previous alphavirus phylogeny based on the E1 coding region. We found four distinct BCRV/FMV clades, one each unique to Nebraska, Colorado, and Oklahoma and one containing isolates from both Nebraska and Colorado. BCRV isolates within the two clades from Nebraska showed 5.7 to 6.2% nucleotide divergence and 0.7 to 1.9% amino acid divergence, and within these clades, we found multiple subclades. Nebraska subclades tended to be confined to one or a few cliff swallow colonies that were close to each other in space, although in some cases, near-identical isolates were detected at sites up to 123 km apart. Viral gene flow occurs when cliff swallows move (bugs) between colony sites, and the genetic structure of BCRV may reflect the limited dispersal abilities of its insect vector.
Buggy Creek virus (BCRV) is a poorly known alphavirus (Togaviridae) that is part of the western equine encephalomyelitis antigenic complex, which includes other alphaviruses such as Sindbis virus (SINV), western equine encephalomyelitis virus (WEEV), Highlands J virus (HJV), and Fort Morgan virus (FMV) (9, 17, 29, 37). Like other members of the WEEV complex, BCRV is a natural recombinant virus derived from Old World SINV and New World eastern equine encephalomyelitis (EEEV) (29, 41). BCRV (and the closely related FMV) is apparently widely distributed in North America, having been found in Texas, Oklahoma, Nebraska, Colorado, South Dakota, and Washington state (7, 8, 17). It was first isolated in 1980 at Buggy Creek in Grady County, Oklahoma (17, 22), although the ecologically very similar FMV was discovered in the 1970s in Colorado (8, 14).
Like most alphaviruses, BCRV is a vector-borne virus with transmission cycles involving an invertebrate vector and a vertebrate host. BCRV is unusual among alphaviruses, however, in that it is routinely vectored by an invertebrate other than mosquitoes: this virus is commonly associated with the cimicid swallow bug (Hemiptera: Cimicidae: Oeciacus vicarius). The bug is an ectoparasite of the colonially nesting cliff swallow (Petrochelidon pyrrhonota) and, to a lesser extent, the introduced house sparrow (Passer domesticus), and both birds serve as hosts to BCRV (14, 17, 34). FMV is also associated with swallow bugs, cliff swallows, and house sparrows. Because the wingless swallow bugs are sedentary and confined to active and inactive cliff swallow nests during most of the year (5, 21), the spatial foci for virus occurrence are predictable and allow the study of how genetic divergence of BCRV potentially varies from site to site over relatively small spatial scales without making assumptions as to the vectors' geographic origin. There is a close antigenic relationship between BCRV and the other WEEV complex alphaviruses (8, 9, 14, 17) that have medical and veterinary significance, and consequently, the study of how BCRV may undergo spatial structuring and evolutionary change may provide important information on this virus group that could not be obtained in studies where volant arthropods (e.g., mosquitoes) are the primary vectors.
To understand more about the evolution of BCRV, we performed a phylogenetic analysis of BCRV isolates collected from bugs in cliff swallow colonies in (primarily) southwestern Nebraska in summer 2001. Our analysis also included the first described strains of BCRV, collected from Oklahoma in the 1980s, and FMV isolates from Colorado in the 1970s. One of our objectives was to determine the phylogenetic relationship of FMV to BCRV. These viruses share ecological, antigenic, and nucleotide sequence similarities (9, 29; R. M. Kinney and M. Pfeffer, unpublished data) and may be strains of the same virus.
We based our phylogeny on the majority of the structural protein genes encoded by the viral subgenomic 26S RNA, including the entire PE2 gene (which encodes the E2 and E3 viral envelope proteins). Most of the major neutralizing epitopes are located on the E2 glycoprotein (29, 37). Under the pressure of a vertebrate immune system, mutations on this gene may help the virus escape destruction. Thus, if there are functional differences among virus isolates, they are likely to be expressed in the E2 gene, and consequently, this region should be highly informative for the study of virus diversification and evolution. This work represents the first phylogenetic survey of BCRV within a population, and from the results, we describe four separate clades of BCRV in the swallow bug vectors of the western Great Plains. We also analyze how genetic similarity of the isolates is related to colony of origin. The results should help us understand patterns of evolutionary change in this alphavirus and how these changes may be related to ecological variables.
MATERIALS AND METHODS
Study organisms.
The swallow bug is an ectoparasite primarily of cliff swallows and is found throughout the bird's wide geographic range (4, 24). Swallow bugs are nest-based parasites that overwinter in cliff swallows' nests or in the cracks and crevices of the nesting substrate near the nests. They are hematophagous, feeding on the birds mostly at night, and they travel on the adult birds relatively rarely (5, 6, 12, 21). Infestations can reach up to 2,600 bugs per nest, and the bugs affect many aspects of cliff swallow life history (2, 3, 5, 10, 23). Swallow bugs are long-lived ectoparasites that begin to reproduce as soon as they feed in the spring. The bugs have access to hosts only when cliff swallows occupy a colony site or reuse existing nests. The birds do not use all of the colony sites in a given year (5), and the bugs seem to be adapted to withstanding long periods of host absence, in some cases persisting at a site not used by cliff swallows for up to perhaps four consecutive years (21, 23, 30, 35). The bugs appear capable of parasitizing introduced house sparrows (Passer domesticus) that occupy nests in some cliff swallow colonies (7, 17; C. Brown et al., personal observation).
