Abstract
In order to gain an understanding of the molecular mechanisms dictating production of the siderophore and dechlorination agent pyridine-2,6-bis(thiocarboxylic acid) (PDTC), we have begun characterization of a gene found in the pdt gene cluster of Pseudomonas stutzeri KC predicted to have a regulatory role. That gene product is an AraC family transcriptional activator, PdtC. Quantitative reverse transcription-PCR and expression of transcriptional reporter fusions were used to assess a role for pdtC in the transcription of pdt genes. PdtC and an upstream, promoter-proximal DNA segment were required for wild-type levels of expression from the promoter of a predicted biosynthesis operon (PpdtF). At least two other transcriptional units within the pdt cluster were also dependent upon pdtC for expression at wild-type levels. The use of a heterologous, Pseudomonas putida host demonstrated that pdtC and an exogenously added siderophore were necessary and sufficient for expression from the pdtF promoter, i.e., none of the PDTC utilization genes within the pdt cluster were required for transcriptional signaling. Tests using the promoter of the pdtC gene in transcriptional reporter fusions indicated siderophore-dependent negative autoregulation similar to that seen with other AraC-type regulators of siderophore biosynthesis and utilization genes. The data increase the repertoire of siderophore systems known to be regulated by this type of transcriptional activator and have implications for PDTC signaling.
The bacterial strain Pseudomonas stutzeri KC excretes the secondary metabolite and transition metal chelator pyridine 2,6-bis(thiocarboxylate) (PDTC) under iron-limited growth conditions. PDTC is also the active agent of the cometabolic dechlorination of carbon tetrachloride (CCl4) displayed by that organism (21). In order to reliably exploit this activity for remediation of CCl4 contamination, an understanding of factors affecting PDTC production is necessary. PDTC is a siderophore, a class of compounds whose production has evolved among bacteria and other organisms for the acquisition of iron (14). The regulation of siderophore production is a component of iron homeostasis, which involves balancing avid uptake capabilities with the avoidance of iron-catalyzed oxidative damage (3).
The production of siderophores is known to be regulated at the level of transcription, being repressed via Fur and its corepressor, ferrous iron, and activated by the cognate siderophore and any of a number of effector systems (40). These integrated control mechanisms allow the organism to avoid or minimize wasteful energy expenditure either when adequate intracellular iron has been acquired or when no siderophore is sensed due to diffusion or other losses to the extracellular milieu. Many bacteria, including P. stutzeri KC, are capable of producing more than one siderophore. It is likely that some form of coordination between alternative siderophore systems occurs through the evolved regulatory mechanisms, but exactly how this may be affected is not clear. If only the Fur repression and various activation systems participate, this may be solely passive; a siderophore that is not providing iron would not be maximally produced, whereas one that does provide iron activates its own production or transport.
Two main types of regulatory system achieve positive transcriptional regulation of siderophore biosynthesis in response to a cognate siderophore. For the ferric citrate system of Escherichia coli and the pyoverdine systems of fluorescent pseudomonads, outer membrane receptors transmit the signal through an allosteric regulatory cascade that increases the availability of specific RNA polymerase sigma subunits to interact with core polymerase and actively transcribe the associated genes (32, 40). For the pyochelin (Pch), yersiniabactin (Ybt), rhizobactin (Rbt), alcaligin (Alc), and quinolobactin (Qbs) systems, araC-type regulatory genes are known to be necessary for maximal expression of the respective siderophore-biosynthetic and transport genes (7, 10, 15, 24, 26). For the AraC paradigm, the activator binds specific DNA repeat sequences as a dimer, and the DNA binding affinity and topology are altered allosterically through the additional capacity to bind the small-molecule effector arabinose (25). Like AraC, it is expected that each of the homologous siderophore regulators requires a coeffector, most likely the cognate siderophore (11). For pyochelin/PchR, this has been demonstrated (29). Models integrating AraC-type signaling with siderophore physiology have included transport of the ferrisiderophore into the cytoplasm by inner and outer membrane transporters to allow interaction with the protein activator (10, 16). However, in two cases, it is known that outer membrane receptor mutants that are defective in utilization of the cognate siderophore as an iron source still activate transcription in response to an exogenously added siderophore (2, 10, 20).
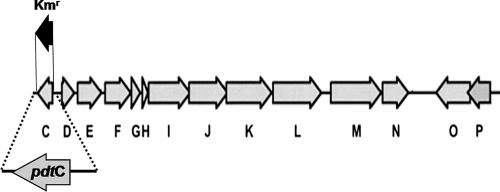
An araC homolog is found within the P. stutzeri KC pdt gene cluster, which encodes functions necessary and sufficient for PDTC production (22). The gene encoding the AraC homolog, pdtC, is found as a single transcriptional unit oriented in a direction opposite that of adjacent pdt genes (Fig. 1), a strong indication that there is a regulatory association, since genes found to be linked in this fashion are usually controlled by the divergently transcribed regulator (19). Other functions encoded within the pdt gene cluster include a cytoplasmic membrane permease (PdtE) (20, 31) and an outer membrane transporter (PdtK) (20). Predicted biosynthetic enzymes include sulfur transferase homologs, a putative dehydratase/oxidoreductase, and an acyladenyltransferase homolog (PdtFGHIJ) (22, 38). PdtP is predicted to be a soluble S-adenosylmethione-dependent O-methyltransferase. Strains bearing cosmid-borne pdt subclusters that lack pdtP show decreased PDTC production (22). Together with the unlikely necessity for an O-methyltransferase in PDTC biosynthesis, the dispensability of PdtP suggested a role in regulation rather than biosynthesis (22, 38); however, this remains speculative.
FIG. 1.
The pdt gene cluster of P. stutzeri KC. Single-letter designations for pdt genes are used for brevity. The position and extent of the pdtC deletion of strain SEMΔpdtC, and the DNA fragment used to test complementation in trans are shown. Predicted functions inferred from similarity searches are given in reference 22 and the text.
Transcriptional activation of the pdt gene cluster in response to exogenously added PDTC has been demonstrated for another PDTC-producing strain, Pseudomonas putida DSM 3601 (23). In this work, we have tested the role of pdtC in the transcriptional regulation of pdt genes and defined the minimal set of genes sufficient for reconstitution of expression from a pdt promoter in an alternative host.
MATERIALS AND METHODS
Media and culture conditions.
