Abstract
Clostridium difficile is the most frequent cause of nosocomial diarrhea worldwide, and recent reports suggested the emergence of a hypervirulent strain in North America and Europe. In this study, we applied comparative phylogenomics (whole-genome comparisons using DNA microarrays combined with Bayesian phylogenies) to model the phylogeny of C. difficile, including 75 diverse isolates comprising hypervirulent, toxin-variable, and animal strains. The analysis identified four distinct statistically supported clusters comprising a hypervirulent clade, a toxin A− B+ clade, and two clades with human and animal isolates. Genetic differences among clades revealed several genetic islands relating to virulence and niche adaptation, including antibiotic resistance, motility, adhesion, and enteric metabolism. Only 19.7% of genes were shared by all strains, confirming that this enteric species readily undergoes genetic exchange. This study has provided insight into the possible origins of C. difficile and its evolution that may have implications in disease control strategies.
Clostridium difficile is a gram-positive, spore-forming anaerobic bacterium that is responsible for a variety of gastrointestinal diseases in humans and other animals, collectively referred to as C. difficile-associated disease (CDAD) (17, 29). The pathogen is frequently associated with antibiotic treatment, and the severity of CDAD ranges from antibiotic-associated diarrhea to the life-threatening pseudomembranous colitis (17). Beyond the morbidity and mortality, CDAD is a severe economic burden, estimated to cost the U.S. health care system in excess of $1 billion annually (21). More disturbingly, the reported incidence of CDAD has increased significantly in the last decade and a new highly virulent strain is causing outbreaks of increased severity in North America and Europe (23, 24, 32). The origin of this strain is uncertain, although it has been proposed that increased use of fluoroquinolones may provide a selective advantage for this epidemic strain that is resistant to the newer fluoroquinolones, gatifloxacin and moxifloxacin (25).
C. difficile is known to produce a number of factors that contribute to its virulence, including two related exotoxins, toxin A (TcdA) and toxin B (TcdB), which are part of a 16-kb pathogenicity locus (PaLoc) where toxin production is negatively controlled by TcdC (30). A minority of strains produce a binary toxin (CdtA/CdtB), but its role in disease is unclear (10, 11). However, production of these toxins alone cannot explain C. difficile pathogenesis. In recent years, increasing numbers of strains have been reported from several countries with truncated versions of toxin A and/or toxin B (10, 31).
A plethora of techniques has been used to type C. difficile, many of which have confirmed the transmission of the organism in hospital environments (1). Commonly used methods are toxinotyping based upon variations in the PaLoc sequence (28), pulsed-field gel electrophoresis (PFGE) (9), PCR ribotyping (26) and restriction endonuclease analysis (REA) (4). These methods have generally been efficient at grouping strains and in particular have been used to distinguish the recently emerged hypervirulent strains as toxinotype III, North American PFGE type 1, REA group BI, or PCR ribotype 027 (generally referred to as BI/NAP1/027) (20, 24, 32). However, these methods have limited discriminatory potential to elucidate the phylogenetic relationships of all strains in a given study. For example, the discriminatory power of PCR ribotyping is not absolute; ribotype 001, the most commonly occurring ribotype in humans, can be subtyped by PFGE (8), and 20 distinct BI group types have been found by REA (24). Additionally, traditional typing systems do not provide information on the genes/genetic loci specific to strains from different sources.
Microarray technology, allied to complex mathematical analysis to determine phylogeny, has provided a sensitive and robust method to examine the genetic relatedness of bacterial populations (2, 6). The genetic relationships described by Bayesian phylogeny of a DNA-DNA microarray data set can then be correlated with the known phenotype and ecological behavior of each bacterial strain in the analysis; this is particularly useful in studying the epidemiology and host association of pathogens (6, 16). Comparative genomic DNA microarray analysis has been used to investigate several bacterial species in relation to pathogenesis and host specificity. Comparison of strains isolated from different hosts as well as virulent and avirulent strains can reveal predicted coding DNA sequences (CDSs) that may be important for virulence, pathogen-host interactions, and transmission (2, 6, 16). To date, microarray analysis of defined cohorts of strains to determine genetic relatedness has not been undertaken for C. difficile.