Cliff swallows are highly colonial passerines that breed throughout most of western North America (4). They build gourd-shaped mud nests and attach them to the vertical faces of cliff walls, rock outcrops, or artificial sites such as the eaves of buildings or bridges. Cliff swallows are migratory, wintering in southern South America, and have a relatively short breeding season in North America from late April to late July.
House sparrows were introduced into North America from Europe in the late 1800s and are found in all parts of the United States (20). They are nonmigratory. House sparrows usurp active cliff swallow nests and will occupy them until the nests fall from the substrate. Numbers of sparrows vary among colony sites, with many colonies having none. Sparrows are resident around the swallow colonies throughout the year, but the extent of their use of nests (and thus their exposure to bugs) in the nonbreeding season (e.g., for winter roosting) and the degree to which they move between colony sites are unknown.
Study sites.
Our study site is centered at the Cedar Point Biological Station (41°13′N, 101°39′W) near Ogallala, in Keith County, along the North and South Platte Rivers and also includes portions of Deuel, Garden, and Lincoln counties, southwestern Nebraska. Cliff swallows have been studied there since 1982 (5). Approximately 160 cliff swallow colony sites are in our 150- by 50-km study area, and about a third of them are not used in a given year. Colony size (the number of active nests) varies widely; in our study area, it ranges from 2 to 3,700 nests, with some birds nesting solitarily. Over a 20-year period, the mean (± standard error) colony size (n = 1,363) was 363 (±16) nests. Each colony site tends to be separated from the next nearest site by 1 to 10 km but in a few cases by ≥20 km.
We also collected bug samples in July 2001 from a cliff swallow colony on a bridge 15 km east of Ault, Weld County, Colorado (40°38′21"N, 104°38′20"W).
Reference strains of BCRV and FMV in the diagnostic collection of the Centers for Disease Control and Prevention were isolated from swallow bugs and sera of house sparrows from 1981 to 1983 in Caddo and Grady counties, Oklahoma, and from 1974 to 1975 in Morgan and Logan counties, Colorado, respectively. The typing of these reference strains and their original assignments to either virus species were apparently done based on titer differences using the monovalent, polyclonal sera routinely used as typing fluids in alphavirus diagnostics in that era (9, 18).
Field collections of bugs.
In 2001, all swallow bugs were collected from the outsides of cliff swallow nests. Bugs generally either clustered just inside the tubular entrances of the nests in a relatively dense mass or were distributed in typically lower densities across the bottom and sides of the nests and below the entrance. We brushed bugs off nests into a wide-mouthed collecting jar using a wire brush. We collected bugs from throughout a colony site (in parts where nests were accessible) but took samples only from nests where bugs were visible to us (i.e., no nests were collected, and thus, no bugs from inside or behind the nests were included). Bugs were transferred from the jar to plastic bags, transported to the Cedar Point Biological Station, and sorted into pools of 100 individuals while alive. Pools were frozen immediately at −70°C. Multiple pools often came from bugs taken from a single nest.
Virus isolation and sequencing.
Pools of 100 bugs were macerated by mortar and pestle and suspended in 2.0 ml of BA-1, a growth medium containing M-199 Hanks' salts, 1% bovine serum albumin, 0.05 M Tris-HCl (pH 7.5), 0.35 g/liter sodium bicarbonate, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 μg/ml Fungizone (Gibco-BRL, Gaithersburg, Md.), and sterile distilled water. Homogenates were clarified by centrifugation and passing through 0.45-μm filters (Millipore, Billerica, Mass.). We added 100 μl of the supernatant in duplicate to a monolayer of Vero cells in a six-well cell culture plate (Corning Costar Corp., Cambridge, Mass.), incubated it for 1 h at 37°C in 5% CO2, and then overlaid it with 3 ml 0.5% agarose in M-199 medium supplemented with 350 mg/liter sodium bicarbonate, 29.2 mg/liter l-glutamine, and antibiotics and returned it to the incubator. A second overlay containing 0.004% neutral red dye was added after a 2-day incubation for plaque visualization. Plaques were scored daily for three additional days. Cultures with plaques (but not single plaques) were harvested by scraping the cell layer into 1 ml of BA-1 supplemented with 20% filter-sterilized fetal bovine serum. For some isolates, a 100-μl aliquot of this suspension was used for inoculating 25-cm2 cell culture flasks with confluent Vero cells held in 8 ml M-199 maintenance medium and incubated at 37°C until a virus-induced cytopathic effect became evident (between 18 and 24 h on average).
Viral RNA was extracted from 140 μl of the infectious precleared supernatant of the second Vero cell passage using the RNeasy extraction kit as recommended by the manufacturer (QIAGEN, Hameln, Germany). Five microliters of the eluted RNA suspension was used as a template in an alphavirus reverse transcription-PCR (RT-PCR) (27) to amplify a 2,075-bp region within the subgenomic 26S RNA spanning a 192-bp segment of the 3′ end of the capsid, the entire E3 (180 bp), E2 (1269 bp), and 6K (165 bp) genes, and the first 269 bp of the E1 gene. Details of the RT-PCR have been described previously (19). Gel-purified amplicons (QIAEX II PCR purification kit) were subjected to cycle sequencing using the ABI Prism BigDye Terminator cycle sequencing kit, version 3.1 (Applied Biosystems, Foster City, Calif.), and the primers listed in Table 1. Sequences were resolved on a 3130-Avant model genetic analyzer (Applied Biosystems, Darmstadt, Germany).
