Abstract
Methionine is produced by methylation of homocysteine. Sinorhizobium meliloti 102F34 possesses only one methionine synthase, which catalyzes the transfer of a methyl group from methyl tetrahydrofolate to homocysteine. This vitamin B12-dependent enzyme is encoded by the metH gene. Glycine betaine can also serve as an alternative methyl donor for homocysteine. This reaction is catalyzed by betaine-homocysteine methyl transferase (BHMT), an enzyme that has been characterized in humans and rats. An S. meliloti gene whose product is related to the human BHMT enzyme has been identified and named bmt. This enzyme is closely related to mammalian BHMTs but has no homology with previously described bacterial betaine methyl transferases. Glycine betaine inhibits the growth of an S. meliloti bmt mutant in low- and high-osmotic strength media, an effect that correlates with a decrease in the catabolism of glycine betaine. This inhibition was not observed with other betaines, like homobetaine, dimethylsulfoniopropionate, and trigonelline. The addition of methionine to the growth medium allowed a bmt mutant to recover growth despite the presence of glycine betaine. Methionine also stimulated glycine betaine catabolism in a bmt strain, suggesting the existence of another catabolic pathway. Inactivation of metH or bmt did not affect the nodulation efficiency of the mutants in the 102F34 strain background. Nevertheless, a metH strain was severely defective in competing with the wild-type strain in a coinoculation experiment.
Sinorhizobium meliloti is a soil bacterium that can fix nitrogen in association with the legume Medicago sativa. As for all soil bacteria, it is subjected to fluctuations of water availability. To cope with reduced water activity, S. meliloti accumulates compatible solutes in order to preserve its turgor. Under hyperosmotic conditions, potassium ions are taken up and glutamate and N-acetylglutaminylglutamine amide are accumulated by de novo synthesis during the exponential phase of growth (39), whereas trehalose is produced at the end of exponential growth (42, 43). As observed for other bacteria (23, 28), the osmotic tolerance of S. meliloti is enhanced when osmoprotectants are present in the growth medium. These compounds are transported into the cell by active uptake systems (23). In most bacteria, osmoprotectants are accumulated as compatible solutes. S. meliloti uses not only accumulated compounds, like dimethylsulfoniopropionate (DMSP) and homobetaine (HB) (32), as osmoprotectants, but also compounds such as sucrose (16), trehalose (15), pipecolate (14), and ectoine that never accumulate but instead are avidly metabolized (18, 42). Glycine betaine is transiently accumulated (43), and dimethylsulfonioacetate (DMSA) is catabolized only in medium of low osmolarity (32). Some betaines, like trigonelline, have been shown to be involved in the nodulation process and are catabolized through a specific pathway (4).
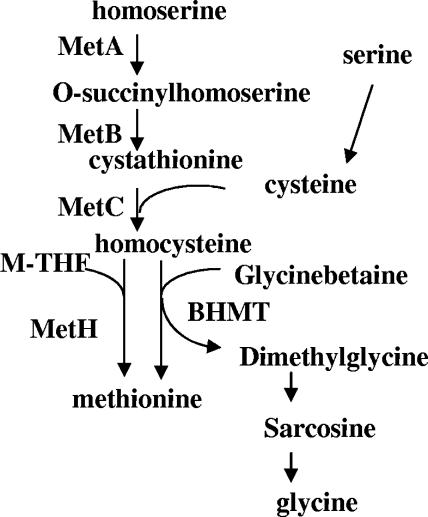
In S. meliloti, the catabolism of glycine betaine proceeds by successive demethylations that lead to glycine (38) (Fig. 1). The first step is catalyzed by betaine homocysteine methyl transferase (BHMT), which transfers a methyl group from glycine betaine to homocysteine, yielding methionine and dimethylglycine (DMG). Subsequent catabolism of DMG leads to the end product glycine (Fig. 1). To date, this is the only glycine betaine catabolic pathway that has been described in S. meliloti. Repression of this degradative pathway in hyperosmotic media allows glycine betaine to accumulate (38). BHMT-dependent catabolism of glycine betaine is widespread in nature and constitutes an important pathway for methionine synthesis in the kidney and liver (10). While the existence of BHMT activity has been suspected in many bacteria, the corresponding gene has never been identified; all the studies of betaine catabolism in bacteria have concerned only dimethylglycine and sarcosine catabolism (26, 41). The only glycine betaine transmethylase to have been described in a bacterium was identified in Pseudomonas aeruginosa (36), but it does not share any homology with mammalian BHMT proteins.
FIG. 1.
Methionine biosynthesis in S. meliloti. The probable methionine biosynthetic pathway as annotated previously (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/) is shown. MetA, homoserine O-succinyltransferase; MetB, cystathionine gamma-synthase; MetC, cystathionine beta-lyase; MetH, 5-methyltetrahydrofolate-homocysteine methyltransferase; BHMT, betaine-homocysteine methyl transferase, as proposed in this study. M-THF represents methyltetrahydrofolate.
Most organisms that have been studied encode more than one enzymatic mechanism for the methylation of homocysteine to generate methionine. For example, Escherichia coli uses two methionine synthases, a cobalamin-dependent (MetH) and a cobalamin-independent (MetE) enzyme, both of which use methyl tetrahydrofolate as a cosubstrate with homocysteine. Only MetH activity was found in S. meliloti (35), and no ortholog of the metE gene has been identified in the S. meliloti strain Rm1021 genome (11) (http://www.genome.ad.jp/kegg/genes.html), suggesting that methionine biosynthesis proceeds only via MetH. Nevertheless, the existence of MetE activity has been postulated in a different S. meliloti strain (1). Thus, the existence of MetE in S. meliloti strains must be tested for each isolate. In minimal medium, methionine synthesis is due to methionine synthase activity, but alternative pathways can operate if appropriate substrates are present in the growth medium. In organisms that have BHMT, glycine betaine is an efficient alternative methyl donor for methionine biosynthesis. Thus, methionine production in an organism is dependent on its metabolic abilities as defined not only by its genome, but also by the availability of the precursors of each pathway. For a symbiotic bacterium like S. meliloti, the pathways of methionine biosynthesis in free-living bacteria or bacteroids could be different due to differences in the presence or absence of methionine precursors between the free-living and symbiotic states.