Bacterial strains and plasmids are listed in Table 1. E. coli strains were routinely grown on LB, while tryptic soy agar or broth was used for Pseudomonas. PM medium (23) was used as a low-transition metal minimal medium for growth under iron-limited conditions. A modified PM medium was formulated to allow growth of an rpoN mutant of P. putida KT2440 and was used in all experiments with P. putida KT2440; succinate and ammonium sulfate were replaced with 10 mM glucose and 10 mM glutamine. Solid media contained 15 g liter−1 agar (Difco, Detroit, MI). Antibiotics were used at the following concentrations: chloramphenicol (Cm) (50 μg ml−1), kanamycin (Km) (50 μg ml−1), tetracycline (Tc) (15 μg ml−1), streptomycin (Str) (100 μg ml−1), and ampicillin (100 μg ml−1). PDTC was synthesized by the method of Hildebrand et al. (17). Pseudomonas strains were grown at 30°C and E. coli at 37°C. Small cultures (3 to 5 ml) of Pseudomonas were incubated in culture tubes on a roller drum. Larger cultures (≥35 ml) were grown using Erlenmeyer flasks filled to 20% of capacity on a rotary shaker at 250 rpm. E. coli cultures were incubated in a rotary shaker.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| BW20767 | Pir+mob+ | S. Minnoch, University of Idaho |
| JM109 | recA endA | Promega, Madison, WI |
| S17.1 | Smrmob+ | S. Minnoch |
| P. stutzeri | ||
| KC | Wild-type PDTC producer | C. Criddle, Stanford University |
| SEMΔpdtC | KC ΔpdtC::Km | This study |
| P. putida | ||
| KT2440 | Wild type | Estacion Exp. Zaidin (EEZ), Granada, Spain |
| PSC 48 | KT2440 rpoS null mutant | EEZ (33) |
| PSC 39 | KT2440 rpoN null mutant | EEZ (18) |
| Plasmids | ||
| pEP185.2sacB1 | Cmrtra+oriR6KsacB vehicles for allele replacement and counterselection | 23 |
| pEP185.2sacB1Δsal | pEP185.2sacB1 lacking SalI-XhoI of cloning site | This study |
| pSEMΔC | pEP185.2sacB1Δsal with ΔpdtC::Km allele | This study |
| pGEM T-easy | AproricolE1cloning vector | Promega |
| pJB3Tc20 | Tcr RK2-based IncP broad-host-range vector | 6 |
| PSEMC | pJB3Tc20 with pdtC gene inserted between KpnI and SphI sites | This study |
| pMP77 | Strr IncQ broad-host-range promoter-probe vector with promoterless xylE | 39 |
| pFpro1 (etc.) | Series of constructs: pMP77 with pdtF promoter fragments of various lengths inserted upstream of xylE using BglII and HindIII sites (Fig. 4) | This study |
| pCpro | pMP77 with pdtC promoter fragment inserted upstream of xylE using BglII and HindIII sites | This study |
Pseudomonas strains carrying promoter-reporter constructs were first grown to exponential phase in PM medium and used to inoculate larger cultures for cell extract preparation. For growth curves, a starting optical density at 600 nm (OD600) of 0.001 was approximated by diluting late-exponential-phase cultures based on their turbidity. For single-point reporter assays, late-exponential-phase cells were diluted to an OD600 of 0.33 and incubated with or without PDTC for 6 to 8 h before being harvested for cell extract preparation. All experiments were repeated at least three times.
DNA and RNA manipulations.
The primers used are listed in Table 2. PCR products were cloned into pGEM-T easy (Promega, Madison, WI) and verified by sequencing at the Vermont Cancer Center DNA Analysis Facility at the University of Vermont. Plasmid DNA was isolated using a Wizard Plus SV miniprep kit (Promega). Genomic DNA was isolated using the cetyltrimethylammonium bromide extraction method (5). Probe synthesis for Southern hybridization used the AlkPhos Direct probe labeling kit (Amersham, Piscataway, NJ). Conjugal transfer of plasmids was achieved by the filter-mating technique (9) with E. coli strains S17.1 and BW20767 as donors.
TABLE 2.
Primers used in this study
| Primer name | Sequencea | Target | Positionb |
|---|---|---|---|
| pdtCLl2 | ATCTCGAGGTTGCCTGCCCTTGACTG | pdtC downstream flanking region | 1972-1990 (+) |
| pdtCLr | ATGAGCTCGTCGACCGCTCCAGCCTAATGTACTG | pdtC downstream flanking region | 3301-3320 (−) |
| pdtCRl | ATCAGTCGACGTGATGACCATGACCCGAC | pdtC upstream flanking region | 4101-4118 (+) |
| pdtCRr2 | GAAGATCTCCAATCAGCAGTCCGACC | pdtC upstream flanking region | 5573-5590 (−) |
| pdtCpro4066Xba | TCTAGACCGTCGTAGCAGGAG | pdtC promoter | 4066-4080 (+) |
| pdtCpro4502Bgl | AGATCTCGGGCGTGCAGTGC | pdtC promoter | 4502-4518 (−) |
| pdtC3958 | GCGAAACCGTAACGTCCG | pdtC cDNA (RACE) | 3958-3975 (+) |
| pdtC3803 | GGCTGATCAACGCGTTG | pdtC cDNA (RACE) | 3808-3824 (+) |
| pdtCrev3391 | GCGATTTGCCGTGGTAGCGAGTAA | pdtC mRNA | 3391-3414 (+) |
| pdtCfwd3638 | CAGGCACGGCAGATGATGGATGAG | pdtC cDNA | 3638-3661 (−) |
| pdtproFwd6184 | ATAGATCTACACCGGCATGC | pdtF promoter | 6184-6196 (+) |
| pdtproFwd6266 | ATAGATCTTCGGCACAGTCG | pdtF promoter | 6266-6278 (+) |
| pdtproFwd6302 | ATAGATCTAAGGCTGAAGTGACCGGC | pdtF promoter | 6294-6319 (+) |
| pdtproFwd6328 | GCCATAGATCTTCGGACAATG | pdtF promoter | 6328-6338 (+) |
| pdtproFwd6356 | GCGCAGATCTTTTGCATAG | pdtF promoter | 6356-6366 (+) |
| pdtproFwd6377 | TAGATCTCACGTCATATGAAAGAACAGCC | pdtF promoter | 6377-6399 (+) |
| pdtpro6400 | AAGATCTAACGGCAATTGCTATAG | pdtF promoter | 6400-6416 (+) |
| pdtproR | ATTCTAGACCTGTTTGATTTCAATGGTAACG | pdtF promoter | 6443-6465 (−) |
| pdtpro6410 | GATCTAGACAATTGCCGTTGGCTGTTC | pdtF promoter | 6392-6410 (−) |
| pdtFFwd | CGGTGTGTTCGGCATTCTTT | pdtF cDNA | 7086-7105 (+) |
| pdtFRev | CCCGATAGCGCATATCCAAA | pdtF mRNA | 7200-7219 (−) |
| AlMoeZR | ACGGGTTGCGAAGTTGTC | pdtF mRNA | 6889-6906 (−) |
| pdtEfwd | CGTTCCGCAGCGCTGTTCGTAG | pdtE cDNA | 5475-5496 (+) |
| pdtErev | CGCCCGGTGGAGGATCTGTAGTCT | pdtE mRNA | 5653-5676 (−) |
| pdtP22937 | CTGGCGCCAATGGAAACAAGTCG | pdtP mRNA | 22937-22959 (+) |
| pdtP23114 | GGGCGGGGTTTTCGTGAGCA | pdtP cDNA | 23114-23133 (−) |
| KC16sFwd | GCGGTGGAGCATGTGGTT | 16S cDNA | |
| KC16sRev | CTGGAAAGTTCTCTGCATGTCAA | 16S rRNA | |
| Abridged anchor | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | Poly(C)-tailed cDNA | |
| Abridged universal | GGCCACGCGTCGACTAGTAC | Amplified cDNA |
Boldface letters indicate restriction sites.