TABLE 1.
Primers used in RT-PCR and sequencing reactions for Buggy Creek virus
| Primer | Direction | Sequencea | Function |
|---|---|---|---|
| α-8187 | + | TGGCAYCAYGGNGCNGTNCARTA | Amplification/sequencing |
| cαc-10262 | − | RTCRCARAARCANTRNGCNCCNCCCCACAT | Amplification/sequencing |
| 8170 | + | GCATGTTTGGGACGAGTC | Sequencing |
| 8171 | − | AATGGCTGATTATGACTCC | Sequencing |
| 8256 | − | GATGTGAGGTGTAAAGTG | Sequencing |
| 8257 | + | TGTATTACCCCTTATGCG | Sequencing |
| 8258 | − | ATCGGACTCGTCCCAAAC | Sequencing |
Y = C or T; R = A or G; N = A, C, G, or T.
Sequences were aligned against the corresponding region in a 1981 BCRV reference sequence (strain 81V1822 [GenBank accession no. AF339474]), and fragments were combined for a given isolate using SeqMan 6.1 (DNAStar; Lasergene) to obtain a continuous nucleotide sequence for each sample. Isolates from Nebraska are labeled here as “colony number-sample number-year,” and thus, the site (colony) of sampling is indicated by the first two digits of the isolate label.
Phylogenetic analysis.
Sequences were aligned in MEGALIGN v. 6.1.2 (Lasergene) using the Clustal algorithm (15, 16). An initial neighbor-joining (NJ) tree was constructed with our 33 BCRV isolates and other representative alphaviruses available at PubMed (GenBank accession numbers AF339474 to AF339488, AF093102, AF093103, AF103728, AF079454, AF079456, AF214040, AF229608, AF429428, AF448539, AJ316244, AJ316246, AY112987, AY604236, DQ189086, DQ241304, K00046, M20162, M69094, M69205, and NC001924). Also included in the NJ tree was the corresponding region from 10 additional unpublished BCRV (strains 81V1823-6 and 84S217) and FMV (75V7468, 77V654, CM4-110, CM4-368, and CM4-976) isolates. A maximum-likelihood (ML) tree of all isolates that grouped with the BCRV isolates was generated using PAUP 4.0b10 (38). We based tree calculations on models of sequence evolution whose parameters were estimated by maximum likelihood using Modeltest 3.07 PPC (28). We assessed the reliability of the tree topology by bootstrapping the data set 1,000 times to generate NJ trees. Internal nodes of particular interest were labeled with percent times obtained, if greater than 50.
Each BCRV isolate is actually a sample of multiple and potentially variable virus particles within the host (11, 40); in this case, a “sample” refers to a pool of bugs, and an isolate from a given sample represents the dominant genotype present. In one case, an isolate had evidence of a sequence polymorphism, indicating the presence of multiple viral genotypes, and it was excluded from the analysis. We use the term “clade” to describe groups of similar and presumably related individuals that share a common ancestor (monophyletic) when that relationship is supported at least 75% of the time by bootstrap analysis and exhibit similar amino acid substitutions (equivalent in some usage to “lineage”). We use the term “subclade” to describe individuals within a given clade that form subgroups that are also supported at least 75% of the time by bootstrap analysis and amino acid similarity. Both terms may refer to groups of individuals that share common silent and nonsilent nucleotide substitutions. We use the term gene flow to refer to the movement of viral genes and the movement of BCRV between discrete bird colony sites containing vectors and hosts.
Nucleotide sequence accession numbers.
All sequences generated from this study can be accessed in GenBank (accession numbers DQ451557 to DQ451599).
RESULTS
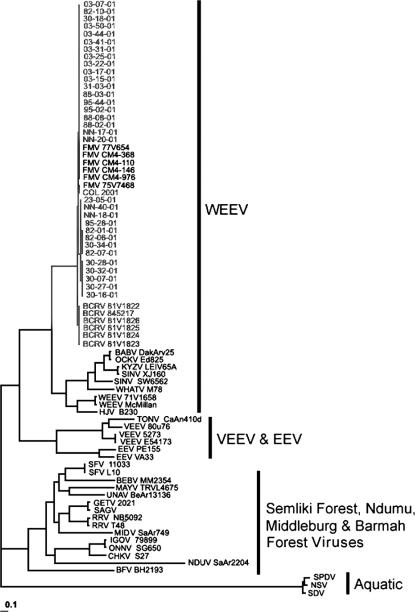
We analyzed sequences from 32 BCRV isolates collected from eight different cliff swallow colonies in southwestern Nebraska in 2001 and 1 isolate from a colony in northeastern Colorado in 2001 (Fig. 1). The NJ phylogenetic tree shows that all the sequences analyzed belong to the WEEV complex branch of alphaviruses (Fig. 1). There is close association with other alphaviruses within that group, such as HJV, SINV, WEEV, and FMV. The BCRV sequences are a distinct group within the WEEV complex and include the six FMV sequences (Fig. 1).
FIG. 1.