In this study, we analyzed the production of methionine in S. meliloti strain 102F34, searched for genes required for the putative pathways in the S. meliloti genome, made mutants defective in these pathways, and analyzed the phenotypes of the mutants. We confirmed that MetH is the only methionine synthase expressed in free-living S. meliloti strain 102F34 and identified the first bacterial gene encoding a protein related to mammalian BHMT.
MATERIALS AND METHODS
Bacterial strains and media.
The S. meliloti parental strain we used was 102F34 Smr, a spontaneously streptomycin-resistant (Smr) derivative of S. meliloti 102F34 obtained in our laboratory. The E. coli strains used were DH5α (17) and SM10 (37). For growth investigations, S. meliloti was grown aerobically in complex medium MSY (29) or minimal medium (3) at 30°C to an optical density at 570 nm of 1.5 to 1.8, prepared, and inoculated in minimal lactate aspartate salts (LAS) medium (33) as previously described (16). The minimal medium described by Vincent (46) was used for the BHMT assay. E. coli strains were grown aerobically in LB medium (27) at 37°C. For the selection of E. coli strains, ampicillin and kanamycin were added at 50 μg/ml and tetracycline at 10 μg/ml. For the selection of S. meliloti strains, streptomycin was used at 100 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 5 μg/ml.
DNA manipulations and analysis.
Chromosomal and plasmid DNA isolation and manipulations were carried out according to standard procedures (2, 34).
Construction of metH and bmt mutants.
DNA fragments located within the coding regions of metH (from bases 1614 to 2431) and SMc04235 (from bases 183 to 866) open reading frames (http://sequence.toulouse.inra.fr/meliloti.html) were amplified by PCR using the oligonucleotides metH1 (5′-GAATATCTTCGCGGTCGC-3′) and metH2 (5′-TTCGAGGATCTTAGCCGAG-3′) for metH and bhmt1 (5′-GCCGACATCATTCTCACC-3′) and bhmt2 (5′-AGATGGTTGCAGGAGGTG-3′) for SMc04235. The PCR products were introduced into the pCR2.1 Topo (Invitrogen) vector and subsequently transferred into the mobilizable plasmid pVO147 (30), which is nonreplicative in S. meliloti. The resulting plasmids were transferred into S. meliloti 102F34 Smr by triparental mating, and recombinants were selected on kanamycin-containing plates. φM12-mediated transductions of the mutations into the parental strain were performed in order to ensure a clean background.
BHMT assay.
[methyl-14C]Glycine betaine (2.07 GBq mmol−1) and [methyl-14C]DMSA (2.0 GBq mmol−1) were obtained as previously described (32, 43). Cells were grown in Vincent medium to mid-exponential growth phase, collected by centrifugation (15,000 × g; 10 min), washed twice, resuspended in 10 mM phosphate buffer, pH 7.5, and permeabilized with toluene (1:100 [vol/vol]). [14C]Glycine betaine or [14C]DMSA (3 kBq) and homocysteine (7 mM) were added to 0.4 mg of proteins of cell extract in a final volume of 50 μl. The mixture was incubated overnight at 37°C and extracted with ethanol (80% vol/vol). The extract was dried under vacuum, resuspended in water, and then fractionated by two-dimensional chromatography, using phenol (80%) in the first dimension and butanol-acetic acid-H2O (12:3:5) in the second dimension. After chromatography, radioactive spots were visualized and quantified using an instant imager (Packard). Glycine betaine, methionine, DMG, and sarcosine were used as internal standards to identify the radioactive spots. They were visualized as previously described (43).
Uptake and intracellular fate of glycine betaine.
The uptake and metabolism of [14C]glycine betaine were analyzed as described previously (16, 19, 43). Cells were grown in LAS medium containing 1 mM [14C]glycine betaine (0.2 MBq/mmol) in the presence or absence of 0.5 M NaCl. CO2 produced during growth was trapped on a strip of filter paper (0.5 by 3 cm) moistened with 30 μl of 5 M KOH. This filter was changed at each sampling time. At regular intervals, 1 to 2 ml of cell suspension was harvested by centrifugation (12,000 × g; 2 min). The pellets were immediately extracted with 80% ethanol as described previously (43). The radioactivities of the ethanol-soluble fraction, the ethanol-insoluble fraction, and CO2 were measured by scintillation counting. The ethanol-soluble fraction was analyzed by paper chromatography (43). In all cases, [14C]glycine betaine represented more than 90% of the total radioactivity of this fraction. The results are the means of at least three independent experiments, and the standard deviation was less than 10%.
Plant assays.
Medicago sativa L. var. Europe (alfalfa) was used as a host plant for testing the nodulation and N2 fixation of S. meliloti strains. Surface-sterilized germinating seedlings were grown in test tubes on nitrogen-free Jensen broth medium (22). One-week-old plants were inoculated with 109 cells of 102F34 Smr or metH and bmt mutants. Plants were grown aerobically at room temperature. Four weeks postinoculation, the number of nitrogen-fixing nodules was assessed. Samples of nodules were randomly harvested and crushed in order to recover the bacteria and to ensure that the bacteria present inside the nodule were the inoculated strain and not revertants.
RESULTS
MetH is the only methionine synthase expressed in free-living S. meliloti 102F34.
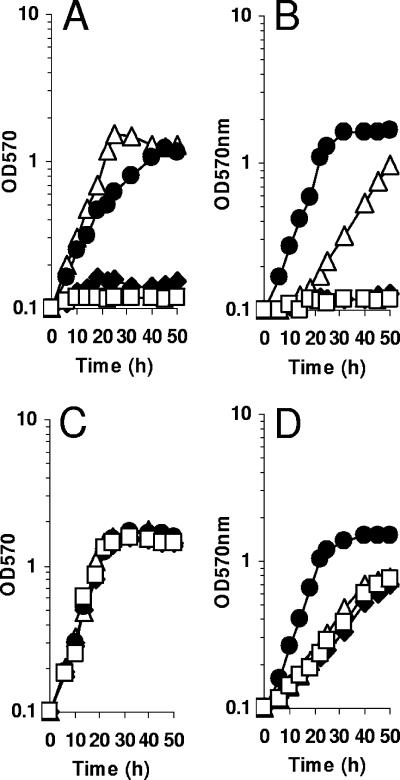
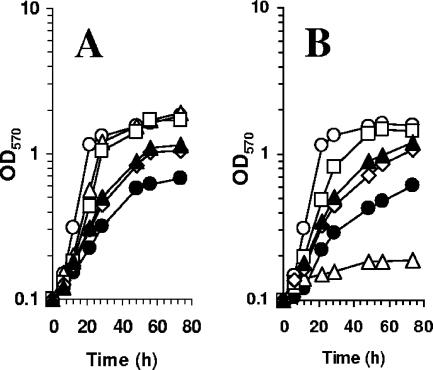
MetH activity enables S. meliloti to synthesize methionine from methyl tetrahydrofolate and homocysteine (47). Methionine can also be obtained from glycine betaine and homocysteine by a BHMT (38). Sequence analysis of the sequenced S. meliloti strain Rm1021 genome revealed the presence of all the putative genes allowing the biosynthesis of methionine (Fig. 1). This analysis indicated that the last step is carried out by the single B12-dependent methionine synthase encoded by metH. Since our strain of the sequenced S. meliloti Rm1021 was unable to use glycine betaine as an osmoprotectant, the metH gene was instead inactivated in S. meliloti strain 102F34 by recombination of a suicide plasmid containing an internal fragment of the coding region. The corresponding metH mutant was unable to grow in LAS medium unless methionine was provided to the cells (Fig. 2A), confirming that metH encodes the only methionine synthase in S. meliloti 102F34.
