Abstract
The expression of many virulence-associated genes in Streptococcus pyogenes is controlled in a growth phase-dependent manner. Unlike the model organisms Escherichia coli and Bacillus subtilis, such regulation is apparently not dependent upon alternative sigma factors but appears to rely on complex interactions among several transcriptional regulators, including Rgg. The purpose of this study was to identify changes in gene expression associated with inactivation of the rgg gene in S. pyogenes strain NZ131 (serotype M49). To this end, the transcriptomes of wild-type and rgg mutant strains were analyzed during both the exponential and postexponential phases of growth using Affymetrix NimbleExpress gene chips. Genomewide differences in transcript levels were identified in both phases of growth. Inactivation of rgg disrupted coordinate expression of genes associated with the metabolism of nonglucose carbon sources, such as fructose, mannose, and sucrose. The changes were associated with an inability of the mutant strain to grow using these compounds as the primary carbon source. Bacteriophage transcript levels were also altered in the mutant strain and were associated with decreased induction of at least one prophage. Finally, transcripts encoding virulence factors involved in cytolysin-mediated translocation of NAD-glycohydrolase, including the immunity factor IFS and the cytolysin (streptolysin O [SLO]), were more abundant in the mutant strain, which correlated with the amount of NADase and SLO activities in culture supernatant fluids. The results provide further evidence that Rgg contributes to growth phase-dependent gene regulation in strain NZ131.
In order to survive, bacteria must be able to respond to changes in the environment and tolerate a variety of stressors. Such adaptation typically involves genomewide changes in transcription, as well as posttranscriptional and posttranslational changes. Escherichia coli and Bacillus subtilis use alternative sigma factors, such as RpoS and SigH, respectively, to coordinate changes in transcription upon entry into the stationary phase of growth. The mechanism facilitates rapid changes in expression without the need to assemble transcriptional complexes de novo. In contrast, the human pathogen Streptococcus pyogenes does not require alternate sigma factors to adapt to the stationary phase of growth (20, 21). Rather, S. pyogenes is thought to rely on interactions among multiple transcriptional factors, including Mga, RofA-like proteins (RALP), two-component regulators (CovRS/CsrRS, FasBCAX, and Ihk/Irr), and Rgg (12). Importantly, these regulators also control the expression of many virulence factors, which are typically expressed in a growth phase-dependent manner. Identifying how such regulatory networks function is important in understanding the regulation of gene expression in S. pyogenes and in designing therapeutic strategies aimed at inhibiting virulence factor expression.
Members of the Rgg family of transcriptional regulators (TIGR01716) are encoded in the genomes of a subset of low-G+C gram-positive bacteria. These include S. pyogenes (5, 14), Streptococcus gordonii (31), Streptococcus oralis (10), Streptococcus sanguinis (35), Streptococcus mutans (24), Lactococcus lactis (27), Listeria monocytogenes, and lactobacilli. Orthologues have not been identified for Staphylococci, Bacillus, or gram-negative bacteria. Rgg-like proteins have a putative helix-turn-helix motif, which is likely to facilitate binding to the promoter regions of Rgg-regulated genes (19, 25, 27, 34). The first member to be characterized was designated Rgg because it is a positive regulator of the adjacent glucosyltransferase G gene (gtfG) in S. gordonii; GtfG catalyzes the formation of extracellular glucans (30, 31). In L. lactis, the Rgg-like regulator GadR activates transcription of the adjacent gadBC operon, which is associated with glutamate-dependent acid tolerance (27). In S. mutans, the Rgg-like protein MutR activates transcription of the adjacent mutAMTFEG operon, which encodes the synthesis of the lantibiotic mutacin (23). Finally, in Lactobacillus sakei, the Rgg-like regulator LasX activates adjacent genes, including the lantibiotic lactocin S (28). Thus, members of the Rgg family typically influence the expression of proximal genes encoding functionally heterogeneous proteins.
The rgg locus of S. pyogenes, also known as ropB (15), is adjacent to speB and mf-1, which encode a secreted protease and nuclease, respectively. Inactivation of rgg abrogates SpeB expression and elevates Mf-1 expression (7). Rgg binds to the speB promoter to activate transcription directly (19). In strain NZ131, Rgg influences the expression of a variety of secreted virulence factors, such as the M protein, Mac, SagA, SpeB, and streptolysin O (SLO) (7). Inactivation of rgg also alters the expression of known regulatory proteins, including Mga, CsrRS/CovRS, FasBCAX, and Ink/Irr (7), which implies that at least some of the changes in virulence gene expression associated with the mutant strain are probably due to the perturbation of other regulatory circuits.
Spotted DNA microarrays were previously used to screen an rgg mutant strain for differences in virulence and regulatory gene transcript levels during the exponential phase of growth. Potential changes were then examined further with TaqMan reverse transcription (RT)-PCR. Using this strategy, 25 genes were identified as being differentially transcribed in the mutant strain during the exponential phase of growth (7). Subsequently, a proteomic approach was used to identify Rgg-regulated proteins in both the exponential and stationary phases of growth. Significant differences in the abundances of growth phase-regulated proteins involved in amino acid metabolism and the response to thermal and oxidative stress were identified (4). Although the results suggested that Rgg was a global regulatory protein, only 10% of the predicted proteome (based on the number of open reading frames identified in the genome sequence of S. pyogenes strain SF370) was detected by using two-dimensional gel electrophoresis. Such coverage is typical and is due to a variety of factors, including protein insolubility and the inability to detect proteins present at low levels. Nevertheless, the proteomic analyses emphasized the importance of assessing growth phase-dependent patterns of gene expression in the rgg mutant strain.