Neighbor-joining tree based on a 2,075-bp region of the viral envelope glycoprotein genes showing the relationships of BCRV and FMV isolates to other known alphaviruses. The eight alphavirus antigenic complexes are shown and include the following viruses: Babanki virus (BABV), Barmah Forest virus (BFV), Bebaru virus (BEBV), Chikungunya virus (CHKV), EEEV, Getah virus (GETV), HJV, Igbo Ora virus (IGOV), Kyzylagach virus (KYZV), Mayaro virus (MAYV), Middleburg virus (MIDV), Norwegian salmonid virus (NSV), Ockelbo virus (OCKV), O'nyong-Nyong virus (ONNV), Ross River virus (RRV), Ndumu virus (NDUV), Sagiyama virus (SAGV), salmon pancreas disease virus (SPDV), Semliki Forest virus (SFV), sleeping disease virus (SDV), Tonate virus (TONV), Una virus (UNAV), Venezuelan equine encephalomyelitis virus (VEEV), Whatoroa virus (WHAV), and WEEV. Names of strains, as cited in the literature, are given where appropriate. GenBank accession numbers for these sequences are given in Materials and Methods. Note the high level of nucleotide sequence identity of BCRV and FMV within the WEEV clade.
Among the BCRV and FMV sequences, there were (in total) 137 nucleotide substitutions and 15 amino acid residues that underwent a change relative to the earliest BCRV isolates from Oklahoma (Table 2). There were 2 amino acid changes in the E3 gene, 10 in the E2 gene, 2 in the 6K gene, and 1 in the fragment of the E1 gene examined. The capsid region had no changes in the fragment used in this analysis.
TABLE 2.
Amino acid changes in a 2,075-bp region of the Buggy Creek virus genome
| Virus clade | Amino acid at different positionsa
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E3
|
E2
|
6K
|
E1
|
||||||||||||
| 17 | 18 | 57 | 60 | 62 | 119 | 218 | 222 | 243 | 270 | 372 | 378 | 33 | 53 | 60 | |
| Okl | D | Q | D | D | K | D | M | R | I | L | T | A | L | V | I |
| Col | N | — | — | G | Q | — | — | — | L | — | — | — | — | — | — |
| NebA-C | N | — | — | — | Q | G | T | — | L | — | — | — | — | — | — |
| NebA-N1 | N | — | — | N | Q | G | — | K | L | — | — | — | — | — | — |
| NebA-N2 | N | — | — | — | Q | G | — | — | L | — | — | V | — | — | — |
| NebA-N3 | N | — | N | — | Q | G | T | — | L | — | — | — | — | — | — |
| NebB-N1 | — | — | — | — | Q | — | — | — | L | — | A | — | F | — | — |
| NebB-N2 | — | — | — | — | Q | — | — | — | L | M | A | — | F | A | — |
| NebB-N3 | G | R | — | — | Q | — | — | — | L | M | A | — | F | A | V |
All isolates of each clade (Fig. 2) are compared to the Oklahoma clade; residues identical to those present in Okl are represented by dashes.
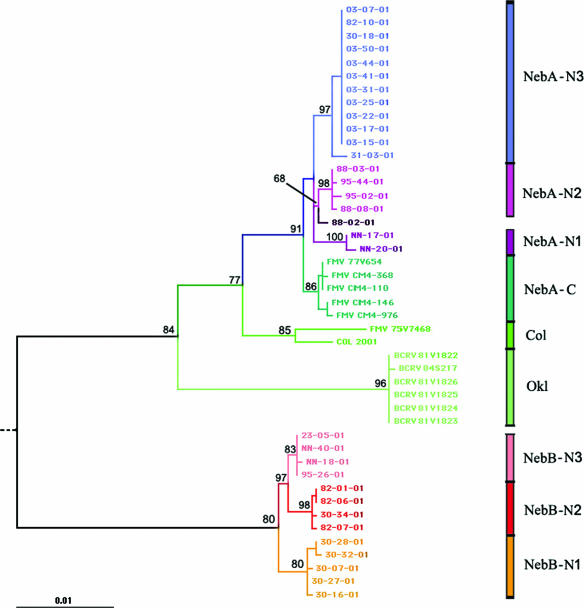
Based on the ML tree of the 2,075-bp region in the structural genes that we examined, there were four BCRV clades (Fig. 2). One is a clade composed solely of Oklahoma isolates from 1981 to 1983, designated clade Okl (Fig. 2). There were two distinct BCRV clades (designated NebA and NebB) circulating in swallow bugs during 2001 at the Nebraska study site (Fig. 2). These two clades both share two amino acid changes at residues E2-62 (K→Q) and E2-243 (I→L), which distinguish them from the Oklahoma isolates (Table 2 and Fig. 2). The substitution at E2-62 is nonconservative, with a change from a basic residue (lysine) to a neutral but polar one (glutamine), while the change at E2-243 is conservative. These substitutions are also shared with the FMV isolates (all from Colorado) and the only BCRV isolate from Colorado (COL 2001) (Table 2 and Fig. 2).
FIG. 2.