GenBank accession number AF196567.
For RNA extractions, cells were harvested and RNA was extracted immediately using the Trizol Max Bacterial RNA Isolation Kit (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. The extracted RNA was stored at −80°C until it was used. All RNA samples were DNase treated. RNA was assessed for DNA contamination by its use as a template for PCRs, and it was assessed for integrity/degradation by electrophoresis through denaturing agarose gels and observation of rRNA after ethidium bromide staining.
For quantitative real-time reverse transcription-PCR (qRT-PCR), cDNA was prepared from 1 μg total RNA using Superscript II (Invitrogen) and random-hexamer primers, according to the manufacturer's protocol. qRT-PCR employed the following primer pairs: pdtC, pdtCfwd3638 and pdtCrev3391; pdtE, pdtEfwd and pdtErev; pdtF, pdtFFwd and pdtFRev; pdtP, pdtP22937 and pdtP23114; 16S rRNA, KC16sFwd and KC16sRev. cDNAs (diluted 1/100 for 16S rRNA) were then used as template for real-time PCR using SYBR Green Jump Start Taq Ready Mix (Sigma, Saint Louis, MO) and SYBR Green to monitor double-stranded-DNA synthesis; 25-μl reaction mixtures included 1 μl of template cDNA and 50 nM primers. Reactions were performed in an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA), and data were analyzed using the Sequence Detection System (SDS 2.2 software; Applied Biosystems). Fluorescence intensity values were determined after 40 cycles of PCR with each of the qRT-PCR primer sets. The efficiency of PCR amplification was determined using a standard curve prepared from serial dilutions of cDNA from one RNA extraction (obtained from an exponentially growing culture of P. stutzeri KC; OD600, 0.6). The purity of the amplified product was verified after PCR by running a dissociation curve (60°C to 95°C over 30 min). Control reactions lacking template DNA were run to ensure that primer dimers were not contributing to the overall signal. RNA abundances for all replicate extractions were related to the standard curve by their respective fluorescence intensity values, giving values of relative concentration. Values obtained for mRNA relative concentrations were normalized to the 16S rRNA relative concentration present in each sample.
The 5′ ends of mRNAs were characterized using a modified protocol for rapid amplification of cDNA ends (5′ RACE) (36). Reverse transcription reactions were performed with 3 μg of total RNA using Superscript II or Thermoscript RNase H− reverse transcriptase (Invitrogen) at 37°C or 65°C, respectively. The gene-specific primers were pdtC3803 and pdtFRev. RT products were RNase treated for 30 min at 37°C. To remove excess primer, reaction mixtures were diluted and purified using the Qiaquick PCR purification kit (QIAGEN). Purified cDNA was resuspended in doubly distilled water, the volume was reduced to 10 μl, and the suspensions were used in a tailing reaction using terminal deoxynucleotidyl transferase (Invitrogen) and dCTP according to the manufacturer's protocol. Amplification of the tailed RT products used a primer that annealed with the poly(C) tail created by terminal deoxynucleotidyl transferase (abridged anchor) and the abridged universal primer from Invitrogen (Table 2), along with pdtC3958 or A1MoeZR. PCR products were visualized on an agarose gel, and bands were then excised and purified for sequencing using the Qiaquick gel extraction kit (QIAGEN).
Sequence and statistical analysis.
The DNA and protein sequence analysis software used were Sequencher (Gene Codes, Ann Arbor, MI), BPROM (Softberry, Inc., Mount Kisco, NY), GenomeMatScan (http://www.pdg.cnb.uam.es/icases/promscan), BLASTP (http://www.ncbi.nih.gov/BLAST/), and REPFIND (http://zlab.bu.edu/repfind/form.html). Statistical inferences were based on P values calculated using one-tailed t tests.
pdtC null allele construction.
A pdtC null allele was constructed by PCR amplification of flanking DNA (with P. stutzeri KC genomic DNA as a template) and insertion of a Kmr cassette. A 1,436-bp DNA fragment located downstream of pdtC was generated using primers pdtCLl2 and pdtCLr. A 1,571-bp fragment located upstream of pdtC was generated using pdtCRl and pdtCRr2. The products were sequentially inserted in the pEP185.2sacB1Δsal suicide vector, first using NotI and SalI (downstream fragment), followed by SalI and BglII (upstream fragment), using restriction sites provided by PCR primers and the vector. A Kmr cassette, amplified from pUTKmlacZ with primers engineered to provide SalI sites (KmSalFor and KmSalRev), was inserted between the upstream and downstream flanking DNAs to give plasmid pSEMΔC. After conjugal transfer into P. stutzeri KC, recombinants were selected by resistance to Km. Double recombinants were selected by growth on LB with 6% sucrose and lacking NaCl and further screened for Cm sensitivity. Allelic replacement was verified by Southern analysis.