FIG. 2.
Phenotype of the metH mutant. metH (A and B) and wild-type (C and D) strains were grown in LAS medium (A and C) or in 0.5 M NaCl-LAS medium (B and D). Growth was analyzed in regular medium (open squares) or in medium supplemented with 1 mM methionine (open triangles), 1 mM glycine betaine (closed circles), or 1 mM DMG (closed diamonds). OD570, optical density at 570 nm.
The metH mutant also grew in LAS medium containing 1 mM glycine betaine, although with a lower growth rate than that observed with methionine supplementation (Fig. 2A). This indicates that S. meliloti strain 102F34 has a BHMT activity capable of using glycine betaine as a methyl donor for methionine synthesis. Furthermore, this result suggests that in LAS medium, MetH is more efficient than the BHMT pathway at carrying out the last step in methionine synthesis, providing the cell with methionine. Another possibility is that the uptake of GB may limit the synthesis of methionine by BHMT, especially at low osmolarity. Supplementation of the medium with DMG did not allow growth of the metH strain, showing that no other subsequent demethylation during glycine betaine catabolism is able to support methionine synthesis (Fig. 1). The noncatabolizable betaines DMSP and HB did not support growth of the metH strain in LAS medium (data not shown). Similarly, another betaine, trigonelline, was not used as a methionine precursor by the metH strain; this observation is consistent with the fact that trigonelline is catabolized in S. meliloti by a BHMT-independent pathway (4, 5). Taken together, these results are consistent with the hypothesis that BHMT is the only S. meliloti enzyme that allows methionine production from betaines.
In LAS medium containing 0.5 M NaCl, the metH mutant was unable to grow. Addition of methionine to this medium restored its growth. In contrast to what is observed in LAS medium (Fig. 2A), in the presence of 0.5 M NaCl, the addition of glycine betaine allowed faster growth of the metH mutant than that observed with methionine (Fig. 2B). A reasonable explanation for this phenomenon is that glycine betaine is not only utilized for methionine synthesis, but is also used as an osmoprotectant, as observed for the wild-type strain (Fig. 2D). These physiological observations suggest the presence of an efficient BHMT activity in 0.5 M NaCl LAS medium. A previous study had reported that high osmolarity in the growth medium decreased the activities of the enzymes involved in the degradation of glycine betaine (38), but it remained sufficient to enable continuous catabolism of glycine betaine in hyperosmotic media (43).
Identification of a BHMT-coding gene.
The transfer of a methyl group from glycine betaine to homocysteine catalyzed by BHMT is not only an alternate way to complete methionine biosynthesis; it was described as the first step in glycine betaine degradation (38). Although no BHMT-encoding gene was annotated in the S. meliloti genome, BLAST analysis with the human BHMT sequence revealed that the protein encoded by the SMc04325 open reading frame showed 24% identity with the human BHMT (9). BLAST scanning of the SMc04325 sequence against microbial genomes revealed that it is highly homologous to the amino-terminal domain of the methionine synthase encoded by metH, as previously observed with mammalian BHMTs (8). In addition, 8 of 19 sequenced genomes of alphaproteobacteria carried an apparent ortholog of the Smc04325 protein. Although none of the Smc04325-related proteins have known functions, all of them had two Zn binding motifs identified in the human and rat enzymes [G(A/V)NC and GGCC] (6, 8, 13) at the same locations as in the human BHMT enzyme (Fig. 3). Similarly, the BHMT amino acids interacting with homocysteine (9) are also conserved at the same positions in all these proteins. In contrast, among the three amino acids interacting with glycine betaine in the crystal structure of human BHMT (9), only one amino acid was conserved at the same position in all of the proteins. Although these data suggest that SMc04325 encodes a BHMT, the lack of conservation of some glycine betaine-interacting amino acids could reflect differences in substrate utilization between the human and S. meliloti enzymes, as the former can use DMSP (25) while the latter is unable to do so (32).
FIG. 3.
Alignment of bacterial BHMTs with the human enzyme. BLAST scanning of the SMc04325 sequence against microbial genomes (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) allowed the identification of various putative bacterial BHMT-encoding sequences. The corresponding proteins were aligned with human BHMT using CLUSTALW (http://www.ebi.ac.uk/clustalw/). The amino acids interacting with homocysteine and glycine betaine and zinc binding domains in human BHMT are boxed and annotated as HC, GB, and Zn, respectively. Each organism is identified by a three-letter code: Sme, Sinorhizobium meliloti; Mel, Mesorhizobium loti MAFF303099; Sil, Silicibacter pomeroyi DSS-3 (Silicibacter sp. strain TM1040); Jan, Jannaschia sp. strain CCS1; Rds, Rhodobacter sphaeroides 2.4.1; Prd, Paracoccus denitrificans PD1222; Unc, uncultured alphaproteobacterium EBAC2C11; Hsa, Homo sapiens. Asterisks indicate amino acids identical in all sequences, colons indicate conserved substitutions, and periods indicate semiconserved substitutions.