In this study, we carried out whole-genome analysis of 75 well-characterized isolates of C. difficile from humans with a range of disease outcomes and from several animal sources, using a whole-genome microarray based on the recently sequenced genome of C. difficile 630. Combining DNA microarray data with sensitive Bayesian-based algorithms has yielded new insights into the population structure of C. difficile, revealing information on the evolution and origin of the pathogen as well as several potential determinants of survival and virulence.
MATERIALS AND METHODS
Strains.
The strains investigated in this study were 55 human isolates (including 21 hypervirulent [epidemic BI/NAP1/027 strains], 13 A+ B+, 14 A− B+, and 7 A− B− strains) and 20 animal isolates (7 bovine, 6 swine, 4 equine, and 3 murine strains) (Table 1). Prior to microarray analysis, strains of toxinotype III, PFGE NAP1, REA BI, or PCR ribotype 027 and with heightened disease severity were designated hypervirulent. Strains were designated A− B+ on the basis of toxinotype or PCR/sequencing analysis of tcdA and tcdB. The microarray was designed on the sequenced strain C. difficile 630, a virulent and multidrug-resistant strain that was observed to spread to several other patients in the same ward (33).
TABLE 1.
C. difficile strains
| Strain | Sourcea | Genotype | Motility locus | Motility | Clinical data | Origin | Clade |
|---|---|---|---|---|---|---|---|
| CD1 | DG | A− B− | Full | Asymptomatic | Human | A− B+ | |
| CF1 | DG | A− B+ | Full* | No data | Human | A− B+ | |
| CF2 | DG | A− B+ | Full* | CDAD | Human | A− B+ | |
| CF4 | DG | A− B+ | Full* | CDAD | Human | A− B+ | |
| CF5 | DG | A− B+ | Full* | Asymptomatic | Human | A− B+ | |
| CG3 | DG | A− B+ | Full* | Asymptomatic | Human | A− B+ | |
| JGS6042 | JGS | A+ B+ | Partial | Nonmotile | Diarrhea | Bovine | A− B+ |
| JGS6047 | JGS | A− B− | Full | Nosocomial diarrhea | Equine | A− B+ | |
| JGS6053 | JGS | A+ B+ | Partial | Motile | Nosocomial diarrhea | Equine | A− B+ |
| JGS6057 | JGS | A+ B+ | Partial | Motile | Nosocomial diarrhea | Equine | A− B+ |
| JGS655 | JGS | A+ B+ | Partial | Nonmotile | Neonatal typhlocolitis | Swine | A− B+ |
| M10 | DD | A− B+ | Full* | Outbreak | Human | A− B+ | |
| M13 | DG | A− B− | Partial | Human | A− B+ | ||
| M17 | DD | A− B+ | Full* | Outbreak | Human | A− B+ | |
| M20 | DD | A− B+ | Full* | Outbreak | Human | A− B+ | |
| M23 | DG | A− B− | Partial | Human | A− B+ | ||
| M3 | DG | A− B− | Partial | Human | A− B+ | ||
| M30 | DD | A− B+ | Partial | Outbreak | Human | A− B+ | |
| M65 | DD | A− B+ | Partial | Outbreak | Human | A− B+ | |
| M68 | DD | A− B+ | Partial | Outbreak | Human | A− B+ | |
| M70 | DD | A− B+ | Partial | Outbreak | Human | A− B+ | |
| M9 | DD | A− B+ | Partial | Outbreak | Human | A− B+ | |
| T7 | DG | A− B− | Full | Human | A− B+ | ||
| BI-1 | DG | tcdC | Partial | Human | HY | ||
| BI-10 | DG | tcdC | Partial | Human | HY | ||
| BI-11 | DG | tcdC | Partial | Human | HY | ||
| BI-12 | DG | tcdC | Partial | Human | HY | ||
| BI-14 | DG | tcdC | Partial | Human | HY | ||