ML tree based on a 2,075-bp region of the viral envelope glycoprotein genes for 32 Nebraska BCRV isolates from 2001, a Colorado isolate from 2001 (labeled COL 2001), 6 FMV isolates collected from 1974 to 1975 (75V7468, 77V654, CM4-110, CM4-146, CM4-368, and CM4-976), and 6 Oklahoma BCRV isolates collected from 1981 to 1983 (81V1822, 81V1823, 81V1824, 81V1825, 81V1826, and 84S217). A 1971 WEEV sequence (71V1658 [GenBank accession number AF214040]) was used as an outgroup to root the tree (not shown). GenBank accession numbers are given in Materials and Methods. Nebraskan BCRV subclades are labeled NebA-N1, NebA-N2, NebA-N3, NebB-N1, NebB-N2, and NebB-N3; the Oklahoma BCRV clade is labeled Okl; the Colorado clade is labeled Col; and the Colorado subclade is labeled NebA-C (see Results). Different virus clades and subclades are color coded for illustrative purposes. Bootstrap support was generated using 1,000 replicate neighbor-joining trees reconstructed under the ML substitution model (27).
In addition to these two substitutions, the four subclades of NebA (NebA-N1, NebA-N2, NebA-N3, and NebA-C) (Fig. 2) are characterized by changes at E3-17 (D→N) and E2-119 (D→G), both of which are nonconservative. The three distinct Nebraska subclades within NebA (N1 to N3) are defined by both silent and nonsilent changes. NebA-N1 is made up solely of isolates from one colony (site NN) and has two distinct conservative changes at E2-60 (D→N) and E2-222 (R→K). NebA-N2 is widespread with respect to sampling and has a distinct conservative change at E2-378 (A→V). Isolate 88-02-01 lies outside this and the other subclades within NebA, lacking any of the distinct amino acid changes that define these clades. Based on the ML tree, it is most closely related to NebA-N2, with moderate bootstrap support of 68 (Fig. 2). Subclade NebA-N3 has a unique mutation at E2-57 (D→N) and is comprised mostly of isolates from one colony (site 03). It also has a mutation at E2-218 (M→T), which it shares with only subclade NebA-C (Table 2). Subclade NebA-C consists of FMV isolates from Colorado. It is similar to isolates taken from Nebraska, and it shares the majority of its amino acid changes with the other NebA subclades (Table 2). NebA-C is distinct only by the presence of several silent mutations and a change at E2-218 (M→T) shared with subclade NebA-N3.
NebB, the other Nebraskan clade, is the most distinct of the four. Its isolates have two mutations that separate them from the other sequences in the tree, E2-372 (T→A) and 6K-33 (L→F). The first change is a nonconservative change from a polar to a nonpolar residue, while the second is conservative. As in clade NebA, there are distinct subgroups within NebB (Fig. 2). Subclade NebB-N1 is comprised of isolates solely from one colony (site 30). NebB-N2 and NebB-N3 are widespread with respect to colonies and share two common amino acid changes at E2-270 (L→M) and 6K-53 (V→A), which are not functionally significant.
The fourth clade consisted of one BCRV isolate (COL 2001) and one FMV isolate (75V7468), both collected in Colorado, and was designated Col. This clade was more closely related to clade NebA than to any other clade. Its position in the tree suggests that the other Colorado isolates (NebA-C) and the three NebA subclades from Nebraska have more recently shared a common ancestor than any have with the Col clade (Fig. 2). In addition to a distinct amino acid change at E3-17 (D→N), which it shares with clade NebA, the Col clade also has a unique mutation at E2-60 (D→G) that distinguishes it from the other clades (Table 2).
A number of isolates also had individual mutations not seen elsewhere: E2-141 (Q→R) in FMV 75V7468, E2-143 (P→R) in isolates 82-01-01 and 82-06-01, E2-284 (T→A) and E2-344 (A→T) in isolate NN-20-01, E2-288 (T→A) in isolate 95-02-01, and E2-422 (H→R) in isolate COL 2001.
Overall, Nebraska isolates from the NebA and NebB clades showed 5.7 to 6.2% interclade divergence at the nucleotide level and 0.7 to 1.9% divergence at the amino acid level. Both clades appear to be widespread within the Nebraska study area and coexist at some sites. Five colonies (sites 30, 82, 88, 95, and NN) had isolates in both branches of the phylogeny (Fig. 2). Within the subclades, however, there was greater spatial structuring: NebA-N1 and NebB-N1 each consisted of isolates all from the same colony, NebA-N2 consisted of isolates from two colonies that were only 4.9 km apart, NebA-N3 consisted of nine isolates from the same colony and another colony (site 31) from a site 7.2 km away, and two of the four isolates in NebB-N2 were from the same colony (Fig. 2). However, there was also similarity in virus isolates taken from the two colonies on the extreme eastern and western edges of our study area (sites 82 and 30). These colonies were approximately 123 km apart, and they each contained representatives of clades NebA-N3 and NebB-N2 (Fig. 2).
DISCUSSION
This report is the first detailed phylogenetic analysis of BCRV. The NJ tree constructed from a 2,075-bp region encompassing a significant portion of the E2 viral envelope glycoprotein-coding region resulted in an accurate phylogenetic categorization of the virus as a WEEV complex member, similar to previous analyses using a portion of the E1 gene (29, 41) or the E2 gene (25). This confirms the suitability of the region that we used for phylogenetic analysis and supports that all our isolates were BCRV and not another, closely related WEEV complex virus.