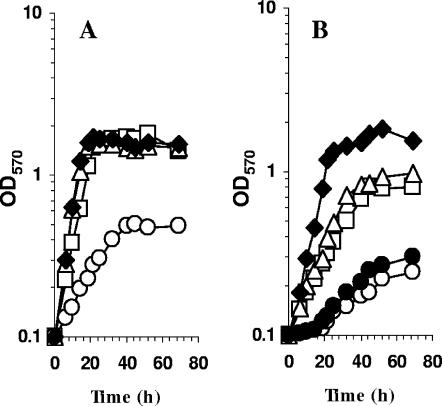
The SMc04325 gene was inactivated by recombination of a suicide vector carrying an internal fragment of the SMc04325 coding sequence. The growth of the wild-type strain and the SMc04325 mutant was analyzed in minimal medium containing lactate, glycine betaine, or DMG as a carbon source (Fig. 4). These three compounds allowed growth of the parental strain. The growth of the mutant on lactate and DMG was identical to that of the wild-type strain. In contrast, the mutant lacking SMc04325 was unable to use glycine betaine as the sole carbon source, suggesting that the SMc04325 protein corresponds to the BHMT activity involved in the catabolism of glycine betaine (38). Thus, we named the corresponding gene bmt (for betaine methyl transferase).
FIG. 4.
Growth of wild-type and bmt strains on glycine betaine as the carbon and energy source. S. meliloti 102F34 (closed symbols) and the bmt mutant (open symbols) were grown in minimal medium AS supplemented with 10 mM of lactate (squares), glycine betaine (circles), or DMG (triangles) as the carbon and energy source. OD570, optical density at 570 nm.
Assay of BHMT activity.
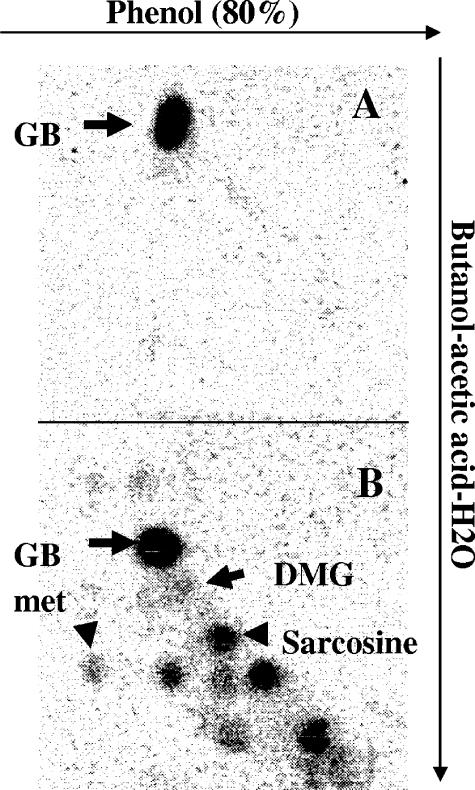
The BHMT activity was analyzed in crude extracts of wild-type and bmt strains (Fig. 5). Methionine, DMG, and sarcosine were produced from glycine betaine in extracts of the wild-type strain, in accordance with the glycine betaine catabolic pathway previously described (38). In contrast, none of these compounds was detected by using extracts of the bmt mutant. This result confirmed that the bmt strain is affected in the first step of glycine betaine catabolism catalyzed by BHMT.
FIG. 5.
BHMT assay. BHMT assays were performed on bmt (A) and wild-type (B) strains grown in Vincent medium to mid-exponential growth phase. Reaction products were separated by two-dimensional chromatography. The positions of spots corresponding to glycine betaine (GB), DMG, methionine (met), and sarcosine are indicated.
As previously described for the mammalian BHMT enzyme (25), DMSA was a more efficient substrate for the S. meliloti enzyme, since the amount of DMSA catabolized after 1 h was similar to the amount of GB catabolized after an overnight incubation with cellular extract (data not shown).
Growth of the bmt mutant is specifically inhibited in the presence of glycine betaine.
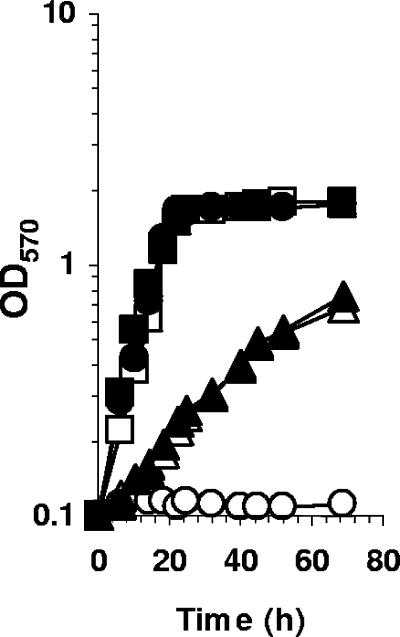
Glycine betaine has different roles in S. meliloti; it can be used as a carbon and nitrogen source, a methionine precursor, or an efficient osmoprotectant. Consequently, the behavior of the bmt mutant was analyzed in LAS medium in the absence or in the presence of 0.5 M NaCl and various osmoprotectants. The bmt mutant grew as efficiently as its parental strain in LAS medium (Fig. 4). The growth of the wild-type strain was not affected by the addition of 1 mM glycine betaine to LAS medium (Fig. 2C). In contrast, the growth rate and the growth yield of the bmt mutant were severely reduced when 1 mM glycine betaine was added to LAS medium. As observed for the wild-type strain (Fig. 6A), addition of 0.5 M NaCl to the growth medium reduced the growth rate of the bmt mutant (Fig. 6B). While glycine betaine improved the growth of the wild-type strain (Fig. 6A), it exerted a strong inhibition of the growth of the bmt mutant when added to hyperosmotic medium (Fig. 6B).
FIG. 6.
Effects of betaines on the growth of the bmt mutant. Wild-type (A) and bmt mutant (B) strains were grown in LAS medium lacking NaCl (open circles) or containing 0.5 M NaCl (closed circles) or 0.5 M NaCl medium supplemented with 1 mM glycine betaine (open triangles), 1 mM trigonelline (closed triangles), 1 mM HB (open squares), or 1 mM DMSP (open diamonds). OD570, optical density at 570 nm.