In this study, Affymetrix NimbleExpress DNA microarrays were used to identify Rgg-dependent changes not only in virulence and regulatory genes but in the transcriptomes of wild-type and rgg mutant strains during both the exponential and postexponential phases of growth. The results define the Rgg regulon in strain NZ131 and confirm that Rgg is a global transcriptional regulator. Many of the differences in gene expression were among growth phase-regulated genes, including genes involved in the catabolism of nonglucose carbohydrates. Another goal was to assess the phenotypes predicted by the expression analysis. Results from these experiments showed the following: (i) Rgg is essential for growth of the pathogen in media containing nonglucose carbohydrates as the primary carbon source; (ii) inactivation of the rgg gene alters prophage gene expression; (iii) inactivation of rgg alters the frequency of prophage induction; (iv) Rgg represses the expression of the virulence-associated NAD-glycohydrolase operon in the exponential phase of growth. Together, the information contributes to efforts aimed at deciphering interactions among regulatory circuits that mediate changes in gene expression in response to changing environmental conditions.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The wild-type S. pyogenes strain NZ131 (serotype M49) and the NZ131 rgg mutant were previously described (5). S. pyogenes was grown at 37°C in a 5% CO2 atmosphere without agitation in either Todd-Hewitt broth (Becton Dickinson, Sparks, MD) containing 0.2% (wt/vol) yeast extract (THY) or chemically defined medium (CDM). The composition of CDM was similar to that previously described (33) and consisted of the following: glycine, 200 mg/liter; l-histidine, 200 mg/liter; l-leucine, 200 mg/liter; l-lysine, HCl, 250 mg/liter; l-proline, 200 mg/liter; l-hydroxyproline, 200 mg/liter; l-aspartic acid, 200 mg/liter; dl-alanine, 200 mg/liter; l-cystine, 2HCl, 2 mg/liter; l-glutamic acid, 200 mg/liter; l-glutamine, 400 mg/liter; l-isoleucine, 200 mg/liter; l-methionine, 200 mg/liter; l-phenylalanine, 200 mg/liter; l-threonine, 400 mg/liter; l-tryptophan, 200 mg/liter; l-tyrosine, 288 mg/liter; l-valine, 200 mg/liter; p-aminobenzoic acid, 0.4 mg/liter; biotin, 0.4 mg/liter; folic acid, 1.6 mg/liter; niacinamide, 2 mg/liter; β-NAD, 5 mg/liter; pantothenate calcium salt, 4 mg/liter; pyridoxal HCl, 2 mg/liter; pyridoxamine HCl, 2 mg/liter; riboflavin, 4 mg/liter; thiamine HCl, 2.0 mg/liter; vitamin B12, 0.2 mg/liter; adenine sulfate, 2H2O, 40 mg/liter; guanine hydrochloride, 40 mg/liter; uracil, 40 mg/liter; CaCl2, 10.2 mg/liter; NaHCO3, 5 g/liter; NaC2H3O2, 5.4 g/liter; NaH2PO4, 6.4 g/liter; Na2HPO4, 14.7 g/liter; FeSO4, 10 mg/liter; Fe(NO3)2, 2 mg/liter; K2HPO4, 400 mg/liter; KH2PO4, 2 g/liter; MgSO4, 682 mg/liter; and MnSO4, 10 mg/liter. l-arginine, l-serine, and l-cysteine were added to freshly prepared liquid CDM in final concentrations of 640 mg/liter, 200 mg/liter, and 1 g/liter, respectively. Sucrose, glucose, fructose, or mannose was added at a final concentration between 0.05 and 2.0% (wt/vol). For experiments using nonglucose carbohydrates, the strains were cultured with THY agar plates overnight. Bacteria were suspended in 5 ml of carbohydrate-free CDM by using a sterile swab, and the A600 was determined. A fraction of the suspension (approximately 200 μl) was then used to inoculate 10 ml of CDM containing sucrose, fructose, or mannose to an A600 of 0.1. The cultures were incubated at 37°C in a 5% CO2 atmosphere. A similar approach was used to inoculate medium containing 0.15% glucose. Growth with different carbon sources was determined with four to six independent experiments. DNase activity was assessed by stab inoculating DNase test agar plates containing methyl green (Becton Dickinson, Sparks, MD) and incubating the plates overnight at 37°C in 5% CO2.
RNA isolation.
An overnight culture of S. pyogenes was inoculated into 40 ml of THY broth in 50-ml tubes to an A600 of 0.08. The cultures were grown to either the exponential or postexponential phase of growth in the absence of antibiotics. The heterologous DNA that disrupts rgg is stable under these conditions, as determined by replicate plating and PCR. Cultures were centrifuged, and the bacteria were suspended in 300 μl of RNAlater (Ambion, Austin, TX) and frozen in liquid nitrogen. RNA was isolated by using an RNeasy Mini kit (QIAGEN, Valencia, CA), according to the procedure recommended by the manufacturer. The concentration and quality of RNA were assessed with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) using an RNA 6000 Nano LabChip kit (Agilent).
cDNA synthesis and biotinylation.
Ten micrograms of total RNA was mixed with 500 ng of random hexanucleotide primers and 1,200 U of Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) for 90 min at 42°C. Following cDNA synthesis, 1 U each of RNase H (Invitrogen) and RNase A (EpiCentre, Madison, WI) was added to the reaction mix and incubated at 37°C for 10 min to degrade any remaining RNA. The cDNA was purified with MinElute PCR purification columns (QIAGEN), and 2 μg of cDNA was fragmented with 0.2 U of DNase I (EpiCentre) for 8 min at 37°C. Fragmentation was stopped by heating for 10 min at 99°C. The 3′ terminus of the fragmented cDNA was labeled with Biotin-N6-ddATP (ENZO, New York, NY) at 37°C for 2 h using 50 U of terminal deoxynucleotide transferase (New England Biolabs, Ipswich, MA).
DNA microarray hybridization and analysis.
Affymetrix NimbleExpress arrays were purchased from Affymetrix (Santa Clara, CA). The arrays were designed based on the S. pyogenes strain SF370 genome sequence (9). The arrays consisted of 2,543 qualifiers representing 1,694 predicted S. pyogenes open reading frames and 804 intergenic region probes. In addition, 45 control oligonucleotides were used for spike-ins.
The GeneChips were hybridized with 1.86 μg of biotin end terminus labeled cDNA in 1× hybridization buffer (100 mM morpholineethanesulfonic acid [MES], 1 M NaCl, 20 mM EDTA, and 0.01% Tween 20 [Pierce Chemical, Rockford, IL], 50 pM control oligonucleotide B2 [Affymetrix], 20 μg herring sperm DNA ′Promega, Madison, WI[, 100 μg acetylated bovine serum albumin [Invitrogen]) for 16 h at 45°C on a rotisserie at 60 rpm. Following hybridization, the hybridization cocktail was removed and the GeneChip filled with nonstringent buffer A (0.9 M sodium chloride, 60 mM sodium phosphate, 6 mM EDTA, and 0.01% Tween 20). GeneChips were postprocessed on an automated Affymetrix GeneChip Fluidics Station 450 using the following protocol: wash 1, 10 cycles of 2 mixes/cycle with nonstringent buffer A at 25°C; wash 2, 4 cycles of 15 mixes/cycle with stringent buffer B (100 mM MES, 0.1 M NaCl, and 0.01% Tween 20) at 45°C; first stain, 10 min at 25°C in streptavidin-phycoerythrin (SAPE) solution (1× MES buffer [100 mM MES, 1 M NaCl, and 0.05% Tween 20], 2 mg/ml acetylated bovine serum albumin, and 10 μg/ml SAPE [Molecular Probes, Eugene, OR]); poststain, wash, 10 cycles of 4 mixes/cycle with nonstringent buffer A at 30°C; second stain, 10 min in antibody solution [1× MES stain buffer, 2 mg/ml acetylated bovine serum albumin, 0.1 mg/ml normal goat immunoglobulin G (Sigma-Aldrich, St. Louis, MO), and 3 μg/ml biotinylated antibody (Vector Laboratories, Burlingame, CA); third stain, 10 min in SAPE solution at 25°C; final wash, 15 cycles of 4 mixes/cycle with nonstringent buffer A at 30°C. The arrays were scanned at 570 nm at a resolution of 1.56 μm using a confocal GC3000 laser scanner (Affymetrix). Gene expression levels were calculated with GeneSpring 7 software and normalized with the per-chip algorithm (Silicon Genetics, Redwood City, CA). For each strain and growth condition, two independently isolated RNA samples were analyzed using 10 Affymetrix chips. The average signal intensity value of each gene was transformed to a log2 (log base 2) value. The change between two experimental conditions (n-fold) was calculated by taking the ratio of the signal intensity (difference of the log2 value) between experimental conditions. Present and absent calls were assessed, and genes with a twofold difference in RNA levels were considered to be differentially expressed (see Tables S1 and S2 in the supplemental material). Statistically significant (t test; P ≤ 0.05) differences with changes in transcript levels of fivefold or greater are summarized in Tables 2 and 3.
TABLE 2.