Construction of xylE promoter fusion constructs.
Nine different PCR fragments were generated for promoter analysis. To derive a nested 5′ truncation series of the pdtF promoter, the reverse primer pdtproR was used with pdtproFwd6184, pdtproFwd6266, pdtproFwd6302, pdtproFwd6328, pdtproFwd6377, or pdtproFwd6400. The products were then inserted between BglII and XbaI sites of pMP77. For 3′ truncation, the reverse primer pdtpro6410 was used with pdtproFwd6184, and the amplification product was inserted at the BglII site of pMP77. The resulting inserts were characterized by sequencing them, and plasmid pFpro8 was chosen as having the correct orientation. For the pdtC promoter, the pdtCpro4066Xba and pdtCpro4502Bgl primers were used, followed by insertion into pMP77 between BglII and XbaI.
To obtain a functional copy of the pdtC gene, a PCR fragment was generated using primers pdtCLl2 and pdtCRr2. An SphI-KpnI subfragment was excised from that product and inserted into the pJB3Tc20 vector to create pSEMC.
Analytical procedures.
PDTC was assayed from culture supernatants as described previously (23). The detection limit for PDTC in culture supernatants was approximately 2.5 μM. Cell extract preparation and catechol dioxygenase assays were performed as described by Klecka and Gibson (17a). The protein concentration was determined by using the bicinchoninic acid method (Pierce, Rockford, IL).
RESULTS
The AraC-type regulator PdtC is necessary for maximal expression of pdt genes.
The pdt gene cluster of P. stutzeri KC is arranged in at least three, and more likely four, operons; the relatively short intergenic regions or overlapping reading frames within pdtDE, pdtFGHIJKL, pdtMN, and pdtPO suggested that they were each operons (Fig. 1). The pdtC open reading frame would constitute an additional transcription unit. In order to assess the predicted role of PdtC as a transcriptional activator, a null allele that lacked the entire pdtC coding region was constructed and used for allelic replacement in strain KC (Fig. 1). The resulting strain, SEMΔpdtC, was grown in minimal medium under iron-limited conditions and compared with the parental strain for its ability to produce siderophore. PDTC production by SEMΔpdtC was sevenfold lower than wild-type levels (Fig. 2). The Pdt− phenotype (a PDTC production defect) of SEMΔpdtC could be complemented in trans with only pdtC on a multicopy plasmid (Fig. 2). The data indicated that PdtC had a role in allowing wild-type levels of siderophore production.
FIG. 2.
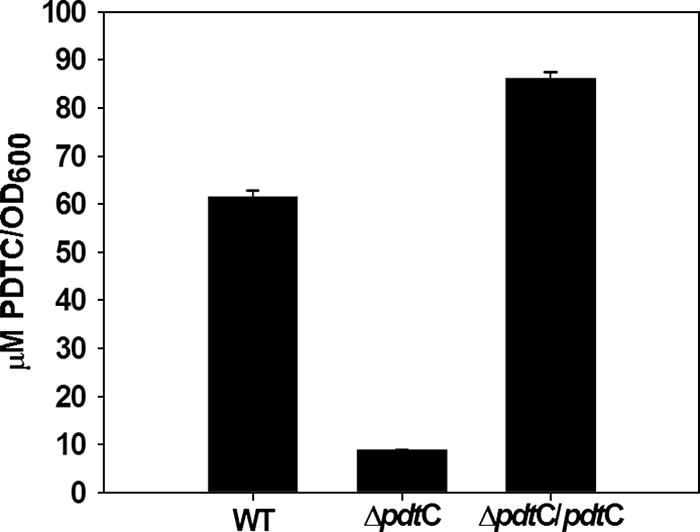
PDTC production by P. stutzeri KC and the ΔpdtC strain, SEMΔpdtC. The bars represent the averages of triplicate determinations for the wild type, a pdtC null mutant, and complemented mutant (ΔpdtC/pdtC). The error bars represent standard errors of the mean for three replicate determinations.
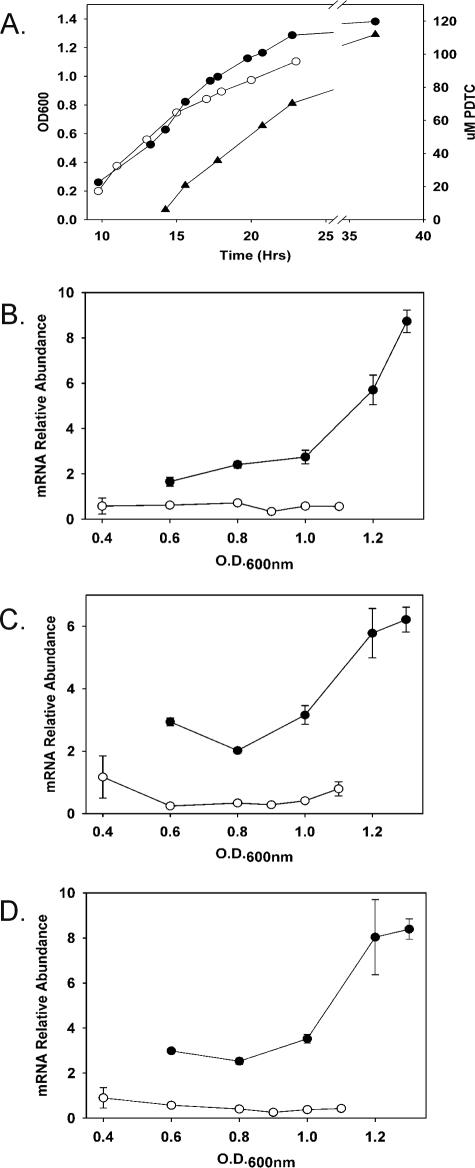
To test a role for PdtC in the transcription of pdt genes, we quantitated mRNA abundances for pdtF, pdtE, and pdtP transcripts by qRT-PCR for both wild-type and ΔpdtC strains grown under iron-limited conditions. Comparisons of growth and relative transcript abundances for the wild type and the ΔpdtC mutant are shown in Fig. 3. The gene chosen for normalization (16S rRNA) is also subject to variation over the culture cycle; therefore, differences between mutant and wild-type normalized expression levels, rather than absolute values, are the most informative comparison in this type of experiment.