S. meliloti uses a wide variety of osmoprotectants. Among the nonaccumulated osmoprotectants, ectoine, pipecolate, sucrose, and trehalose had no effect on the growth of the bmt strain in LAS medium, and these compounds allowed identical improvements of growth under hyperosmotic conditions for the wild-type and bmt strains (data not shown). Among the diversity of trimethylammonium compounds used as osmoprotectants by S. meliloti, two, DMSP and homobetaine, are accumulated but are not catabolized (32). These compounds were not used as methionine precursors by the metH strain, suggesting that they are not substrates of BHMT. The plant-derived betaine, trigonelline, is catabolized by S. meliloti independently of the action of BHMT (4). S. meliloti 102F34 used these betaines as powerful osmoprotectants in hyperosmotic media (Fig. 6A). In LAS medium, the growth of the bmt mutant was not affected by the presence of DMSP, homobetaine, or trigonelline, but these compounds improved growth in the presence of 0.5 M NaCl (Fig. 6B). In summary, these results showed that glycine betaine specifically inhibits the growth of the bmt strain in hyperosmotic media.
Growth inhibition by glycine betaine in the bmt strain is relieved by methionine.
The inhibitory effect of glycine betaine on the growth of the bmt strain was relieved when methionine was added to LAS medium containing glycine betaine either in the presence or in the absence of 0.5 M NaCl. The growth pattern in LAS medium containing glycine betaine and methionine was identical to that observed in the absence of glycine betaine (Fig. 7A). In hyperosmotic media, growth inhibition in the presence of glycine betaine was also relieved by methionine supplementation (Fig. 7B), allowing improvement of the growth rate to a level identical to that of the wild-type strain (Fig. 6A). Addition of methionine to the hyperosmotic medium lacking glycine betaine did not improve growth. In order to analyze the defect induced by glycine betaine on the biosynthetic pathway of methionine, growth of the bmt mutant was monitored in 0.5 M NaCl medium containing 1 mM glycine betaine supplemented with the methionine precursor homocysteine (Fig. 7B). The addition of homocysteine had no effect on growth. Since only methionine supplementation allowed growth recovery, we conclude that glycine betaine affects the conversion of homocysteine to methionine by MetH. Therefore, glycine betaine affects either the production or the activity of MetH.
FIG. 7.
Influences of glycine betaine and methionine on the growth of the bmt mutant. The bmt mutant was grown in LAS medium lacking NaCl (A) or containing 0.5 M of NaCl (B) without any supplementation (open triangles) or in the presence of 1 mM glycine betaine (open circles), 1 mM methionine (open squares), 1 mM glycine betaine plus 1 mM methionine (closed diamonds), or 1 mM glycine betaine plus 1 mM homocysteine (closed circles). The growth rates of the bmt mutant in LAS medium were identical in the absence and in the presence of 1 mM homocysteine. OD570, optical density at 570 nm.
The absence of BHMT decreases but does not abolish glycine betaine catabolism.
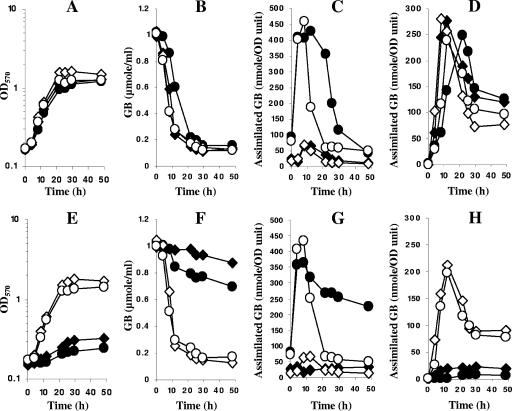
Glycine betaine catabolism was analyzed in vivo in wild-type and bmt mutant strains grown in LAS medium containing 1 mM [14C]glycine betaine in the absence or presence of 0.5 M NaCl (Fig. 8). Compared to the wild-type strain (Fig. 8B), the assimilation of glycine betaine, analyzed by its disappearance from the growth medium, was reduced in the bmt mutant grown in LAS medium (Fig. 8F). The levels of intracellular glycine betaine and glycine betaine-derived radiocarbons in the ethanol-insoluble fraction were very low, showing an inefficient catabolism of glycine betaine. These results demonstrate that in the bmt mutant, glycine betaine is catabolized, but more slowly that in the parental strain, since only 10% of external glycine betaine was catabolized after 50 h of culture (Fig. 8F), while the wild-type cells depleted glycine betaine from the medium within the first 20 h of growth (Fig. 8B) (43).
FIG. 8.
Fate of glycine betaine in the wild-type strain and the bmt mutant. The wild type (A, B, C, and D) and the bmt mutant (E, F, G, and H) were grown in LAS medium containing 1 mM [C14]glycine betaine in the absence (diamonds) or in the presence (circles) of 0.5 M NaCl. Growth was in the absence (closed symbols) or in the presence of 1 mM methionine (open symbols). Growth (A and E) was monitored, and aliquots were periodically harvested and extracted in 80% ethanol. The radioactivities remaining in the growth medium (B and F) and in the ethanol-soluble fraction (C and G) and incorporated in insoluble material (D and H) were quantified by scintillation counting. OD570, optical density at 570 nm.
The addition of 0.5 M NaCl to the growth medium slightly improved glycine betaine assimilation in the bmt strain, since 30% of the external glycine betaine level disappeared after 50 h of growth (Fig. 8F). The initial uptake velocity of glycine betaine was increased in comparison to LAS medium and reached a value similar to that obtained with wild-type cells (43), allowing a high intracellular level (Fig. 8G) similar to that reached in wild-type cells (Fig. 8C). In wild-type cells, glycine betaine accumulation is only transitory (Fig. 8C). In contrast, glycine betaine accumulation was maintained in the bmt mutant at the end of the exponential phase of growth (Fig. 8G), which resulted from the reduction of catabolism, as shown by the low level of detection of radiolabel molecules in the ethanol-insoluble material. These data suggest that glycine betaine catabolism in the bmt mutant is also reduced in the presence of NaCl.
Glycine betaine resulted in drastic growth inhibition of the bmt mutant (Fig. 8E). The addition of methionine relieved this inhibition and allowed increased catabolism of glycine betaine in either LAS or hyperosmotic medium (Fig. 8). The behavior of glycine betaine in the presence of methionine (Fig. 8F to H) was identical to that observed with the wild-type strain (Fig. 8B to D), and external glycine betaine was depleted within the first 20 h of growth in LAS medium in both strains. Glycine betaine was actively catabolized in both strains, and its accumulation in stressed cells was abolished as soon as external glycine betaine was depleted from the medium.
These results showed that BHMT is not the sole pathway for glycine betaine catabolism in S. meliloti strain 102F34. At least one alternative pathway exists and can be detected in the bmt mutant when the medium is supplied with methionine.