Transcriptome changes (fivefold or greater and P values of <0.05) associated with rgg inactivation during exponential growth
| Category and SPy no.a,b | Genec | Description | Fold change(s) (rgg/wt)d |
|---|---|---|---|
| More abundant in rgg mutant strain | |||
| Metabolism | |||
| 1541-1542-1543-1544-1546-1547 | arcC/-/-/arcB/-/arcA | Arginine deiminase operon | 43, 47, 43, 47, 20, 16 |
| 1057-1058-1059-1060 | Hypothetical protein, PTS system | 8, 14, 30, 26 | |
| 1111 | Alcohol dehydrogenase | 10 | |
| 258 | Glucose kinase | 8 | |
| 44 | adhA | Alcohol dehydrogenase | 5 |
| Transport | |||
| 252-254-255 | Sugar transport | 5, 9, 7 | |
| Exoproteins/virulence associated | |||
| 165-166-167 | spn/ifs/slo | NAD-glycohydrolase, SPN immunity factor, streptolysin O | 53, 32, 28 |
| 1436-1437 | mf-3/- | Putative DNase; hypothetical protein | 27, 25 |
| 1983 | scl | Collagen-like surface protein | 13 |
| 2010 | scpA | C5a peptidase precursor | 5 |
| 1979 | ska | Streptokinase A precursor | 5 |
| Regulatory | |||
| 1596 | Putative transcriptional regulator | 6 | |
| 1549 | ahrC.2 | Putative arginine repressor | 5 |
| Hypothetical | |||
| 428 | Hypothetical protein | 9 | |
| 1698 | yitS | Hypothetical protein | 7 |
| 256, 1339, 1563 | Hypothetical proteins | 6 | |
| 136 | Hypothetical protein | 5 | |
| Less abundant in rgg mutant strain | |||
| Metabolism | |||
| 1136 | xpt | Xanthine phosphoribosyltransferase | −17 |
| 900 | pyrF | Putative orotidine-5 -decarboxylase PyrF | −6 |
| Stress responsive | |||
| 1856 | norA | Antibiotic resistance protein NorA | −57 |
| 1205 | Putative antimicrobial resistance factor | −7 | |
| Transport | |||
| 1270 | Putative amino acid symporter | −16 | |
| 1137 | Putative purine permease | −16 | |
| 323 | braB | Putative branched-chain amino acid transport protein | −10 |
| 324 | Putative sodium dicarboxylate symporter | −6 | |
| 1506-1507 | Putative amino acid ABC transporters | −6, −5 | |
| 1414 | kup | Putative cation (K+) transport protein | −5 |
| Exoproteins/virulence associated | |||
| 2039 | speB | Pyrogenic exotoxin B | −35 |
| 1357 | graB | GRAB | −8 |
| Regulatory | |||
| 216 | ralp | Regulatory protein; RofA related | −10 |
| Hypothetical | |||
| 227, 993 | Hypothetical proteins | −7 | |
| 943, 706, 1340 | Hypothetical proteins | −6 | |
| 917, 1736 | Hypothetical proteins | −5 | |
| Miscellaneous | |||
| 991 | Structural protein; phage associated | −7 |
SPy numbers designate open reading frames based on the SF370 S. pyogenes genome annotation (9).
Contiguous genes likely to be cotranscribed are separated with a dash.
Hyphen indicates an unnamed gene.
Change in transcript level for rgg mutant compared to that for the wild type.
TABLE 3.
Transcriptome changes (fivefold or greater and P values of <0.05) associated with rgg inactivation during postexponential growth
| Category and SPy no.a,b | Genec | Description | Fold change(s) (rgg/wt)d |
|---|---|---|---|
| More abundant in the rgg mutant | |||
| Metabolism | |||
| 1058-1059-1060 | Hypothetical protein-PTS system | 8, 10, 15 | |
| 2081, 2083-2084, 2087 | hutI, -/-, - | Histidine metabolism | 9, 11, 5, 6 |
| 392 | upp | Uracil phosphoribosyltransferase | 5 |
| Stress responsive | |||
| 123 | Putative heat shock protein HSP33 | 5 | |
| Transport | |||
| 831 | pyrP | Putative uracil permease | 5 |
| Exoproteins/virulence associated | |||
| 165 | spn | NAD-glycohydrolase precursor | 14 |
| 2010 | scpA | C5a peptidase precursor | 7 |
| 1436-1437 | mf-3/- | DNase | 6, 5 |
| Regulatory | |||
| 1202 | Putative transcription regulator, GntR family | 7 | |
| Hypothetical | |||
| 2191 | Hypothetical protein | 18 | |
| 843 | Hypothetical protein | 13 | |
| 1562-1563, 1565-1566-1567 | Hypothetical proteins | 10, 9, 7, 7, 7 | |
| 305 | Hypothetical protein | 8 | |
| 844 | Hypothetical protein | 7 | |
| 1046 | Hypothetical protein | 6 | |
| 306-307 | Hypothetical proteins | 6, 6 | |
| 726, 733 | Hypothetical proteins | 5 | |
| Miscellaneous | |||
| 937 | int3 | Putative integrase, phage associated | 11 |
| 1391 | Putative methyltransferase | 6 | |
| 469 | Putative 42-kDa protein | 6 | |
| 1629 | priA | Primosomal replication factor Y | 5 |
| 870 | fms | Putative polypeptide deformylase | 5 |
| Less abundant in the rgg mutant strain | |||
| Metabolism | |||
| 2172-2173 | -/yoxJ | NUDIX protein | −14, 13 |
| 1541, 1544 | arcC, arcB | Arginine deiminase operon | −7, 5 |
| 1183-1184 | Decarboxylase complex | −5, 7 | |
| Stress responsive | |||
| 1856 | norA | Putative antibiotic resistance protein NorA | −28 |
| Transport | |||
| 230 | Putative ABC transporter (ATP-binding protein) | −5 | |
| Exoproteins/virulence associated | |||
| 2040 | SpeB maturation protein | −192 | |
| 2039 | speB | Pyrogenic exotoxin B | −59 |
| 1357 | graB | GRAB | −6 |
| 212 | speG | Exotoxin G precursor | −5 |
| Regulatory | |||
| 216 | ralp | Regulatory protein, RofA related | −16 |
| 946 | P1-type antirepressor, phage associated | −12 | |
| 1994 | pai1 | Pai1 protein (theoretical repressor) | −6 |
| Hypothetical | |||
| 943 | Hypothetical protein | −26 | |
| 238 | Hypothetical protein | −12 | |
| 1956 | Hypothetical protein | −8 | |
| 706, 940, 1959 | Hypothetical proteins | −7 | |
| 219, 917, 981, 989, 998, 2215 | Hypothetical proteins | −6 | |
| 1024 | Hypothetical proteins | −5 | |
| Miscellaneous | |||
| 707 | Putative holin, phage associated | −6 | |
| 1073 | rplL | 50S ribosomal protein L7 L12 | −5 |
SPy numbers designate open reading frames based on the SF370 S. pyogenes genome annotation (9).
Contiguous genes likely to be cotranscribed are separated with a hyphen.
Hyphen indicates an unnamed gene.
Change in transcript level for rgg mutant compared to that for the wild type.
Quantitative RT-PCR.