FIG. 3.
Growth, PDTC production, and pdt gene expression by iron-limited cultures of P. stutzeri KC and the ΔpdtC strain, SEMΔpdtC. (A) Growth curves and PDTC production. •, P. stutzeri KC, OD600; ○, SEMΔpdtC, OD600; ▴, PDTC production by P. stutzeri KC. (B, C, and D) Relative mRNA abundances for pdtF, pdtE, and pdtP. Filled symbols, P. stutzeri KC; open symbols, SEMΔpdtC. The data are from the experiment shown in panel A with RNA extracted from samples obtained during the interval prior to the 24-h time point. The error bars are standard errors of the means of duplicate RNA extractions analyzed in duplicate.
The ΔpdtC mutant was reproducibly impaired in growth relative to the wild type in the later stages of the culture cycle. It is not clear whether this impairment was due directly to the defect in PDTC production, especially since the cells should still have been capable of producing the alternative siderophore deferrioxamine E. Very low pdt transcript abundances were also observed for the ΔpdtC mutant. The differences were greatest as the cultures approached stationary phase; the relative abundance of each of the transcripts increased three- to sevenfold for the wild-type culture prior to entry into stationary phase, whereas the ΔpdtC mutant showed no significant changes over the same period. The data supported a transcriptional-activator role for PdtC and indicated that multiple pdt transcripts are affected by a lack of PdtC, in the manner of a PdtC-controlled regulon.
PdtC and upstream promoter-proximal sequences are necessary for maximal transcription from the pdtF promoter.
AraC-type regulators bind to their respective operator sites, usually with a requirement for a small-molecule coeffector (11). A predicted σ70 promoter with a Fur box is located in a region of DNA upstream of pdtF, and Fur-dependent iron regulation of a promoter located within that segment of DNA has been demonstrated using an E. coli/lacZ reporter system (38). The transcriptional start site for pdtF was assessed using the 5′ RACE RT-PCR technique. The result indicated a start site 20 bp downstream of that predicted (nucleotide 6444 of GenBank AF196567). However, this putative start site lay within a potential hairpin comprising the transcribed Fur box inverted repeat and additional sequence. As RNA secondary structure may have impeded reverse transcription, a thermostable enzyme protocol was applied in addition to the standard protocol, but it yielded the same result, supporting an unconventional promoter rather than the well-conserved, predicted σ70 promoter found in this region.
Deletion analysis was used to identify regions necessary for wild-type expression from the pdtF promoter and to determine whether there was a dependence on PdtC corresponding to the existence of an operator site. For those experiments, segments of DNA of various lengths found in the 5′ region of pdtF were inserted into the promoter-probe construct pMP77 and introduced into the wild-type PDTC-producing strain P. stutzeri KC. The largest promoter construct used in the experiments (pFpro1) contained an insert whose ends extended into the upstream reading frame (pdtE) and beyond the downstream end of the predicted Fur box (Fig. 4). Truncation by 55 bp at the 3′ end of that insert resulted in no detectable expression. Deletion of 82 bp from the 5′ end of the pFpro1 insert resulted in a significant increase in expression (P < 0.001), indicating the presence of repressor activity. All further 5′ truncations (i.e., larger than 80 bp) resulted in significant decreases in expression (P < 0.05); successive 36-, 26-, 28-, and 21-bp deletions decreased activity 2-fold, 3.6-fold, 5.2-fold, and 50-fold, respectively. Deletion of the −35 hexamer corresponding to the predicted σ70 promoter, but still 45 bp upstream of the transcription start site, abolished promoter activity. Growth under iron-replete conditions (50 μM FeSO4) led to 77-fold or greater repression of reporter activity for strains carrying pFpro1, pFpro2, or pFpro3 (data not shown). The data were consistent with the presence of an extended promoter that was sufficient for basal levels of expression and also demonstrated that an approximately 80-bp upstream region contributed positively to expression. The fact that a 27-bp segment of DNA gave a 10-fold increase in expression (reporter activity from pFpro5 versus pFpro6) indicated that a critical portion of the positive-acting sequence lay in that segment of DNA.
FIG. 4.
Deletion analysis of the pdtF promoter. On the left are diagrams of the pdtEF intergenic region (top) and fragments used to construct transcriptional fusions (below). The open and filled boxes represent −35 and −10 hexamers, respectively, identified by software analysis. The angled arrow represents the pdtF transcriptional start site determined by 5′ RACE, and the double oval represents the predicted Fur binding site. The numbers on the left indicate the positions of deletion endpoints (in bp) relative to the transcription start site. Data from reporter assays derived in the strain KC (WT) and SEMΔpdtC (ΔpdtC) backgrounds are shown to the right of each corresponding fragment. The data are means and standard errors of at least three independent experiments. ND, no activity detected.
To test whether the ability of upstream DNA to enhance transcription required PdtC, the largest reporter construct (pFpro1) and the construct having only the predicted minimal promoter (pFpro6) were tested in the ΔpdtC mutant. The inclusion of the upstream DNA segment did not lead to increased expression by that strain (Fig. 4). The levels of expression seen for the ΔpdtC mutant with either pFpro1 or pFpro6 were not significantly different from expression by the wild type harboring pFpro6 (P = 0.2), consistent with a dependence on PdtC for transcriptional activation through binding at an operator site located upstream and adjacent to the minimal promoter.
Reconstitution of pdt transcriptional activation in the surrogate host P. putida KT2440: dependence on pdtC and siderophore.
The results described above indicated that production of PDTC and transcriptional regulation of the pdt cluster were dependent on the product of the pdtC gene. However, it was unclear whether the cognate siderophore was necessary for the observed effects. It seemed plausible that PdtC alone was sufficient for transcriptional activation in response to the cognate siderophore. This would mean that many organisms might serve as hosts for investigating the dependence of transcriptional activation upon siderophores. We chose P. putida strain KT2440 for these tests based on the genome sequence and previous work with a related strain. The complete genome sequence of KT2440 has been determined (30), and we found no pdt orthologs by sequence alignment searches (1). It has also been demonstrated that pdt genes are expressed in that background, since PDTC production was observed when a cosmid containing the entire P. stutzeri KC pdt gene cluster was introduced into the clonally related strain mt-2 (22, 34). P. putida KT2440 grown in PM minimal medium does not show iron uptake from 55Fe-PDTC (20), and the clonally related strain mt-2 has been shown to be sensitive to the bacteriostatic effect of PDTC seen for other nonutilizing bacteria (37), i.e., even under conditions that would have induced a cryptic transport system, this strain showed severe growth inhibition in the presence of PDTC. Such growth inhibition can be abated by plasmid-borne transporter genes (20). For this reason, in our experiments using plasmid-bearing KT2440 derivatives, PDTC exposure was imposed after exponential growth had been achieved and using an amount that minimized growth retardation (10 μM).