Nodulation efficiency of the metH and bmt mutants.
In various rhizobia, the formation of nitrogen-fixing nodules by methionine-deficient mutants is compromised unless methionine is provided to the plant growth medium (1, 24, 44, 45). Therefore, we analyzed the nodulation behavior of the metH and bmt mutants of S. meliloti 102F34. After 4 days of germination, Medicago sativa seedlings were inoculated with either the bmt or metH mutant strain or the 102F34 Smr parent and grown in nitrogen-deprived medium. Plants were visually screened for nodulation by observing the root system 4 weeks after bacterial inoculation. Nodulation was effective and efficient on plants inoculated with either of the mutants, as well as with the 102F34 parental strain. All plants which were inoculated with the wild-type or either of the mutant strains had green leaves and comparable numbers of effective nodules, whereas the uninoculated control plants were smaller, with yellow leaves. The numbers of pink nodules per root were similar in plants inoculated with any of the three strains. Bacteria from mutant-generated nodules were streaked on MSY plates and analyzed for kanamycin resistance, and their phenotypes were characterized. All the analyzed colonies had the phenotype of the mutant used for inoculation. Thus, N2-fixing nodules formed on alfalfa were due to infection by the metH or bmt mutants and not by revertants.
Plants were also inoculated with a mixture of the wild-type strain and the bmt or metH derivative. After 4 weeks, nodules were harvested and crushed to recover the bacteria on appropriate MSY medium, and the numbers of Kanr (mutant strains) and Kans (wild type strain) bacteria were determined. The ratio of bmt to wild-type bacteria isolated from nodules was identical to that used for inoculation. In contrast, when the metH strain was coinoculated with the wild-type strain in identical proportions, the number of metH colonies recovered from nodules was 4 orders of magnitude lower than that of the wild-type colonies. This proportion was not improved when the ratio of metH over the wild-type strain was increased in the inoculation mixtures. This observation clearly indicates that metH is required for the competitiveness of S. meliloti during the establishment of symbiosis with alfalfa.
DISCUSSION
Methionine synthesis.
In bacteria, the last step in methionine synthesis can be carried out either by a cobalamine-dependent (MetH) or a cobalamine-independent (MetE) enzyme. In S. meliloti, a MetE activity was postulated (20), but only MetH activity has been observed experimentally (35). Moreover, a careful analysis of the S. meliloti strain Rm1021 genome revealed only the presence of a metH gene. The inactivation of this gene in S. meliloti strain 102F34 abolished growth in minimal media, proving that in S. meliloti strain 102F34, only one methionine synthase exists. Inactivation of metH in S. meliloti Rm1021 also results in methionine auxotrophy (unpublished data), suggesting that this property is shared by various S. meliloti strains.
MetH is not the sole pathway allowing methionine synthesis in S. meliloti. A previous study showed the existence of a BHMT (38), which allows glycine betaine to be used as the methyl donor for the last step in methionine bioynthesis (38). In this study, we have characterized the BHMT coding sequence (named bmt). Interestingly, BHMT of S. meliloti is related to the human BHMT and to the amino-terminal domain of MetH enzymes, which carries the Zn- and homocysteine-binding domains (8, 9, 12). While betaine methyl transferase activity has been suspected in many bacteria, the corresponding genes have never been characterized. A glycine betaine transmethylase that can carry out the last step in methionine synthesis has been identified in Pseudomonas aeruginosa (36). However, this protein has no homology with S. meliloti BHMT or S. meliloti MetH and no homolog exists in the S. meliloti strain Rm1021 genome. Conversely, a BLAST scan of the P. aeruginosa genome with the S. meliloti BHMT sequence revealed only the existence of MetH. Thus, P. aeruginosa and S. meliloti use distinct enzymes for glycine betaine demethylation. BLAST searches revealed the presence of bmt orthologs only in the genomes of some alphaproteobacteria (Mezorhizobium loti, Silibacter pomeroyi, Rhodobacter sphaeroides, Jannaschia sp. strain CCS1, Paracoccus denitrificans, and the uncultured alphaproteobacterium EBAC2C11).
Glycine betaine catabolism.
Previous work on glycine betaine catabolism had suggested that only the BHMT pathway allowed glycine betaine catabolism in S. meliloti. The present work shows that the situation might be more complex, since glycine betaine catabolism was observed in vivo in bmt strains. Thus, another glycine betaine catabolic pathway exists in bmt strains, but this catabolic activity is reduced in hyperosmotic media. Methionine supplementation enabled improved glycine betaine catabolism and its utilization as a growth substrate. This was unexpected, since the bmt mutant possesses a wild-type copy of the metH gene. These observations suggest two conclusions: (i) glycine betaine compromises MetH activity in the bmt strain and (ii) the alternative glycine betaine catabolic pathway does not produce methionine.
Inhibition of MetH at the transcriptional and enzymatic levels by glycine betaine was documented in Aspergillus nidulans (21). When glycine betaine is provided to A. nidulans, BHMT is the preferred pathway for methionine supply (21). A similar phenomenon could exist in S. meliloti. The presence of glycine betaine has no consequences for the methionine supply when BHMT is functional, but in its absence, the rate of methionine synthesis appears to be reduced, as suggested by the reduction of the growth rate of the bmt mutant when glycine betaine was added to the growth medium. Inhibition of methionine synthesis was greater in media containing 0.5 M of NaCl. This is probably due to the increase of the intracellular concentration of glycine betaine in response to the hyperosmotic stress, leading to greater inhibition of MetH activity.
Our results suggest that MetH activity can be affected by the presence of glycine betaine in the growth medium, involving an efficient BHMT activity to provide the cell with methionine. Surprisingly, the mutant lacking BHMT activity was able to catabolize glycine betaine through an alternate catabolic pathway which remains to be characterized. However, to be optimal, this pathway requires the presence of methionine and therefore an efficient BHMT activity, as MetH is inhibited by the presence of glycine betaine. This suggests that the alternative pathway is not of minor importance for glycine betaine catabolism. In vivo glycine betaine catabolism through the alternative pathway is dependent on the functioning of BHMT.
Nodulation.