Oligonucleotide primers and TaqMan probes (Table 1) were designed with Primer Express 2.0 software (ABI Prism; PE Biosystems, Framingham, Mass.) and purchased from Sigma-Genosys (The Woodlands, TX). Amplification and detection were done with the ABI Prism 7700 sequence detection system (PE Applied Biosystems) using TaqMan One-Step RT-PCR Master Mix reagents (Roche, Indianapolis, Ind.), as recommended by the manufacturer. Each assay was done in triplicate with at least two independently isolated RNA samples and analyzed as previously described (6).
TABLE 1.
Primers and TaqMan probes used for quantitative RT-PCR
| SPy no.a | Designationb | Forward (5′-3′) | Reverse (5′-3′) | Fluorescent probe (5′-3′)c |
|---|---|---|---|---|
| 165 | spn (NAD-glycohydrolase) | CACCTACACTAAAAAACCGCATCA | CAAAAGTGACCTCTGACAAGGCTAA | TAACAGAGGTCCATCAGGGACAAAACAAGGAT |
| 216 | None (putative RofA-related regulator) | ACCCGCTTGTTTGGTTTGAG | GGTGCAAGGCTAAGATGGTTTT | ACTGTATCCAACAGCAGCGCGAAGATTT |
| 901 | pyrE (putative orotate phosphoribosyltransferase) | CCCTTTTACGTGGGCATCTG | CTTCTGGGAAATGAGCCTTGAT | CACTGATAATCGTGTCACCCTTTCTTATCCTAAGACTC |
| 1059 | None (putative PTS enzyme II, component C) | TTGGAATGTTGGCAGCCTTT | CCAAGTGCCATAGTGGAGCAT | TTCATGAATATCTTTGCTCCAATGGTTGATAAAGC |
| 1547 | arcA (arginine deiminase) | GGTGGTGGTAAAGTGCCTATGGT | CAAGTTCGTCCCCACCTTCA | TATGACCGTAATGAAACCACTCGCA |
| 1856 | norA (putative antibiotic resistance protein NorA) | GAGTGGATGAGCACCGCTTT | AAAGGAAGCCACACCACTACCA | TCTTGATTTAGGTTTAGGTGCTGGGCCTTACC |
| 1981 | relA ((p)ppGpp synthetase, GTP pyrophosphokinase) | CCGAAAACCATCGCAAAATG | CGAAATGCGCTCTTGTTTGTC | TGAAATTGGCTGACCGCCTGCATA |
| 2039 | speB (pyrogenic exotoxin B) | CGCACTAAACCCTTCAGCTCTT | ACAGCACTTTGGTAACCGTTGA | TACTGGTGGCGGCGCACC |
| 2042 | rgg (transcription regulator) | AAGTCAACAAAGGAAAAGAACCTTTT | AAATAAGTCCGCTCTGTCAGACAGT | TTGGTCAAGGTGTTATTAGCAACTCTTACTGAGGAA |
| 2043 | mf (mitogenic factor) | CACAGGTCTCAAATGATGTTGTTCT | CTGTCATTGAATGTCCAAGCTAATG | ATGATGGCGCAAGCAAGTACCTAAACGA |
| 2084 | None (putative serine cycle enzyme) | ACCTGGTTTCTGCCGTTTTTG | TTCGACCATGAGGGTCATCA | CCAAAAGAGACTGACGAAGACAAGGCTGCT |
| none | NZ131.1 prophage integrase | TCCGGAGATCAATTTGTTCGA | CATGGTTCTTGCTTTTACAGTTTTTG | TTGGACAATTGGTAGAAGAGTGGAAGACATCACA |
SPy numbers are based on annotation of the SF370 S. pyogenes strain complete genome (9).
Gene designations are in italics, and the corresponding protein function, if known or predicted, is in parentheses.
Covalently linked at the 5′ end to 5-carboxyfluorescein and at the 3′ end to N,N,N-tetramethyl-6-carboxyrhodamine.
Determination of SLO cytolytic activity.
SLO activity was determined as previously described (3). Briefly, sterile culture supernatant fluids were prepared from 10-ml cultures of the wild-type and rgg mutant strains. One milliliter of fluid was reduced with 4 mM dithiothreitol for 10 min at room temperature. A 0.5-ml amount of 2% (vol/vol) rabbit erythrocytes (Colorado Serum, Denver, CO) suspended in phosphate-buffered saline (PBS) (pH 7.4) was added, and the mixture was incubated for 30 min at 37°C. Following centrifugation to pellet intact erythrocytes, the A541 value of the supernatants was determined. To confirm the activity was due to SLO, supernatant fluids were pretreated with 25 μg/ml of cholesterol, which inhibits SLO. The mean and standard error of the means from the analysis of four independently isolated culture supernatant fluids are reported.
Determination of NADase activity.
NADase activity was determined as previously described (3). Briefly, sterile culture supernatant fluids were prepared from 10-ml cultures of the wild-type and rgg mutant strains. One hundred microliters of serial twofold dilutions of the fluids were added to 96-well microtiter plates. NAD+ (Sigma) was added to each well at a final concentration of 0.67 mM, and the plates were incubated at 37°C. After 1 h, 40 μl of 5 N NaOH was added to each well and the plate was incubated in the dark for 1 h. Fluorescent NAD+ was detected by excitation at 365 nm. Each result is reported as the greatest dilution resulting in an absence of fluorescence. The mean and standard error of the means are reported for four independently isolated culture supernatant fluids.
Analysis of SF370.2-related phage lysogenic and lytic conditions.
Analysis of the NZ131.1 prophage was done by using PCR as described in the text. Primers cpsFQ (5′-AATTGCGTGAAGAACTCGGCTG-3′ ), int3 (5′-TGGACATGCTGACTCGAAAACTACTC-3′ ), speH (5′-CGGGAACAAAAATACTCTAAAGGAACTG-3′), and mutX (5′-GCTTACCCCCAACTGAAATCCACTT-3′) were designed with Primer Express 2.0 software and purchased from Sigma-Genosys. DNA templates were prepared from S. pyogenes grown in both THY broth and on THY agar plates. Several colonies grown on agar plates were transferred into 50 μl of deionized water, incubated for 7 min at 97°C, and centrifuged. Five microliters of the supernatant fluid was used as a template for PCR. For S. pyogenes grown in broth, DNA was prepared from the cell pellet, as described above. PCR was carried out with an initial denaturation step of 2 min at 93°C followed by 35 cycles of amplification steps of 30 s at 93°C, 1 min at 55°C, and 1 min at 72°C and a final extension step of 10 min at 72°C. PCR products were visualized in 1.5% agarose gel containing ethidium bromide (0.5 μg/ml).
Microarray data accession number.
All of the microarray data are available through the Gene Expression Omnibus data repository at NCBI (http://www.ncbi.nlm.nih.gov/geo/) via accession number GSE 3989.
RESULTS
DNA microarray analysis.