The placement of the FMV sequence within the BCRV branch indicates that these two virus are the most closely related to each other (compared to other WEEV complex viruses). This phylogenetic analysis (also see reference 29) demonstrates that Buggy Creek virus and Fort Morgan virus are strains of the same virus, a conclusion that is also consistent with the close ecological similarity between the two. The FMV strains available to us are more evolutionarily derived than the original BCRV strains from Oklahoma reported previously by Hopla et al. (17) (Fig. 2).
Several of the amino acid changes observed in the BCRV isolates can be mapped to regions with pathogenic and immunogenic properties in related viruses. The change at E2-62 (K→Q), common to clades NebA, NebB, and Col, is a change from a basic to a polar residue, and research with monoclonal antibodies has shown this residue to be the component of a pathogenesis site in SINV associated with virulence and host cell penetration (26, 32). E2-243 (I→L) is also common to all three non-Oklahoma clades and is found within an antigenic determinant of the E2 ectodomain in Ross River virus (39) and Semliki Forest virus (13). The changes at E2-372 (T→A) and 6K-33 (L→F) are unique to clade NebB, and the former mutation falls within the transmembrane domain of the E2 glycoprotein (31, 36). Whether any of these changes influence the fitness of the virus is unknown. However, the finding that they are linked with distinct clades found in nature indicates that some form of purifying or positive selection might be operating, and these data would be appropriate for comparative functional genome analyses. The nucleotide and amino acid divergences between the clades are within or close to the range of differences seen among recognized subtypes of other alphaviruses (e.g., SINV and EEEV) (25, 41).
Clades NebA and NebB were widely distributed in our Nebraska study area, with near-identical isolates detected in cliff swallow colonies up to 123 km apart. However, the BCRV isolates also showed a relatively high degree of spatial structure, with some of the subclades confined to one or a few colonies that were quite close to each other in space. Interpreting these spatial patterns requires a knowledge of the extent to which viral gene flow occurs between sites. Although the transmission dynamics of BCRV are poorly understood, the virus presumably is moved from site to site whenever an infected bug moves and possibly also when an infected cliff swallow or house sparrow transfers between colony sites (assuming that birds maintain viremias of sufficient titers to infect bugs that feed on them). Bug movement correlates strongly with cliff swallow movement, because bugs disperse only by clinging to the feet of a swallow that is moving between sites (6). The relatively high degree of spatial structure among BCRV subclades probably reflects the birds' greater likelihood of moving between closely spaced colonies (5) and the fact that successful dispersal by a bug is more likely when the distance moved (and thus the time spent clinging to a bird's feet) is short. Formal analyses of the genetic similarity of isolates in relation to colony spatial position await a larger sample of sequences from additional years.
If viral gene flow is largely dependent on cliff swallows' moving of bugs, one would predict some genetic divergence of the virus (i) over distances greater than the typical movement radius of cliff swallows selecting colonies and (ii) when gaps in the distribution of swallow colony sites, due to unsuitability of habitat, serve to isolate colonies in one area from bird movement from another area. Consistent with this, we found that the FMV isolates collected in Colorado approximately 250 km from the western edge of the Nebraska study area occupied basal positions to the NebA branch (Fig. 2) and showed some amino acid substitutions not seen in the Nebraska samples (Table 2). Similarly, the Oklahoma BCRV isolates from 1981 to 1983, collected approximately 750 km south of the Nebraska study area, showed some amino acid divergence from both the Nebraska and Colorado clades (Table 2). Although the Oklahoma and (most of the) Colorado results are potentially confounded by different sampling years, the distances from Nebraska to both the Colorado and Oklahoma sites are well beyond the typical movement radius of a cliff swallow when assessing and choosing colony sites to occupy (and thus when potentially moving infected bugs) (5). This is supported by the more recently isolated Colorado isolate (COL 2001), which, although collected 25 years after the “Fort Morgan” viruses from the CDC's reference collection, was more similar to those isolates than to recent ones from Nebraska. Because cliff swallow breeding distribution is typically concentrated along east/west-flowing rivers in most of the Great Plains (C. Brown, personal observation), the distribution of colonies is not continuous from the Nebraska study area to the Colorado or Oklahoma sampling sites. Thus, the genetic divergence of these isolates is probably attributable in part to reduced viral gene flow between these areas.
An unresolved question is whether the two Nebraska BCRV clades are functionally different in terms of effects on hosts, vectors, or ecology (e.g., overwintering persistence). More samples and additional studies will be necessary to address these questions. However, we note that clade NebB was not found in the two largest cliff swallow colonies sampled (860 and 240 nests from sites 03 and 88, respectively). The other colonies, including all the ones with clade NebB, varied in size between 1 and 90 nests. Colony size could affect virus prevalence from year to year (and thus whether a particular clade is detected) because the number of active nests correlates directly with bug population size (7), which may affect the likelihood of local virus persistence over the winter. Perhaps one clade is more susceptible to extreme seasonal fluctuations in bug population size that are brought about by small colonies being less likely to be occupied by birds in consecutive years (5).
The relatively high degree of spatial structure among the BCRV subclades detected in this study suggests that WEEV complex alphaviruses have the capacity to show sequence variation that can be associated with particular geographic locations. If so, this may make it possible to identify a given isolate's probable geographic origin simply by sequencing and may assist in determining the movement characteristics and the source of epizootics in the medically important WEEV complex viruses. Documentation of nucleotide variation among the spatially distinct BCRV clades could be especially important, given that mutations at only one or few nucleotide positions in other alphaviruses can change enzootic strains to virulent, epidemic variants (1, 33).