| BI-15 | DG | tcdC | Partial | Human | HY | ||
| BI-16 | DG | tcdC | Partial | Human | HY | ||
| BI-2 | DG | tcdC | Partial | Human | HY | ||
| BI-3 | DG | tcdC | Partial | Human | HY | ||
| BI-4 | DG | tcdC | Partial | Human | HY | ||
| BI-5 | DG | tcdC | Partial | Human | HY | ||
| BI-6 | DG | tcdC | Partial | Human | HY | ||
| BI-6p | DG | tcdC | Partial | Human | HY | ||
| BI-6p2 | DG | tcdC | Partial | Human | HY | ||
| BI-7 | DG | tcdC | Partial | Human | HY | ||
| BI-8 | DG | tcdC | Partial | Human | HY | ||
| R12087 | JB | A+ B+ | Partial | Human | HY | ||
| R20291 | JB | A+ B+ | Partial | Human | HY | ||
| R20352 | JB | A+ B+ | Partial | Human | HY | ||
| R20928 | JB | A+ B+ | Partial | Human | HY | ||
| BI-9 | DG | tcdC | Partial | Human | HA1 | ||
| B-one | DG | A+ B+ | Partial | Human | HA1 | ||
| J9 | DG | A+ B+ | Partial | Human | HA1 | ||
| JGS355 | JGS | A+ B+ | Full | Diarrheic SCID | Mouse | HA1 | |
| JGS356 | JGS | A+ B+ | Full | Diarrheic SCID | Mouse | HA1 | |
| JGS360 | JGS | A+ B+ | Full | Diarrheic SCID | Mouse | HA1 | |
| JGS6041 | JGS | A+ B+ | Partial | Nosocomial diarrhea | Equine | HA1 | |
| JGS692 | JGS | A− B− | Full | Nonmotile | Neonatal typhlocolitis | Swine | HA1 |
| K14 | DG | A+ B+ | Partial | Human | HA1 | ||
| M124 | DD | A+ B+ | Partial | Nonoutbreak | Human | HA1 | |
| M134 | DD | A+ B+ | Partial | Nonoutbreak | Human | HA1 | |
| M135 | DD | A+ B+ | Partial | Nonoutbreak | Human | HA1 | |
| M151 | DD | A+ B+ | Partial | Nonoutbreak | Human | HA1 | |
| M47 | DD | A+ B+ | Partial | Outbreak | Human | HA1 | |
| R10459 | JB | A+ B+ | Partial | Human | HA1 | ||
| R8366 | JB | A+ B+ | Partial | Human | HA1 | ||
| VPI 10463 | DG | A+ B+ | Partial | Human | HA1 | ||
| Y2 | DG | A+ B+ | Partial | Human | HA1 | ||
| AA1 | DG | A− B− | Absent | Human | HA2 | ||
| AA2 | DG | A− B+ | Absent | Human | HA2 | ||
| JGS647 | JGS | A+ B+ | Absent | Nonmotile | Neonatal typhlocolitis | Swine | HA2 |
| JGS652 | JGS | A+ B+ | Absent | Nonmotile | Neonatal typhlocolitis | Swine | HA2 |
| JGS673 | JGS | A+ B+ | Absent | Nonmotile | Diarrhea | Bovine | HA2 |
| JGS674 | JGS | A+ B+ | Absent | Nonmotile | Diarrhea | Bovine | HA2 |
| JGS675 | JGS | A+ B+ | Absent | Nonmotile | Diarrhea | Bovine | HA2 |
| JGS676 | JGS | A+ B+ | Absent | Nonmotile | Diarrhea | Bovine | HA2 |
| JGS677 | JGS | A+ B+ | Absent | Nonmotile | Diarrhea | Bovine | HA2 |
| JGS679 | JGS | A+ B+ | Absent | Nonmotile | Diarrhea | Bovine | HA2 |
| JGS688 | JGS | A+ B+ | Absent | Nonmotile | Neonatal typhlocolitis | Swine | HA2 |
| JGS691 | JGS | A+ B+ | Absent | Nonmotile | Neonatal typhlocolitis | Swine | HA2 |
| M120 | DD | A+ B+ | Absent | Nonoutbreak | Human | HA2 | |
| M133 | DD | A+ B+ | Absent | Nonoutbreak | Human | HA2 |
DG, Dale Gerding; DD, Denise Drudy; JB, Jon Brazier; JGS, Glenn Songer; HY, hypervirulent clade strains; HA, human and animal isolates (two clades); A− B+, defective toxin clade; tcdC, A+ B+ but with an 18-bp deletion in tcdC; full, CD0226-CD0271 present; *, full motility-associated loci except a CD0253-CD0254 deletion; partial, CD0245-CD0271 present only; absent, all loci absent.