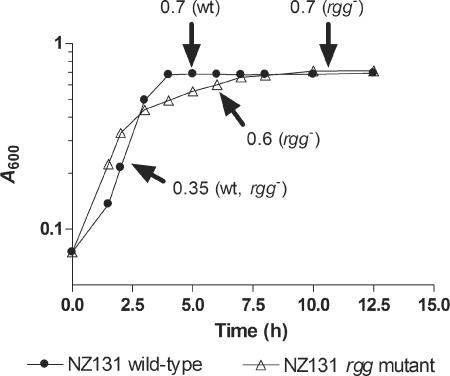
To identify changes in the S. pyogenes transcriptome associated with rgg inactivation, the transcript profiles of the wild-type and rgg mutant strains were analyzed with Affymetrix NimbleExpress whole-genome chips. RNA was purified from wild-type cultures during exponential and postexponential phases of growth (A600 = 0.35 and 0.70, respectively) (Fig. 1). RNA was isolated at three points during growth of the mutant strain (A600 = 0.35, 0.60, and 0.70) (Fig. 1). To assess the microarray results, quantitative RT-PCR was used to measure the transcripts of selected genes during exponential (A600 = 0.35 for both strains) and postexponential (A600 = 0.70 for both strains) growth. Ten genes were selected, which represented the magnitude of interstrain differences identified with DNA microarrays. During both phases of growth, the results obtained with the two methods correlated (r2 = 0.92 and 0.88, for exponential and postexponential phases of growth, respectively) (see Fig. S1 in the supplemental material).
FIG. 1.
Growth of wild-type and rgg mutant strains in THY broth. RNA samples were collected at various times during growth of both the wild-type (•) and rgg mutant (▵) strains. Arrows designate points of RNA isolation, and the A600 of the culture at the time of collection is shown.
Genomewide changes in transcription in the rgg mutant strain.
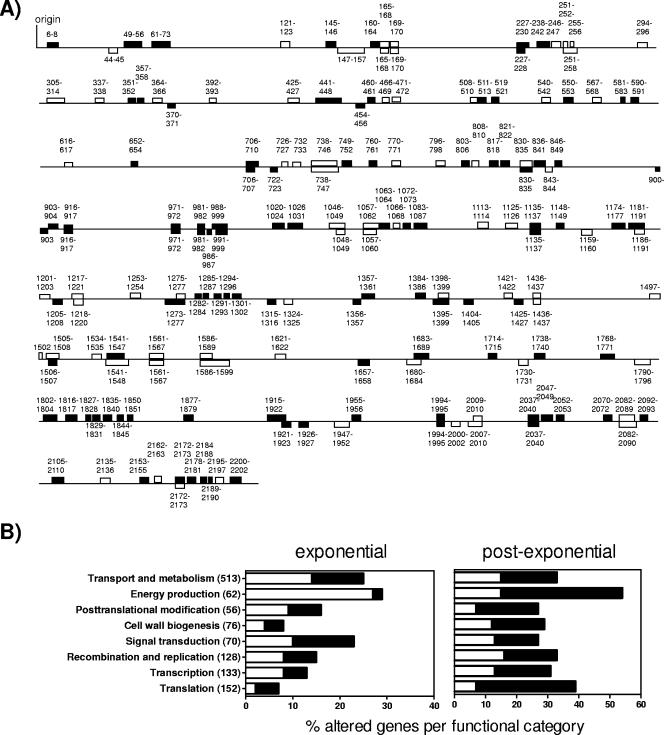
The gene expression profiles of the wild-type and rgg mutant strains were compared. The transcriptome of the rgg mutant strain at an A600 of 0.65 (Fig. 1) was similar to that at an A600 of 0.35 (data not shown). Thus, we focused on comparing wild-type and mutant strain transcript levels in the exponential (A600 = 0.35) and postexponential (A600 = 0.70) phases of growth. In the exponential phase of growth, 165 gene transcripts were more abundant while 134 were less abundant in the mutant strain (see Table S1 in the supplemental material); statistically significant (P < 0.05) differences of fivefold or more are reported in Table 2. During the postexponential phase of growth, 227 gene transcripts were more abundant and 340 were less abundant in the rgg mutant strain (see Table S2 in the supplemental material); statistically significant (P < 0.05) differences of fivefold or more are reported in Table 3. Taken together, a total of 706 genes were differentially expressed in the rgg mutant strain compared to expression in the wild-type strain during the exponential and postexponential phases of growth. Many of the genes are proximal to one another in the chromosome (Fig. 2A), indicating that rgg inactivation altered the expression of polycistronic operons encoding proteins with related functions. In general, the results indicate that Rgg represses gene transcription in the exponential phase of growth and promotes transcription in the postexponential phase of growth (Fig. 2B). Moreover, the majority of differences in both phases of growth were among genes involved with energy production and metabolism (Fig. 2B).
FIG. 2.
Genomewide location and function of Rgg-regulated genes in strain NZ131. (A) Polycistronic operons regulated by Rgg during exponential and postexponential phases of growth are indicated by boxes below and above the line, respectively, which represents the chromosomal gene arrangement in strain SF370. Open blocks and filled blocks indicate more- or less-abundant transcripts in the rgg mutant strain, respectively. Numbers correspond to the annotation of the SF370 genome (9). (B) Changes in functionally categorized gene transcripts in the rgg mutant strain during exponential and postexponential growth. The percentages of more-abundant (open bars) and less-abundant (closed bars) gene transcripts in the mutant strain compared to those in the wild-type strain in each functional category are shown. The number of genes within each category is indicated in parentheses.
Differences in the expression of several transcriptional regulators, including two-component regulatory systems, were associated with rgg inactivation. For example, during exponential-phase growth of the mutant strain, transcripts encoding LytR and LytS were each threefold more abundant than for the wild-type strain. In the postexponential phase of growth, transcript levels encoding YesNM (SPy1061 and -1062), CsrS/CovS (SPy337), LytRS (SPy1587 and -1588), and SPy1621 and -1622 were all approximately twofold more abundant for the mutant strain, while those encoding YufM (SPy1106), FasB (SPy242), and CpsY (SPy898) were approximately twofold less abundant for the mutant strain (see Table S2 in the supplemental material). In addition, the expression of a RALP regulator (SPy216) was 10- and 16-fold less abundant in the mutant strain during the exponential and postexponential phases of growth, respectively (Tables 2 and 3). A greater number of regulatory gene transcripts were altered for the rgg mutant strain during the postexponential phase of growth than during the exponential phase, which may be responsible, at least in part, for the greater number of changes in structural gene expression associated with this growth phase (see Tables S1 and S2 in the supplemental material).
Rgg-associated changes in the expression of nonglucose catabolic operons.
The growth phase-dependent expression of genes associated with amino acid metabolism was markedly influenced by rgg inactivation. For example, exponential phase repression of the arginine deiminase operon (SPy1547, -1546, -1544, -1543, -1542, and -1541) was relieved in the rgg mutant strain during the exponential phase of growth (Table 2). The results correlated with those of previous studies that showed increased synthesis of the corresponding enzymes in the mutant strain (4, 6). In addition, nine genes (SPy2081, -2082, -2083, -2084, -2085, -2087, -2088, -2089, and -2090), which are adjacent to each other in the chromosome and involved in histidine metabolism, were more abundant in the mutant strain during both the exponential and postexponential periods of growth (see Tables S1 and S2 in the supplemental material).
Transcripts encoding enzymes involved in citrate metabolism (citCDEFX and oadA) were also influenced by rgg inactivation (see Tables S1 and S2 in the supplemental material). In the wild-type strain, these genes were induced an average of 15-fold upon entry into the postexponential phase of growth (data not shown). In contrast, the mutant strain expressed the genes during the exponential phase of growth (see Table S1 in the supplemental material), which is consistent with the idea that Rgg represses postexponential phase-induced genes during the exponential phase of growth.