In reconstitution experiments using strain KT2440/pFpro3, there was no detectable reporter expression, with or without added PDTC (Table 3), indicating that strain KT2440 lacked components present in P. stutzeri KC necessary for expression from the pdtF promoter. When the pdtC gene was also introduced into the KT2440 background on a second plasmid, giving strain KT2440/pFpro3 pSEMC, there was again no detectable expression from control cells (Table 3), indicating that pdtC alone was insufficient for correcting the apparent deficiency. However, when KT2440/pFpro3 pSEMC cells were exposed to the siderophore, expression was reconstituted to a level approximately 24% of that seen in P. stutzeri KC (Table 3). Therefore, siderophore-dependent transcriptional activation of the pdtF promoter could be reconstituted with only the pdtC gene.
TABLE 3.
Reconstitution of pdtF promoter expression in the heterologous host P. putida KT2440 by pdtC (in trans) plus cognate siderophore
| Culturea | XylE activity (nmol min−1 mg−1)b |
|---|---|
| KT2440 | ND |
| KT2440/pFpro3 | ND |
| KT2440/pFpro3 + PDTCc | ND |
| KT2440/pFpro3 pSEMC | ND |
| KT2440/pFpro3 pSEMC + PDTC | 7.2 ± 1.8 |
Cultures were grown in PM glucose/glutamine.
Mean ± standard error of three independent experiments. ND, not detected.
PDTC was added to 20 μM.
Expression of pdtC.
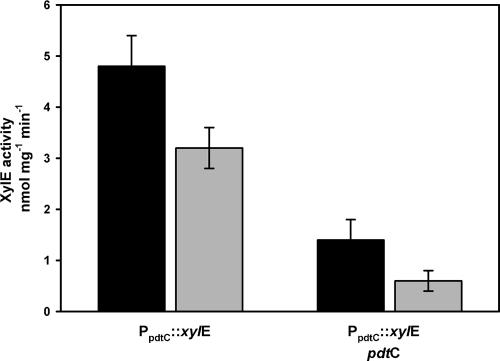
AraC-type regulator genes associated with siderophore systems are often found in opposing orientation to genes regulated by the respective proteins. Autoregulation effected by promoter overlap has been described for some of those regulators (4, 10, 29). The dependence of pdtC on its gene product and/or siderophore for expression was tested using the surrogate host P. putida KT2440 (Fig. 5). The experiments used pCpro, a transcriptional reporter construct with a 462-bp insert cloned upstream of the xylE reporter gene of pMP77. The inserted DNA included sequence beginning immediately upstream of the pdtC initiation codon, spanning the pdtC/pdtD intergenic region and including 37 bp of the pdtD coding region. Expression from pCpro was observed in all strains tested, i.e., no additional genes were required for reconstitution of expression from the cloned pdtC promoter. However, introduction of the pdtC gene led to significantly decreased expression relative to the reporter alone (P < 0.005). This repression without the addition of siderophore is reminiscent of XylS, which, when expressed in multicopy, shows regulatory effects in the absence of effectors (11).
FIG. 5.
Expression from the pdtC promoter by the pdt− host, P. putida KT2440; effects of PdtC and siderophore. The black bars are data from control experiments, and the gray bars are data from experiments in which cells were exposed to 20 μM PDTC. PpdtC::xylE denotes the presence of the reporter plasmid pCpro, and pdtC denotes pSEMC. The data are means and standard errors of at least three independent experiments.
An equilibrium may exist between various states of operator-bound and siderophore-bound activator molecules (4), and therefore, the effective dose of siderophore may be different from that seen for a biosynthetic gene promoter. The effect of added siderophore on the regulation of pdtC expression was measured by exposure of exponentially growing cells to 20 μM PDTC prior to assaying reporter activity (Fig. 5). A higher concentration of PDTC was used for these experiments than with pdtF promoter fusions in order to maximize the potential for effects on expression. Decreased reporter activity was seen in all strains exposed to PDTC relative to activity obtained without PDTC exposure. The decrease was relatively small (33%) in the strain containing the reporter alone. The effect of exogenously added siderophore was greater when the pdtC gene was present; a 57% decrease from the low basal level seen in that strain. The effect seen in the absence of PdtC may be attributed to effects of iron deprivation, whereas the combined effect of PdtC and siderophore exposure (an eightfold decrease in expression) are more likely attributable to specific regulation.
No promoter was identified for the pdtC gene using bacterial-promoter prediction software. This led us to investigate the site of transcription initiation and the requirement for an alternative sigma factor. The transcription start site was mapped and indicated that pdtC is transcribed with a long 5′ untranslated leader (347 bp). Although we were unable to determine the transcriptional start site for pdtD by the same methodology, a −10 hexamer (TATAAT) can be found centered 11 bp upstream from the pdtC transcriptional start site on the opposite strand, predicting overlapping promoters for pdtC and pdtD.
We examined the roles of two alternative sigma factors known to be involved in regulation in response to different nutritional stresses, RpoN and RpoS, as a means of potentially explaining the poor agreement (lack of sequence identity) with the canonical σ70 promoter sequence. For the experiments, we utilized mutants available in the KT2440 genetic background. PDTC production by strains carrying the entire pdt cluster on a cosmid (pT31) was used as one measure of expression, and analysis using the pCpro reporter construct was another. Both rpoS and rpoN strains were capable of PDTC production, although in decreased amounts for the rpoN strain (45% ± 18% [mean ± standard deviation]; n = 4; data normalized to OD600). Reporter expression in the rpoN mutant background was somewhat lower (82% of wild type), but the difference was not significant at a high confidence interval (P = 0.18). The data indicated that neither RpoS nor RpoN was necessary for expression from the pdtC promoter under the conditions used.