Microarray design.
The microarray was constructed using the approach described previously to include all 3,688 chromosomal predicted CDSs from strain 630 (excluding 92 additional CDSs annotated since construction of the microarray) (15). Ten pairs of gene-specific primers were designed to each sequence in the gene pool by using Primer3(27). Primers were 20 to 25 bp and were designed as previously described (14, 27), with a matched Tm of ∼60°C, an amplicon size range from 50 to 800 bp, and an optimum size of 600 bp. Selection was based on BLASTN analysis of the PCR products against genes; all 10 PCR products for each target sequence were compared to the sequence of each gene in the gene pool, and the longest product with the least similarity (or no similarity) to any other sequence in the gene pool was selected. This approach maximizes sensitivity and minimizes cross-hybridizations. Additionally, multiple reporters were designed to some genes, including eight for tcdA, seven for tcdB, three for cdtA, four for cdtB, and two for each gene involved in S-layer formation.
Amplification of microarray reporter elements.
PCR primers were synthesized by MWG Biotech (Ebersberg, Germany) and supplied in a 96-well format to enable high-throughput amplification using a liquid handling and PCR amplification robot (RoboAmp 9600; MWG Biotech). PCRs were performed with 10 ng DNA template, 5 U HotStar Taq DNA polymerase (QIAGEN), 0.5 μM primers, 1.5 mM MgCl2, and 200 mM deoxynucleoside triphosphates. Thermocycling was performed using denaturation of 95°C for 15 min, 40 cycles of 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min, followed by a final extension of 72°C for 5 min. Subsequent rounds of PCR amplification with modified conditions were performed until a single product of predicted size was obtained for all genes that were not amplified under standard conditions. Additional validation was undertaken by sequencing 5% of the amplified genes. Microarrays were constructed by robotic spotting of the PCR products in duplicate on UltraGAPS aminosilane-coated glass slides (Corning), using MicroGrid II (BioRobotics, United Kingdom) (14). The microarrays were postprint processed according to the slide manufacturer's instructions, using hydration and UV irradiation, and stored in a dark, dust-free environment.
Hybridizations.
Hybridizations were performed as previously described (7, 13, 16) with 2 to 3 μg of test genomic DNA labeled with Cy3-dCTP and 2 μg Cy5-dCTP with labeled C. difficile 630 genomic DNA as a common reference for all hybridizations. Microarray slides were prehybridized in 3.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and 10 mg/ml bovine serum albumin at 65°C for 20 min before a wash in distilled water for 1 min and a subsequent wash for 1 min in isopropanol. Test strain-labeled DNA was mixed with reference strain-labeled DNA, purified using a MiniElute kit (QIAGEN), denatured at 95°C, and mixed to achieve a final volume of 23 μl hybridization solution of 4× SSC and 0.3% SDS. Using a 22- by 22-mm LifterSlips (Eyrie Scientific), a microarray was hybridized overnight, sealed in a humidified hybridization chamber (Telechem International), and immersed in a water bath at 65°C for 16 to 20 h. Slides were washed once in 400 ml 1× SSC and 0.06% SDS at 65°C for 2 min and twice in 400 ml 0.06× SSC for 2 min. Microarrays were scanned using a 418 array scanner (Affymetrix) and intensity fluorescence data acquired using BlueFuse (BlueGnome). Test strains were hybridized up to three times on microarrays that have duplicate sets of reporters representing the C. difficile genome.