Methionine synthesis has been described as being essential for efficient nodulation by various rhizobia. In S. meliloti strain Rmd201, metA/metZ, metE, and metF mutants are auxotrophic for methionine. They form ineffective nodules containing a reduced number of infected cells, in which the bacteria fail to undergo complete differentiation into bacteroids (1). Similar results have been described for some methionine auxotrophs of S. meliloti strain 104A14 (24), which formed ineffective nodules. The nodulation efficiencies of methionine auxotrophs of other rhizobia are also affected, as in the cases of a metZ mutant of Rhizobium etli (44) and a cysD derivative of Sinorhizobium sp. strain BR816 (40). In contrast, the cysG derivative of R. etli forms effective nodules (45). All these studies suggest that the plants provide low concentrations of methionine to the bacteria. Variations can be observed, depending on the plant-rhizobium model; the presence of other organic sulfur sources, like cysteine or glutathione, could allow growth inside the plant (45). All of these previous studies concerned mutants whose production of homocysteine was affected. Nodulation by metH mutants had not been analyzed previously.
S. meliloti 102F34 metH and bmt single mutants could proliferate in M. sativa and form effective nodules. This suggests that the plant provides the bacteria with a level of methionine that is sufficient for S. meliloti strain 102F34 or a methionine precursor like glycine betaine. The metH mutant had the ability to proliferate when glycine betaine was present in the growth medium; however, glycine betaine studies have shown that the plant does not provide glycine betaine, homobetaine, or DMSP to the bacteria (28). These compounds were not detected in M. sativa; stachydrine and trigonelline were the only two methylammoniums produced by M. sativa (28). Stachydrine and trigonelline can be used as growth substrates and as osmoprotectants by S. meliloti. The assimilation pathway of stachydrine does not require BHMT activity but uses another pathway that produces proline by two successive demethylations (7, 31). This pathway does not produce methionine, so it could not rescue a metH mutant. In the presence of trigonelline, MetH activity is not impaired in vitro. MetH activity is not expected to be inhibited by plant betaines and could allow growth of the bmt mutant within the plant. The absence of a glycine betaine supply and the noninhibition of MetH by plant betaines suggest that MetH is the main pathway for methionine synthesis during symbiosis.
Since the metH mutant had no obvious defect in nodulation, the plant must provide the bacteria with methionine. However, the amount of methionine supplied by the plant must be limiting, as we observed that a metH mutant competes extremely poorly against its wild-type strain when equally coinoculated onto the plant.
Acknowledgments
We thank S. Georgeault, C. Monnier, M. Uguet, and M. C. Savary for technical assistance.
This work was supported in part by the Centre National de la Recherche Scientifique and the Ministère de la Recherche et de l'Education Nationale and in part by Public Health Service Grant GM31030 from the National Institutes of Health (NIH) to G.C.W. G.C.W. was also supported by an American Cancer Society Research Professorship and an HHMI Professorship. L.B. was supported by Region Bretagne and NIH.
REFERENCES
- 1.Abbas, B. A., K. E. Vineetha, C. K. Prasad, N. Vij, R. Hassani, and G. S. Randhawa. 2002. Symbiotic characteristics of cysteine and methionine auxotrophs of Sinorhizobium meliloti. Indian J. Exp. Biol. 40:1121-1130. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Barra, L., N. Pica, K. Gouffi, G. C. Walker, C. Blanco, and A. Trautwetter. 2003. Glucose 6-phosphate dehydrogenase is required for sucrose and trehalose to be efficient osmoprotectants in Sinorhizobium meliloti. FEMS Microbiol. Lett. 229:183-188. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, C., L. R. Barran, C. A. Malpica, and C. Rosenberg. 1991. Genetic analysis of a region of the Rhizobium meliloti pSym plasmid specifying catabolism of trigonelline, a secondary metabolite present in legumes. J. Bacteriol. 173:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boivin, C., S. Camut, C. A. Malpica, G. Truchet, and C. Rosenberg. 1990. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breksa, A. P., III, and T. A. Garrow. 1999. Recombinant human liver betaine-homocysteine S-methyltransferase: identification of three cysteine residues critical for zinc binding. Biochemistry 38:13991-13998. [DOI] [PubMed] [Google Scholar]
- 7.Burnet, M. W., A. Goldmann, B. Message, R. Drong, A. El Amrani, O. Loreau, J. Slightom, and D. Tepfer. 2000. The stachydrine catabolism region in Sinorhizobium meliloti encodes a multi-enzyme complex similar to the xenobiotic degrading systems in other bacteria. Gene 244:151-161. [DOI] [PubMed] [Google Scholar]
- 8.Evans, J. C., D. P. Huddler, M. T. Hilgers, G. Romanchuk, R. G. Matthews, and M. L. Ludwig. 2004. Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc. Natl. Acad. Sci. USA 101:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, J. C., D. P. Huddler, J. Jiracek, C. Castro, N. S. Millian, T. A. Garrow, and M. L. Ludwig. 2002. Betaine-homocysteine methyltransferase: zinc in a distorted barrel. Structure 10:1159-1171. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein, J. D., and J. J. Martin. 1984. Methionine metabolism in mammals. Distribution of homocysteine between competing pathways. J. Biol. Chem. 259:9508-9513. [PubMed] [Google Scholar]
- 11.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, B., N. Campillo, F. Garrido, M. Gasset, J. Sanz-Aparicio, and M. A. Pajares. 2003. Active-site-mutagenesis study of rat liver betaine-homocysteine S-methyltransferase. Biochem. J. 370:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez, B., M. A. Pajares, M. Martinez-Ripoll, T. L. Blundell, and J. Sanz-Aparicio. 2004. Crystal structure of rat liver betaine homocysteine S-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding. J. Mol. Biol. 338:771-782. [DOI] [PubMed] [Google Scholar]