Inactivation of rgg was associated with changes in the expression of genes involved in the utilization of nonglucose carbohydrates. In general, the transcripts were more abundant in the mutant strain during the exponential phase of growth. For example, SPy1815, -1595, and -1593, which are involved in sucrose metabolism, were more abundant in the mutant strain during the exponential phase of growth (see Table S1 in the supplemental material). Similarly, genes involved in lactose (SPy1708) or fructose and mannose (SPy1057, -1058, -1059, and -1060) metabolism were more abundant in the mutant strain during the exponential phase of growth (see Table S1 in the supplemental material); however, not all the genes in each pathway were similarly expressed in the mutant strain, and some were even less abundant in the mutant strain than in the wild-type strain. For example, SPy1918 and -1921, which are involved in lactose catabolism, were each twofold less abundant in the mutant strain than in the wild-type strain in the exponential phase of growth. In addition, the scrB gene (SPy1816), whose product (sucrose-6-phosphate hydrolase) is necessary for hydrolysis of sucrose to glucose and fructose, was transcribed less in the rgg mutant. The same was true regarding the fructose-bisphosphate aldolase fba gene (SPy1889) and the transaldolase mipB gene (Spy2048), which encode the key enzymes involved in the utilization of fructose and mannose by the rgg mutant (see Table S2 in the supplemental material). The data indicated that rgg inactivation altered the expression of genes involved in the utilization of nonglucose carbohydrates; however, the phenotypic significance of the changes was unclear.
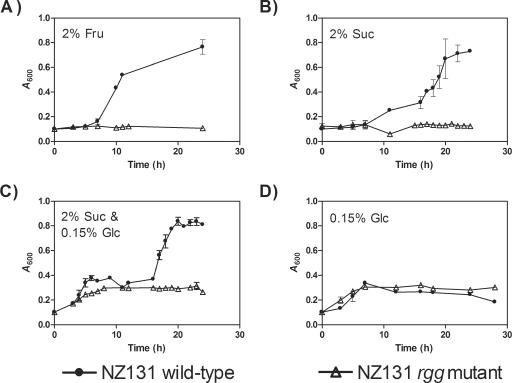
Rgg is essential for utilization of fructose, mannose, and sucrose.
To assess the significance of Rgg-associated changes in the expression of genes involved in the metabolism of nonglucose carbohydrates, growth of the wild-type and mutant strains was assessed in CDM containing these compounds as the primary C source. The wild-type strain had a growth yield of an A600 value of 0.4 after 24 h of incubation in broth containing 2% (wt/vol) mannose; however, the mutant strain was unable to grow (A600 < 0.1). Similar results were obtained using 2% (wt/vol) fructose as the primary C source (Fig. 3A). To determine if the mutant strain was defective in the ability to adapt to growth with different C sources, growth was assessed using CDM containing both glucose and sucrose. A diauxic pattern of growth was characteristic of the wild-type strain in this medium (Fig. 3C), indicating that the strain preferentially degraded glucose in the first period of growth and then adapted to growth with sucrose. In contrast, growth of the mutant strain ceased upon the depletion of glucose (Fig. 3C and D). For these experiments, it was necessary to use a low concentration of glucose; otherwise, the cultures reached a growth yield that was too high to support additional growth with the second carbohydrate, probably due to the accumulation of metabolic end products. Together with results from the expression analysis, the results indicate that Rgg-dependent regulation is essential for growth of the pathogen with fructose, mannose, and sucrose as the primary C sources.
FIG. 3.
Growth of the wild-type (•) and rgg mutant (▵) strain with various C sources. The strains were cultured in CDM containing the following: (A) 2% fructose, (B) 2% sucrose, (C) 2% sucrose and 0.15% glucose, or (D) 0.15% glucose. To detect a diauxic pattern of growth, glucose was used at a limiting concentration. Growth was measured by determining the absorbance of cultures. The results are shown as the means and standard errors of the means from at least four independent experiments.
rgg inactivation alters prophage gene expression and induction.
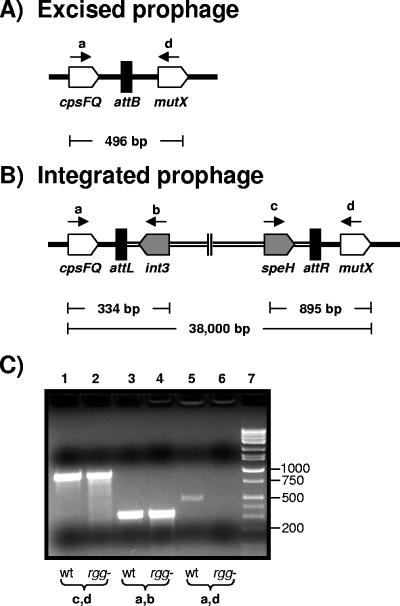
Genome variation among S. pyogenes isolates is largely due to differences in the number and types of prophages present in the chromosome. NZ131 contains two prophages (J. J. Ferretti, personal communication), both of which were partially represented in the array and designated NZ131.1 and NZ131.2. Transcriptome analysis revealed that the expression of several prophage genes was altered in the mutant strain during the exponential and postexponential phases of growth (see Tables S1 and S2 in the supplemental material). The majority of differences were among structural genes, which were less abundant in the mutant strain. In contrast, int3 transcripts (SPy937), encoding a putative integrase of the NZ131.1 prophage, were 11-fold greater in the mutant strain in the postexponential phase of growth (Table 3). The difference in expression implied that rgg inactivation decreased the frequency of NZ131.1 prophage induction. To test this, PCR was used to detect the attB site in wild-type and rgg mutant strain cultures as an indicator of prophage excision (Fig. 4A). As controls, primers were also designed to amplify the prophage-genome junctions attL and attR (Fig. 4B). attB was detected only in samples obtained from the wild-type strain and not from those of the rgg mutant strain (Fig. 4C). The results correlate with the transcriptional data and indicate that rgg inactivation is associated with a decrease in NZ131.1 prophage induction.
FIG. 4.
Detection of attB, attL, and attR of the NZ131.1 prophage. Schematic diagrams of the (A) attB and (B) attL and attR sites are shown. The chromosomal genes (cpsFQ and mutX) and the prophage-associated genes (int3 and speH) are designated. The positions of PCR primers are shown by lettered black arrows, and the expected sizes of the amplicons are indicated. (C) PCR analysis of the wild-type strain (lanes 1, 3, and 5) and the rgg mutant strain (lanes 2, 4, and 6). Lanes 1 and 2, PCR of the attR site with the speH (c) and mutX (d) primers. Lanes 3 and 4, PCR of the attL site with cpsFQ (a) and int3 (b) primers. Lanes 5 and 6, PCR of attB with the cpsFQ (a) and mutX (d) primers.
All strains of S. pyogenes encode at least one secreted nuclease, and additional nuclease genes are often encoded by prophages. Although the function of secreted nucleases is not clear, DNase activity contributes to the virulence associated with S. pyogenes (32). Transcription profiling revealed that Rgg is a repressor of both a chromosomally encoded nuclease (mf-1; SPy2043), which is adjacent to rgg in the chromosome, and a prophage-encoded DNase (mf-3; SPy1436). Transcripts encoding MF-3 were 27-fold higher in the rgg mutant than in the wild-type strain in the exponential phase of growth (Table 2), which correlated with an increase in extracellular DNase activity (Fig. 5). Together the results indicate that rgg inactivation alters the expression of prophage-encoded genes in strain NZ131, including virulence-associated nucleases.
FIG. 5.