Microarray data analysis and comparative phylogenomics.
Data were initially processed and normalized using GeneSpring 7.2 (Silicon Genetics). Values below 0.01 were set to 0.01. The measured intensity for each CDS was divided by its control channel value in each sample; if the control channel was below 0.01, then 0.01 was used instead. If both the control channel and the signal channel were below 0.01, then no data were reported. Data were divided by the 50th percentile of all genes that had a raw measurement above 0.01 and were not flagged as low confidence (P < 0.1). The designation of CDSs in each strain as present, divergent, or absent was determined by the use of GACK software (16). GACK calculated an estimated probability of presence (EPP) value for each gene. A gene was designated present if it had a calculated EPP of 100%, divergent if it had an EPP between 0% and 100%, and deleted if it had an EPP of 0%. 0% EPP indicates a 0% chance of being falsely assigned as a divergent gene, and 100% EPP indicates a minimum assurance that a gene was present (19). The GACK output for all genes was used for phylogeny inference calculated using a Bayesian phylogenetic algorithm (MrBayes v3.1.1, http://mrbayes.CSIT.FSU.EDU). MrBayes requires binary data so divergent genes were reclassified as present. The Bayesian model used four-chain Markov chain Monte Carlo and 16-category gamma distribution with 1 million iterations with a heat of 0.7 as described previously (2). Phylogenetic trees were sampled every 40th iteration, and tree structure convergence was statistically assessed across all potential phylogenies (except an initial 10,000 tree burn-in). The final (1,000,000th) trees produced by separate runs were statistically assessed for convergence. Phylogeny inference was based on a conservative estimation of gene loss.
Microarray data accession numbers.
Fully annotated microarray data have been deposited in BμG@Sbase (accession number E-BUGS-41; http://bugs.sgul.ac.uk/E-BUGS-41) and also ArrayExpress (accession number E-BUGS-41).
RESULTS AND DISCUSSION
Overall phylogeny.
A well-characterized collection of 75 C. difficile isolates was selected for genomic comparisons from diverse geographical origins comprising 21 hypervirulent, 13 A+ B+, 14 A− B+, and 7 A− B− human isolates and 7 bovine, 6 swine, 4 equine, and 3 murine isolates (Table 1). All isolates were competitively hybridized with the C. difficile 630 DNA microarray. From these data, the Bayesian phylogeny of the C. difficile isolates revealed four major clades unequivocally supported by Bayesian probabilities (Fig. 1). This included a hypervirulent clade with 20 of the 21 hypervirulent isolates (HY), a defective toxin clade with all 14 A− B+ variants (A−B+), and two clades that had animal isolates intermixed with human isolates (HA1 and HA2).
FIG. 1.
Phylogenetic relationship of strains associated with different clinical outcomes and animal sources represented as four major clades (HY, A− B+, HA1, and HA2). Strains are designated at the end of the branches and are colored according to the animal source from which the C. difficile strain was isolated. Black, human; blue, mouse; green, bovine; red, swine; light blue, equine. Branches with ** have a P value of 1.0 and represent 100% of all phylogenies showing a given topology. * indicates a P value of ≥0.98.
Hypervirulent clade and microevolution.