- 14.Gouffi, K., T. Bernard, and C. Blanco. 2000. Osmoprotection by pipecolic acid in Sinorhizobium meliloti: specific effects of d and l isomers. Appl. Environ. Microbiol. 66:2358-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouffi, K., N. Pica, V. Pichereau, and C. Blanco. 1999. Disaccharides as a new class of nonaccumulated osmoprotectants for Sinorhizobium meliloti. Appl. Environ. Microbiol. 65:1491-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouffi, K., V. Pichereau, J. P. Rolland, D. Thomas, T. Bernard, and C. Blanco. 1998. Sucrose is a nonaccumulated osmoprotectant in Sinorhizobium meliloti. J. Bacteriol. 180:5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Jebbar, M., L. Sohn-Bosser, E. Bremer, T. Bernard, and C. Blanco. 2005. Ectoine-induced proteins in Sinorhizobium meliloti include an ectoine ABC-type transporter involved in osmoprotection and ectoine catabolism. J. Bacteriol. 187:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jebbar, M., R. Talibart, K. Gloux, T. Bernard, and C. Blanco. 1992. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J. Bacteriol. 174:5027-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, J. Q., W. Wei, B. H. Du, X. H. Li, L. Wang, and S. S. Yang. 2004. Salt-tolerance genes involved in cation efflux and osmoregulation of Sinorhizobium fredii RT19 detected by isolation and characterization of Tn5 mutants. FEMS Microbiol. Lett. 239:139-146. [DOI] [PubMed] [Google Scholar]
- 21.Kacprzak, M. M., I. Lewandowska, R. G. Matthews, and A. Paszewski. 2003. Transcriptional regulation of methionine synthase by homocysteine and choline in Aspergillus nidulans. Biochem. J. 376:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaijalainen, S., M. Schroda, and K. Lindstrom. 2002. Cloning of nodule-specific cDNAs of Galega orientalis. Physiol. Plant 114:588-593. [DOI] [PubMed] [Google Scholar]
- 23.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 24.Kerppola, T. K., and M. L. Kahn. 1988. Symbiotic phenotypes of auxotrophic mutants of Rhizobium meliloti 104A14. J. Gen. Microbiol. 134:913-919. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. B., J. W. Blunt, M. Lever, and P. M. George. 2004. A nuclear-magnetic-resonance-based assay for betaine-homocysteine methyltransferase activity. Anal. Biochem. 330:199-205. [DOI] [PubMed] [Google Scholar]
- 26.Meskys, R., R. J. Harris, V. Casaite, J. Basran, and N. S. Scrutton. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur. J. Biochem. 268:3390-3398. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Miller, K. J., and J. M. Wood. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50:101-136. [DOI] [PubMed] [Google Scholar]
- 29.O'Gara, F., and K. T. Shanmugam. 1976. Control of symbiotic nitrogen fixation in rhizobia. Regulation of NH4+ assimilation. Biochim. Biophys. Acta 451:342-352. [DOI] [PubMed] [Google Scholar]
- 30.Oke, V., B. G. Rushing, E. J. Fisher, M. Moghadam-Tabrizi, and S. R. Long. 2001. Identification of the heat-shock sigma factor RpoH and a second RpoH-like protein in Sinorhizobium meliloti. Microbiology 147:2399-2408. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, D. A., E. S. Sande, J. A. C. Vriezen, F. J. de Bruijn, D. Le Rudulier, and C. M. Joseph. 1998. A new genetic locus in Sinorhizobium meliloti is involved in stachydrine utilization. Appl. Environ. Microbiol. 64:3954-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichereau, V., J.-A. Pocard, J. Hamelin, C. Blanco, and T. Bernard. 1998. Differential effects of dimethylsulfoniopropionate, dimethylsulfonioacetate, and other S-methylated compounds on the growth of Sinorhizobium meliloti at low and high osmolarities. Appl. Environ. Microbiol. 64:1420-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rains, D. W., L. N. Csonka, D. Le Rudulier, T. P. Croughan, S. S. Yang, S. J. Stavarek, and R. C. Valentine. 1982. Osmoregulation by organisms exposed to saline stress: physiological mechanisms and genetic manipulation, p. 283-302. In A. San Pietro (ed.), Biosaline research: a look to the future. Plenum Publishing Corp., New York, N.Y.
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Sato, K., S. Inukai, and S. Shimizu. 1974. Vitamin B12-dependent methionine synthesis in Rhizobium meliloti. Biochem. Biophys. Res. Commun. 60:723-728. [DOI] [PubMed] [Google Scholar]
- 36.Serra, A. L., J. F. Mariscotti, J. L. Barra, G. I. Lucchesi, C. E. Domenech, and A. T. Lisa. 2002. Glycine betaine transmethylase mutant of Pseudomonas aeruginosa. J. Bacteriol. 184:4301-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, R., J. Quandt, and W. Klipp. 1989. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene 80:161-169. [DOI] [PubMed] [Google Scholar]
- 38.Smith, L. T., J. A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, L. T., and G. M. Smith. 1989. An osmoregulated dipeptide in stressed Rhizobium meliloti. J. Bacteriol. 171:4714-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snoeck, C., C. Verreth, I. Hernandez-Lucas, E. Martinez-Romero, and J. Vanderleyden. 2003. Identification of a third sulfate activation system in Sinorhizobium sp. strain BR816: the CysDN sulfate activation complex. Appl. Environ. Microbiol. 69:2006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, H., R. Tamamura, S. Yajima, M. Kanno, and M. Suguro. 2005. Corynebacterium sp. U-96 contains a cluster of genes of enzymes for the catabolism of sarcosine to pyruvate. Biosci. Biotechnol. Biochem. 69:952-956. [DOI] [PubMed] [Google Scholar]
- 42.Talibart, R., M. Jebbar, G. Gouesbet, S. Himdi-Kabbab, H. Wróblewski, C. Blanco, and T. Bernard. 1994. Osmoadaptation in rhizobia: ectoine-induced salt tolerance. J. Bacteriol. 176:5210-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talibart, R., M. Jebbar, K. Gouffi, V. Pichereau, G. Gouesbet, C. Blanco, T. Bernard, and J.-A. Pocard. 1997. Transient accumulation of glycine betaine and dynamics of endogenous osmolytes in salt-stressed cultures of Sinorhizobium meliloti. Appl. Environ. Microbiol. 63:4657-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tate, R., A. Riccio, E. Caputo, M. Iaccarino, and E. J. Patriarca. 1999. The Rhizobium etli metZ gene is essential for methionine biosynthesis and nodulation of Phaseolus vulgaris. Mol. Plant Microbe Interact 12:24-34. [DOI] [PubMed] [Google Scholar]
- 45.Tate, R., A. Riccio, M. Iaccarino, and E. J. Patriarca. 1997. A cysG mutant strain of Rhizobium etli pleiotropically defective in sulfate and nitrate assimilation. J. Bacteriol. 179:7343-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent, J. L. 1970. A manual for the practical study of root nodule bacteria. Blackwell, Oxford, United Kingdom.
- 47.Watson, R. J., R. Heys, T. Martin, and M. Savard. 2001. Sinorhizobium meliloti cells require biotin and either cobalt or methionine for growth. Appl. Environ. Microbiol. 67:3767-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]