Acknowledgments
M.P., J.E.F., and C.R.B. contributed equally to this work.
We thank Jennifer Malfait and Mike Shanahan for field assistance; Gudrun Zöller and Ines Patzwald for technical assistance; the School of Biological Sciences at the University of Nebraska—Lincoln for use of the Cedar Point Biological Station; the Union Pacific Railroad and the Robert Clary, Dave Knight, and Loren Soper families for access to land; D. B. Francy, Cluff Hopla, Gordon Smith, and W. D. Sudia for collecting and isolating the FMV and BCRV strains from the 1970s and 1980s; and Glen Collier, Valerie O'Brien, and Ann Powers for comments on the manuscript.
This work was supported by grants from the National Institutes of Health (R01-AI057569) and the National Science Foundation (DEB-0075199).
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Anishchenko, M., R. A. Bowen, S. Paessler, L. Austgen, I. P. Greene, and S. C. Weaver. 2006. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc. Natl. Acad. Sci. USA 103:4994-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, C. R., and M. B. Brown. 1986. Ectoparasitism as a cost of coloniality in cliff swallows (Hirundo pyrrhonota). Ecology 67:1206-1218. [Google Scholar]
- 3.Brown, C. R., and M. B. Brown. 1992. Ectoparasitism as a cause of natal dispersal in cliff swallows. Ecology 73:1718-1723. [Google Scholar]
- 4.Brown, C. R., and M. B. Brown. 1995. Cliff swallow (Hirundo pyrrhonota), no. 149. In A. Poole and F. Gill (ed.), Birds of North America. Academy of Natural Sciences, Philadelphia, Pa.
- 5.Brown, C. R., and M. B. Brown. 1996. Coloniality in the cliff swallow: the effect of group size on social behavior. University of Chicago Press, Chicago, Ill.
- 6.Brown, C. R., and M. B. Brown. 2004. Empirical measurement of parasite transmission between groups in a colonial bird. Ecology 85:1619-1626. [Google Scholar]
- 7.Brown, C. R., N. Komar, S. B. Quick, R. A. Sethi, N. A. Panella, M. B. Brown, and M. Pfeffer. 2001. Arbovirus infection increases with group size. Proc. R. Soc. Lond. B 268:1833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calisher, C. H., T. P. Monath, D. J. Muth, J. S. Lazuick, D. W. Trent, D. B. Francy, G. E. Kemp, and F. W. Chandler. 1980. Characterization of Fort Morgan virus, an alphavirus of the western equine encephalitis virus complex in an unusual ecosystem. Am. J. Trop. Med. Hyg. 29:1428-1440. [DOI] [PubMed] [Google Scholar]
- 9.Calisher, C. H., N. Karabatsos, J. S. Lazuick, T. P. Monath, and K. L. Wolff. 1988. Reevaluation of the western equine encephalitis antigenic complex of alphaviruses (family Togaviridae) as determined by neutralization tests. Am. J. Trop. Med. Hyg. 38:447-452. [DOI] [PubMed] [Google Scholar]
- 10.Chapman, B. R., and J. E. George. 1991. The effects of ectoparasites on cliff swallow growth and survival, p. 69-92. In J. E. Loye and M. Zuk (ed.), Bird-parasite interactions: ecology, evolution and behaviour. Oxford University Press, Oxford, United Kingdom.
- 11.Domingo, E. 1998. Quasispecies and the implications for virus persistence and escape. Clin. Diagn. Virol. 10:97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George, J. E. 1987. Field observations on the life cycle of Ixodes baergi and some seasonal and daily activity cycles of Oeciacus vicarius (Hemiptera: Cimicidae), Argas cooleyi (Acari: Argasidae), and Ixodes baergi (Acari: Ixodidae). J. Med. Entomol. 24:683-688. [DOI] [PubMed] [Google Scholar]
- 13.Grosfeld, H., B. Velan, M. Leitner, S. Lustig, B. E. Lachi, S. Cohen, and A. Shafferman. 1991. Delineation of protective epitopes on the E2-envelope glycoprotein of Semliki Forest virus. Vaccine 9:451-456. [DOI] [PubMed] [Google Scholar]
- 14.Hayes, R. O., D. B. Francy, J. S. Lazuick, G. C. Smith, and E. P. J. Gibbs. 1977. Role of the cliff swallow bug (Oeciacus vicarius) in the natural cycle of a western equine encephalitis-related alphavirus. J. Med. Entomol. 14:257-262. [Google Scholar]
- 15.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 17.Hopla, C. E., D. B. Francy, C. H. Calisher, and J. S. Lazuick. 1993. Relationship of cliff swallows, ectoparasites, and an alphavirus in west-central Oklahoma. J. Med. Entomol. 30:267-272. [DOI] [PubMed] [Google Scholar]
- 18.Karabatsos, N. 1985. International catalogue of arboviruses, including certain other viruses of vertebrates, 3rd ed. American Society of Tropical Medicine and Hygiene, San Antonio, Tex.