DISCUSSION
Our results confirm the role of the pdtC gene product as a transcriptional activator of genes within the pdt gene cluster and support a model that includes the cognate siderophore as a coeffector for that activation. Data that supported that model include the very low level of abundance of relevant mRNAs in a ΔpdtC mutant and the requirement for both pdtC and a cognate siderophore for reconstitution of expression in a surrogate host. Previous work had indicated that a transposon insertion mapped near the C terminus of pdtC had no effect on PDTC production (22). In light of the present work, it must be concluded that the position of the insertion was either incorrectly located due to the limited resolution of the restriction-mapping technique used or that the particular disruption allele did not affect domains critical to the function of PdtC.
It was also demonstrated that DNA sequences upstream of a putative biosynthetic gene were required for PdtC-dependent levels of transcription. Although our data did not directly demonstrate interaction of PdtC with promoter-proximal DNA, its sequence similarity to the AraC helix-turn-helix motif and sufficiency to act as a regulator in a surrogate host are compelling evidence for that mode of regulation. The possible involvement of other factors in full transcriptional activation was suggested by the relatively low levels of expression from the pdtF promoter in the KT2440 host relative to the native organism (24%). It is unlikely that transporter genes are the missing components, since no loss of transcriptional activation was seen in pdtK (outer membrane receptor) or pdtE (cytoplasmic membrane permease) mutants of P. putida DSM 3601 relative to their isogenic pdtK+ pdtE+ counterpart (20). It will be interesting to investigate whether other genes of the pdt cluster, particularly pdtP, contribute to transcriptional activation.
Positive regulation by an AraC-type transcriptional activator places the PDTC system within a category of siderophore systems known to have this mode of regulation (2, 7, 16, 24, 26). Well-characterized members of that category include the pyochelin system of Pseudomonas aeruginosa PAO1 (35), the yersiniabactin system of Yersinia pestis (10) and Yersina entercolitica (4), and the alcaligin system of Bordetella (7). For pyochelin, detailed characterization of promoters of the pch gene cluster has demonstrated the presence of PchR binding sites upstream of several genes (29). That work also showed that the 32-bp “PchR box” was sufficient for pyochelin-dependent activation. Our characterization of the pdtF promoter indicated that a larger (80- to 110-bp) upstream region was necessary for full transcriptional activity. A smaller promoter-proximal region was sufficient for expression at levels above that seen for a ΔpdtC mutant. The size of the DNA tract contributing to expression is larger than would be expected for the hypothetical footprint of a single PdtC dimer and includes negative- as well as positive-acting elements. The larger regulatory region may include multiple binding sites for PdtC or may indicate that other regulators have a role in expression from that promoter, in addition to PdtC. The main points that can be taken from the data are that PdtC is required to observe transcriptional activation, regardless of the potential involvement of other activators, and that a minimal, or “core,” sequence is present within 27 bp upstream of the minimal promoter that is also required for activation. Despite our efforts, no repeat units were identified within the pdtF upstream DNA that could also be recognized within sequences upstream of the other promoters analyzed (pdtC/pdtD and pdtP), i.e., no “PdtC box” was evident. Though we expected such a site to be present, sequence degeneracy likely obscured its identification.
Very different modes of regulation are found among siderophore systems. In addition to AraC-type activators, examples that include sigma/anti-sigma factors (40) and a two-component sensor/response regulator (8) are known. It seems likely that regulatory systems become linked to particular siderophore systems and are maintained through evolution by how well they allow tuning of the system in response to utilization efficiency. One characteristic of the siderophore system that imposes an important constraint would be whether iron is delivered via intracellular translocation of the iron-siderophore or by extracytoplasmic (e.g., periplasmic) release of iron from the siderophore and subsequent uptake of the released iron. Since AraC family regulators are DNA binding proteins, effectors that interact with them to modulate that binding must be cytoplasmically localized. Models envisioned for siderophore systems regulated by AraC family regulators have incorporated the iron-siderophore complex as the intracellular effector of transcriptional activation (10, 16). That model also fits well into a passive-control mechanism for coordinating alternative siderophore systems within the same cell; the siderophore that returns iron to the cell most efficiently leads to activation of the system, while less efficient alternative systems remain at basal levels of expression. The dispensability of outer membrane receptors for transcriptional activation in the yersiniabactin, enterobactin, and P. putida DSM3601/PDTC systems, all of which include AraC-type regulators (2, 10), and our ability to reconstitute PDTC-dependent activation in a surrogate host with only the transcriptional regulator introduce some apparently paradoxical characteristics into such a model. Those observations indicate that the system is tuned to an effector signal (i.e., a level of iron-siderophore) that does not actually provide nutritional benefit to the cell. The possibility of alternative receptors/transporters cannot be ruled out, but unlike the alternative systems that have been described (12, 13, 28), the affinity of any such system present in strain KT2440 for PDTC-Fe is too low to allow the organism to grow in the presence of exogenously added PDTC (37). However, the dispensability of a specific outer membrane receptor does not appear to be a general characteristic of siderophore systems regulated by AraC family members, since a role for the pyochelin outer membrane receptor (FptA) in transcriptional regulation has been indicated (16). At any rate, it appears that the effector of PDTC-dependent activation (PDTC or Fe-PDTC) can enter cells by an alternative route, resulting in transcriptional regulation that requires only the activator. Another possible effector to be considered is free PDTC, rather than Fe-PDTC. Such a mechanism would be reminiscent of transcriptional activation by another AraC family activator, XylS, which uses freely diffusible aromatic acid effectors to allow transcriptional activation in many host bacteria (6, 27). More details regarding the molecular mechanism underlying PDTC-dependent regulatory phenomena are necessary to resolve the apparent paradox of unlinked nutritional benefit and siderophore signaling.
Acknowledgments
This work was supported by the Vermont Agricultural Experiment Station.