Detection of DNase activity. The wild-type (wt) and rgg mutant strains were stab inoculated into DNase test agar plates containing methyl green and incubated overnight at 37°C with 5% CO2. DNase activity resulted in a zone of clearing around the stab site.
Rgg is a repressor of the virulence-associated NAD-glycohydrolase operon.
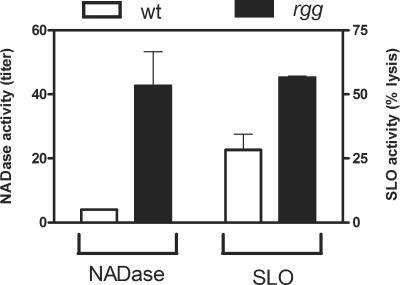
Transcriptome analysis revealed that rgg inactivation increases the expression of an operon that encodes cytolysin-mediated translocation (CMT) of S. pyogenes NAD-glycohydrolase (SPN), which functions similarly to the type III secretion apparatus of gram-negative bacteria. Specifically, toxicity to human host cells can occur via cytolysin (SLO)-mediated translocation of an SPN into host cells, where the enzyme generates ADP-ribose and nicotinamide (2, 3, 15, 17, 18, 29). An immunity protein (IFS) which protects S. pyogenes from the toxicity associated with SPN was recently described (18). In the exponential phase of growth, inactivation of rgg was associated with 53-, 32-, and 28-fold increases in the transcripts encoding SPN, IFS, and SLO, respectively (Table 2). In the postexponential phase of growth, transcripts were also abundant in the mutant strain (see Table S2 in the supplemental material). The increase in slo transcripts in the rgg mutant strain was associated with an increase in hemolytic activity when grown on agar plates containing sheep blood (data not shown). In addition, the enzymatic activities associated with streptolysin O and NADase were significantly higher in culture supernatant fluids obtained from the mutant strain than in those from the wild-type strain (Fig. 6).
FIG. 6.
NADase and SLO cytolytic activity in culture supernatant fluids. The wild-type (open bars) and rgg mutant (closed bars) strains were grown in THY overnight; the amount of NADase activity present in sterile supernatant fluids was determined and is expressed as the highest twofold dilution that had sufficient activity to completely hydrolase NAD+. SLO activity is reported as the percentage of erythrocyte lysis that could be inhibited by preincubation of culture supernatant fluid with cholesterol.
DISCUSSION
Growth phase-dependent regulation of gene expression in S. pyogenes appears to occur independently of alternative sigma factors. Rather, S. pyogenes seems to rely on a complex network of transcriptional regulators (12). Previously, a combination of spotted DNA microarrays and Taqman RT-PCR was used to identify 25 Rgg-regulated genes during the exponential phase of growth (7). In the current study, we assessed Rgg-associated changes in transcription using Affymetrix NimbleExpress arrays and identified 293 and 588 Rgg-regulated genes in the exponential and postexponential phases of growth, respectively. In addition, results of the current study showed that Rgg represses exponential-phase expression of the virulence-associated CMT NAD-glycohydrolase operon and influences prophage gene expression and induction. Finally, using CDM, we discovered that Rgg is essential for the adaptation of strain NZ131 to growth using several nonglucose carbohydrates as the primary C source. Overall, the results indicate that Rgg is a global regulatory protein in S. pyogenes strain NZ131 that contributes to growth phase-dependent changes in gene expression.
Adaptation to nonglucose C sources.
Previous results showed that the mutant strain has a greater growth yield in THY media (6), which implied that rgg inactivation did not decrease the fitness of the organism under in vitro conditions. In contrast, results described here clearly show that rgg is important for growth of the pathogen with nonglucose carbohydrates. Successful colonization of the human host and invasion of normally sterile sites, such as the blood or subcutaneous tissues, is likely to require adaptation to growth with nonglucose C sources. Therefore, chemotherapeutic strategies aimed at disrupting Rgg-mediated regulation, for example, via small molecules (11) or small-inhibiting RNAs, may diminish the ability of the pathogen to invade or colonize human tissue and thereby decrease human disease.
Indirect effects on transcription associated with rgg inactivation.
Several properties associated with the rgg mutant strain are likely to cause Rgg-independent changes in the transcriptome. These include differences in metabolism and growth and altered expression of other regulatory genes (6, 7). The current study confirms that inactivation of rgg alters the expression of other transcriptional regulators, including mga, sagA (pel), and crsS (covS), as previously described (7). Moreover, several additional changes were detected among putative regulatory transcriptional regulators (Spy146, -427, -533, -599, -1062, -1187, -1202, -1259, -1285, -1386, -1395, -1587, -1596, -1602, -1680, -1699, -1870, and -2177) and ahrC.2, ralp, pyrR, and luxS. These differences in regulatory genes are likely to account for at least some of the changes in gene expression associated with rgg inactivation in strain NZ131. Differences in the growth and metabolism of the mutant strain are also likely to alter the transcriptome. For example, the mutant strain excretes NH3 during exponential growth, while the wild-type strain produces lactate. In addition to the compounds themselves, the difference in the pH of the medium may influence gene expression. Finally, differences in the growth of the strains, particularly in the postexponential phase, are also likely to contribute to differential expression.
In strain NZ131, Rgg is an important regulator involved in pathogen adaptation to alternative nutrient sources. Among gram-positive bacteria, CodY is another important global regulatory protein that mediates changes in gene expression in response to the metabolic status of the cell. Recently, differences in the transcription of genes representing several functional categories were identified in a codY mutant strain of NZ131 (16). Interestingly, nearly all the CodY-regulated genes identified are also regulated by Rgg but in an opposite manner. For example, grab, braB, and xpt transcripts are more abundant in a codY mutant strain during exponential growth than in the wild-type strain (16); in contrast, each is less abundant in the rgg mutant strain (see Table S1 in the supplemental material). Similarly, mga, nga, prtS, scpA, mf-1/sdn, slo, and dppA transcript levels are depressed in a codY mutant strain (16) but elevated in the rgg mutant strain. Together, the results are consistent with the idea that global patterns of gene expression in S. pyogenes result from interactions among redundant, or overlapping, regulatory circuits.
Interaction between Rgg and prophage-encoded genes.
Rgg influenced the expression of several prophage genes, including structural genes, integrase, and virulence-associated genes, such as hylP2, which encodes a secreted hyaluronidase, and mf-3, which encodes a secreted nuclease. Integrases can function in both prophage and transposon integration and, in complex with excisionase, in prophage excision (8). An inverse correlation between prophage induction and the abundance of int3 transcripts was identified, suggesting that Int3 functions to promote prophage integration under these conditions. The results are also consistent with those of a previous report, which showed that prophage induction and gene expression levels do not necessarily correlate (1).