- 19.Kinney, R. M., M. Pfeffer, K. R. Tsuchiya, G.-J. Chang, and J. T. Roehrig. 1998. Nucleotide sequences of the 26S mRNAs of the viruses defining the Venezuelan equine encephalitis antigenic complex. Am. J. Trop. Med. Hyg. 59:952-964. [DOI] [PubMed] [Google Scholar]
- 20.Lowther, P. E., and C. L. Cink. 1992. House sparrow, no. 12. In A. Poole, P. Stettenheim, and F. Gill (ed.), Birds of North America. Academy of Natural Sciences, Philadelphia, Pa.
- 21.Loye, J. E. 1985. The life history and ecology of the cliff swallow bug, Oeciacus vicarius (Hemiptera: Cimicidae). Cahiers Off. Rech. Sci. Tech. Outre-Mer Ser. Entomol. Med. Parasitol. 23:133-139. [Google Scholar]
- 22.Loye, J. E., and C. E. Hopla. 1983. Ectoparasites and microorganisms associated with the cliff swallow in west-central Oklahoma. II. Life history patterns. Bull. Soc. Vector Ecol. 8:79-84. [Google Scholar]
- 23.Loye, J. E., and S. P. Carroll. 1991. Nest ectoparasite abundance and cliff swallow colony site selection, nestling development, and departure time, p. 222-241. In J. E. Loye and M. Zuk (ed.), Bird-parasite interactions: ecology, evolution and behaviour. Oxford University Press, Oxford, United Kingdom.
- 24.Myers, L. E. 1928. The American swallow bug, Oeciacus vicarius Horvath (Hemiptera, Cimicidae). Parasitology 20:159-172. [Google Scholar]
- 25.Norder, H., J. O. Lundström, O. Kozuch, and L. O. Magnius. 1996. Genetic relatedness of Sindbis virus strains from Europe, the Middle East, and Africa. Virology 222:440-445. [DOI] [PubMed] [Google Scholar]
- 26.Pence, D. F., N. L. Davis, and R. E. Johnston. 1990. Antigenic and genetic characterization of Sindbis virus monoclonal antibody escape mutants which define a pathogenesis domain on glycoprotein E2. Virology 175:41-49. [DOI] [PubMed] [Google Scholar]
- 27.Pfeffer, M., R. M. Kinney, and O.-R. Kaaden. 1998. The alphavirus 3′-nontranslated region: size heterogeneity and arrangement of repeated sequence elements. Virology 240:100-108. [DOI] [PubMed] [Google Scholar]
- 28.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 29.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75:10118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rannala, B. H. 1995. Demography and genetic structure in island populations. Ph.D. dissertation. Yale University, New Haven, Conn.
- 31.Rice, C. M., J. R. Bell, M. W. Hunkapiller, E. G. Strauss, and J. H. Strauss. 1982. Isolation and characterization of the hydrophobic COOH-terminal domains of the Sindbis virion glycoproteins. J. Mol. Biol. 154:355-378. [DOI] [PubMed] [Google Scholar]
- 32.Schoepp, R. J., and R. E. Johnston. 1993. Sindbis virus pathogenesis: phenotypic reversion of an attenuated strain to virulence by second-site intragenic suppressor mutations. J. Gen. Virol. 74:1691-1695. [DOI] [PubMed] [Google Scholar]
- 33.Schuffenecker, I., I. Iteman, A. Michault, S. Murri, L. Frangeul, M.-C. Vaney, R. Lavenir, N. Pardigon, J.-M. Reynes, F. Pettinelli, L. Biscornet, L. Diancourt, S. Michel, S. Suquerroy, G. Guigon, M.-P. Frenkiel, A.-C. Brehin, N. Cubito, P. Despres, F. Kunst, F. A. Rey, H. Zeller, and S. Brisse. 2006. Genome microevolution of Chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3:e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott, T. W., G. S. Bowen, and T. P. Monath. 1984. A field study of the effects of Fort Morgan virus, an arbovirus transmitted by swallow bugs, on the reproductive success of cliff swallows and symbiotic house sparrows in Morgan County, Colorado, 1976. Am. J. Trop. Med. Hyg. 33:981-991. [DOI] [PubMed] [Google Scholar]
- 35.Smith, G. C., and R. B. Eads. 1978. Field observations on the cliff swallow, Petrochelidon pyrrhonota (Vieillot), and the swallow bug, Oeciacus vicarius Horvath. J. Wash. Acad. Sci. 68:23-26. [Google Scholar]
- 36.Strauss, E. G., E. M. Lenches, and J. H. Strauss. 2002. Molecular genetic evidence that the hydrophobic anchors of glycoproteins E2 and E1 interact during assembly of alphaviruses. J. Virol. 76:10188-10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swofford, D. L. 1998. PAUP* 4.0—phylogenetic analysis using parsimony (*and other methods), ed. 4.0b8. Sinauer Associates, Sunderland, Mass.
- 39.Vrati, S., C. A. Fernon, L. Dalgarno, and R. C. Weir. 1988. Location of a major antigenic site involved in Ross River virus neutralization. Virology 162:346-353. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W.-K., S.-R. Lin, C.-M. Lee, C.-C. King, and S.-C. Chang. 2002. Dengue type 3 virus in plasma is a population of closely related genomes: quasispecies. J. Virol. 76:4662-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weaver, S. C., W. Kang, Y. Shirako, T. Rumenapf, E. G. Strauss, and J. H. Strauss. 1997. Recombinational history and molecular evolution of western equine encephalomyelitis complex alphaviruses. J. Virol. 71:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]