We thank Scott Tighe, Timothy Hunter, Joyce Heckman, and Ken Hampel for technical advice.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. T., and S. K. Armstrong. 2004. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J. Bacteriol. 186:7302-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quiñones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 4.Anisimov, R., D. Brem, J. Heesemann, and A. Rakin. 2005. Molecular mechanism of YbtA-mediated transcriptional regulation of divergent overlapping promoters ybtA and irp6 of Yersinia enterocolitica. FEMS Microbiol. Lett. 250:27-32. [DOI] [PubMed] [Google Scholar]
- 5.Ausebel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 6.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, P. Karunakaran, and S. Valla. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in Gram-negative bacteria. Plasmid 38:35-51. [DOI] [PubMed] [Google Scholar]
- 7.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean, C. R., and K. Poole. 1993. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol. Microbiol. 8:1095-1103. [DOI] [PubMed] [Google Scholar]
- 9.DeLorenzo, V., M. Herrero, U. Jakubzik, and K. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fetherston, J. D., S. W. Bearden, and R. D. Perry. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22:315-325. [DOI] [PubMed] [Google Scholar]
- 11.Gallegos, M.-T., R. F. Schleif, A. Bairoch, K. Hofman, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghysels, B., B. Dieu, S. Beatson, J.-P. Pirnay, U. Ochsner, A. Vasil, and P. Cornelis. 2004. FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150:1671-1680. [DOI] [PubMed] [Google Scholar]
- 13.Ghysels, B., U. Ochsner, U. Möllman, L. Heinisch, M. Vasil, P. Cornelis, and S. Matthijs. 2005. The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiol. Lett. 246:167-174. [DOI] [PubMed] [Google Scholar]
- 14.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs, D. E., and K. Poole. 1993. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J. Bacteriol. 175:5882-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildebrand, U., W. Ockels, J. Lex, and H. Budzikiewicz. 1983. Zur Struktur eines 1:1-Adduktes von Pyridin-2,6-Dicarbothiosäure und Pyridin. Phosphorus Sulfur 16:361-364. [Google Scholar]
- 17a.Klecka, G. M., and D. T. Gibson. 1981. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl. Environ. Microbiol. 41:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köhler, T., S. Harayama, J. L. Ramos, and K. Timmis. 1989. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J. Bacteriol. 171:4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korbel, J. O., L. J. Jensen, C. von Mering, and P. Bork. 2004. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat. Biotechnol. 22:911-917. [DOI] [PubMed] [Google Scholar]
- 20.Leach, L., and T. A. Lewis. 2006. Identification and characterization of Pseudomonas membrane transporters necessary for utilization of the siderophore pyridine-2,6-bis(thiocarboxylic acid) (PDTC). Microbiology 152:3157-3166. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C.-H., T. A. Lewis, A. Paszczynski, and R. L. Crawford. 1999. Identification of an extracellular catalyst of carbon tetrachloride dehalogenation from Pseudomonas stutzeri strain KC as pyridine-2,6-bis(thiocarboxylate). Biochem. Biophys. Res. Commun. 261:562-566. (Erratum, 265:7709.) [DOI] [PubMed] [Google Scholar]
- 22.Lewis, T. A., M. Cortese, J. Sebat, T. Green, C.-H. Lee, and R. L. Crawford. 2000. A Pseudomonas stutzeri gene cluster encoding the biosynthesis of the CCl4-dechlorination agent pyridine-2,6-bis(thiocarboxylic acid). Environ. Microbiol. 2:407-416. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, T. A., L. Leach, S. E. Morales, P. R. Austin, H. J. Hartwell, B. Kaplan, C. Forker, and J.-M. Meyer. 2004. Physiological and molecular genetic evaluation of the dechlorination agent, pyridine-2,6-bis(monothiocarboxylic acid) (PDTC) as a secondary siderophore of Pseudomonas. Environ. Microbiol. 6:159-169. [DOI] [PubMed] [Google Scholar]
- 24.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O. Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 26.Matthijs, S., C. Baysse, N. Koedam, A. K. Tehrani, L. Verheyden, H. Budzikiewicz, M. Schafer, B. Hoorelbeke, J.-M. Meyer, H. DeGreve, and P. Cornelis. 2004. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol. Microbiol. 52:371-384. [DOI] [PubMed] [Google Scholar]
- 27.Mermod, N., J. L. Ramos, P. R. Lehrbach, and K. Timmis. 1986. Vector for regulated expression of cloned genes in a wide range of gram-negative bacteria. J. Bacteriol. 167:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, J.-M. 1992. Exogenous siderophore-mediated iron uptake in Pseudomonas aeruginosa: possible involvement of porin OprF in iron translocation. J. Gen. Microbiol. 138:951-958. [DOI] [PubMed] [Google Scholar]
- 29.Michel, L., N. Gonzales, S. Jagdeep, T. Nguyen-Ngoc, and C. Reimmann. 2005. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 58:495-509. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, K. E., C. Weinei, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. P. Martins dos Santos, et al. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 31.Paszczynski, A., J. Sebat, D. Erwin, and R. L. Crawford. 2004. Biotransformation of carbon tetrachloride by the facultative anaerobic bacterium Pseudomonas stutzeri, p. 317-328. In M. M. Nakano and P. Zuber (ed.), Strict and facultative anaerobes: medical and environmental aspects. Horizon Bioscience, Wymondham, United Kingdom.
- 32.Poole, K., and G. A. McKay. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8:661-686. [DOI] [PubMed] [Google Scholar]
- 33.Ramos-Gonzales, M. L., and S. Molin. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 180:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regenhardt, D., H. Heuer, D. U. Fernandez, C. Strömpl, E. R. B. Moore, and K. Timmis. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 4:912-915. [DOI] [PubMed] [Google Scholar]
- 35.Reimmann, C., L. Serino, M. Beyeler, and D. Haas. 1998. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144:3135-3148. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Rapid amplification of 5′ cDNA ends, p. 8.54-8.6. In Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sebat, J., A. Paszczynski, M. Cortese, and R. L. Crawford. 2001. Antimicrobial properties of pyridine-2,6-dithiocarboxylic acid, a metal chelator produced by Pseudomonas spp. Appl. Environ. Microbiol. 67:3934-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sepulveda-Torres, L. D. C., A. Huang, H. Kim, and C. S. Criddle. 2002. Analysis of regulatory elements and genes required for carbon tetrachloride degradation in Pseudomonas stutzeri strain KC. J. Mol. Microbiol. Biotechnol. 4:151-161. [PubMed] [Google Scholar]
- 39.Spaink, H. P., J. H. Okker, C. A. Wijffelmann, E. Pees, and B. J. J. Lugtenberg. 1984. Promoter in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 40.Visca, P., L. Leoni, M. J. Wilson, and I. L. Lamont. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45:1177-1190. [DOI] [PubMed] [Google Scholar]