Several possible explanations may account for the reduction in prophage induction and changes in prophage gene expression in the mutant strain. For example, carbon starvation increases bacteriophage gene expression in L. lactis (26), while ClpXP and RpoS (22) both alter the frequency of phage mu induction in E. coli (13). Inactivation of rgg in strain NZ131 is associated with both changes in metabolism and an increase in the expression of stress-responsive genes (4, 6). Therefore, rgg inactivation may indirectly alter prophage gene expression and induction. Alternatively, several features of Rgg suggest it is a remnant of a prophage-encoded regulatory gene. For example, the G+C content of rgg is 32%, which is similar to that of prophage open reading frames and markedly lower than that of the chromosome (39%). Furthermore, the genes adjacent to the rgg locus have G+C contents of 37 and 39%. The G+C contents of the orthologues lasX, gadR, and mutR are 25, 29, and 30%, respectively, while those of the corresponding chromosomes are 38, 35, and 37%, respectively. The Rgg protein also shares amino acid similarity to bacteriophage repressor proteins, and paralogues are present in the S. pyogenes chromosome, which may be indicative of previous prophage integration sites. Finally, Rgg orthologues are present in only a subset of low-G+C gram-positive pathogens, which may reflect the host range of the putative ancestral bacteriophage. Thus, Rgg may have retained the ability to control the expression of ancestrally related prophage genes. Interactions between specific prophages and chromosomally encoded regulatory factors, such as Rgg, could contribute to strain-associated patterns of gene expression. Studies are currently under way to determine if Rgg directly interacts with prophage genes to alter transcription and phage induction.
Rgg regulates the expression of the virulence-associated NAD-glycohydrolase operon at transcriptional and posttranslational levels.
Previously, rgg inactivation was shown to increase the expression of several virulence factors localized to the cell wall, including members of the Mga regulon (M49 and C5a peptidase). In the current study, we found that rgg inactivation also increased the expression of genes responsible for cytolysin-mediated translocation of SPN into mammalian cells. Similar to the changes in the expression of Mga-regulated genes in the rgg mutant strain, the transcripts encoding SLO, SPN, and IFS were much more abundant in the mutant strain during the exponential phase of growth than in the wild-type strain (Table 2). Furthermore, the changes correlated with increased enzymatic activities in culture supernatant fluids (Fig. 6). Interestingly, Rgg is also required for stationary phase expression of the secreted SpeB protease, which can degrade several streptococcal proteins including SLO and SPN (19). Thus, in strain NZ131, Rgg represses the transcription of the NAD-glycohydrolase operon in the exponential phase of growth and indirectly regulates the degradation of the proteins in the stationary phase via the activation of speB expression. The results support the hypothesis that Rgg is important in coordinating the phenotypic changes associated with the transition to the stationary phase.
In summary, we identified Rgg-regulated genes in strain NZ131. The results are important in characterizing regulatory networks of S. pyogenes that control gene expression, including virulence-associated operons. The results also show that Rgg is essential for growth with nonglucose carbohydrates. Together, these properties suggest that disrupting Rgg-mediated regulation in vivo could disrupt virulence factor expression and decrease the ability of the pathogen to adapt to growth with alternative C sources. Such an approach could decrease both pathogen dissemination within the host and virulence factor production, thereby minimizing human disease.
Supplementary Material
Acknowledgments
We thank I. Biswas, P. Dunman, and G. Somerville for critical review of the manuscript. We thank S. BonDurant and K. Eyster for assistance with the DNA microarray experiments.
This work was supported by NIAID/NIH grant RO1 AIO52147 to M.S.C. and NIH grant 2 P20 RR016479 from the INBRE Program of the National Center for Research Resources.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Banks, D. J., B. Lei, and J. M. Musser. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71:7079-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bricker, A. L., V. J. Carey, and M. R. Wessels. 2005. Role of NADase in virulence in experimental invasive group A streptococcal infection. Infect. Immun. 73:6562-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bricker, A. L., C. Cywes, C. D. Ashbaugh, and M. R. Wessels. 2002. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol. Microbiol. 44:257-269. [DOI] [PubMed] [Google Scholar]
- 4.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186:7091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, E. H., R. I. Gumport, and J. F. Gardner. 2002. Interactions between integrase and excisionase in the phage lambda excisive nucleoprotein complex. J. Bacteriol. 184:5200-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara, T., T. Hoshino, T. Ooshima, S. Sobue, and S. Hamada. 2000. Purification, characterization, and molecular analysis of the gene encoding glucosyltransferase from Streptococcus oralis. Infect. Immun. 68:2475-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung, D. T., E. A. Shakhnovich, E. Pierson, and J. J. Mekalanos. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670-674. [DOI] [PubMed] [Google Scholar]
- 12.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 13.Lamrani, S., C. Ranquet, M. J. Gama, H. Nakai, J. A. Shapiro, A. Toussaint, and G. Maenhaut-Michel. 1999. Starvation-induced Mucts62-mediated coding sequence fusion: a role for ClpXP, Lon, RpoS and Crp. Mol. Microbiol. 32:327-343. [DOI] [PubMed] [Google Scholar]
- 14.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madden, J. C., N. Ruiz, and M. Caparon. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell 104:143-152. [DOI] [PubMed] [Google Scholar]
- 16.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259-275. [DOI] [PubMed] [Google Scholar]
- 17.Meehl, M. A., and M. G. Caparon. 2004. Specificity of streptolysin O in cytolysin-mediated translocation. Mol. Microbiol. 52:1665-1676. [DOI] [PubMed] [Google Scholar]
- 18.Meehl, M. A., J. S. Pinkner, P. J. Anderson, S. J. Hultgren, and M. G. Caparon. 2005. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathogens 1:362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2003. Expression of the secondary sigma factor σX in Streptococcus pyogenes is restricted at two levels. J. Bacteriol. 185:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495-502. [DOI] [PubMed] [Google Scholar]
- 22.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 23.Qi, F., P. Chen, and P. W. Caufield. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi, F., P. Chen, and P. W. Caufield. 2000. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 66:3221-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlinson, E. L., I. F. Nes, and M. Skaugen. 2005. Identification of the DNA-binding site of the Rgg-like regulator LasX within the lactocin S promoter region. Microbiology 151:813-823. [DOI] [PubMed] [Google Scholar]
- 26.Redon, E., P. Loubiere, and M. Cocaign-Bousquet. 2005. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J. Bacteriol. 187:3589-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 28.Skaugen, M., E. L. Andersen, V. H. Christie, and I. F. Nes. 2002. Identification, characterization, and expression of a second, bicistronic, operon involved in the production of lactocin S in Lactobacillus sakei L45. Appl. Environ. Microbiol. 68:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens, D. L., D. B. Salmi, E. R. McIndoo, and A. E. Bryant. 2000. Molecular epidemiology of nga and NAD glycohydrolase/ADP-ribosyltransferase activity among Streptococcus pyogenes causing streptococcal toxic shock syndrome. J. Infect. Dis. 182:1117-1128. [DOI] [PubMed] [Google Scholar]
- 30.Sulavik, M. C., and D. B. Clewell. 1996. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J. Bacteriol. 178:5826-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumby, P., K. D. Barbian, D. J. Gardner, A. R. Whitney, D. M. Welty, R. D. Long, J. R. Bailey, M. J. Parnell, N. P. Hoe, G. G. Adams, F. R. DeLeo, and J. M. Musser. 2005. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune system. Proc. Natl. Acad. Sci. USA 102:1679-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Rijn, I., and R. E. Kessler. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vickerman, M. M., P. E. Minick, and N. M. Mather. 2001. Characterization of the Streptococcus gordonii chromosomal region immediately downstream of the glucosyltransferase gene. Microbiology 147:3061-3070. [DOI] [PubMed] [Google Scholar]
- 35.Vickerman, M. M., M. C. Sulavik, and D. B. Clewell. 1995. Oral streptococci with genetic determinants similar to the glucosyltransferase regulatory gene, rgg. Infect. Immun. 63:4524-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.