Previous studies using multilocus sequence typing (MLST) analysis on human isolates recovered from antibiotic-associated disease and pseudomembranous colitis found that strains did not cluster into a hypervirulent lineage (22). However, in this study, 20/21 hypervirulent strains clearly clustered into a distinct lineage. The HY clade consisted of 20 isolates, all of which were classified as hypervirulent. The 20 strains were from diverse locations in the United States, Canada, and the United Kingdom, confirming their transcontinental spread. The United Kingdom isolate (R20291) was a particularly aggressive strain isolated from Stoke Mandeville Hospital, while the Canadian strain (R20352) was a highly transmissible strain from Quebec.
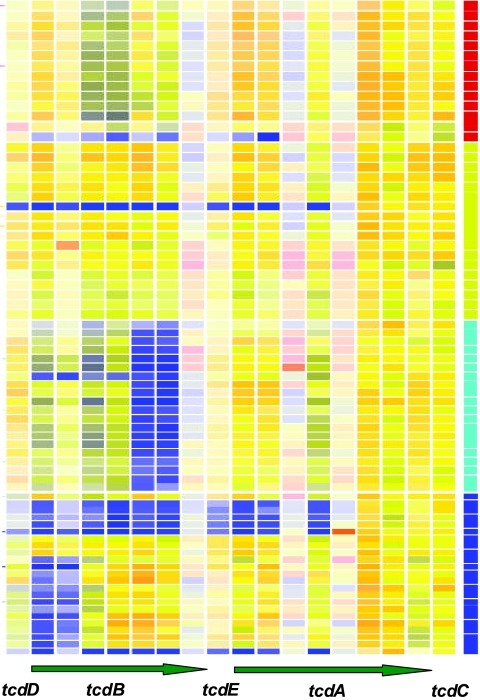
The genomes of strains in the hypervirulent clade characteristically had a number of deletions compared to those of strain 630, with the exception of BI-9, which appears in clade HA1. BI-9 does not have a characteristic apparent deletion at the end of tcdB, specific to the hypervirulent strains and previously unreported (Fig. 2). Alternatively, substantial divergence in gene sequence can result in loss of hybridization signal and therefore appear as a deletion on the microarray. The microarray results may indicate a novel 3′ end for tcdB in these strains. Interestingly, the hypervirulent strains have been described as high expressors of toxins A and B. This has been ascribed to a point mutation in tcdC (24). However, this would not be detected by the microarrays used in this study. Table 2). shows the genes absent from all hypervirulent strains (by GACK and McClade analyses), with the exception of BI-14 (HY outlier) and BI-9 (HA1). Given the recent recognition that gene loss or “black holes” may contribute to increased virulence in pathogens (pathoadaptation) (5), these deletions may be significant in terms of the increased virulence of these strains and therefore are worthy of further investigation.
FIG. 2.
Selected gene map on toxin PaLoc (tcdD, tcdB, tcdE, tcdA, and tcdC). A horizontal bar indicates array competitive genomic hybridization of a single strain, and a vertical color bar represents the presence (yellow lines) or absence/high divergence (blue lines) of each gene from CD0659 (tcdD), on the left, to CD0664 (tcdC), on the right. In the clade blocks, dark blue represents strains in the A− B+ clade, light blue represents strains in the HY clade, yellow represents strains in the HA1 clade, and red represents strains in the HA2 clade.
TABLE 2.
HY-specific deletions
| Loci | Deletion(s)/divergent genes |
|---|---|
| CD0630-CD0719 | Methenyltetrahydrofolate cyclohydrolase |
| CD0630-CD0720 | Putative FolD bifunctional protein (includes methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase) |
| CD0630-CD0721 | Conserved hypothetical protein |
| CD0630-CD0722 | Putative methylenetetrahydrofolate reductase |
| CD0630-CD0723 | Putative carbon monoxide dehydrogenase/acetyl coenzyme A synthase complex, dihydrolipoyl dehydrogenase subunit |
| CD0630-CD0724 | Putative carbon monoxide dehydrogenase/acetyl coenzyme A synthase complex, nickel-inserting subunit |
| CD0630-CD1744 | Two-component sensor histidine kinase |
| CD0630-CD1745 | Hypothetical protein |
| CD0630-CD2013 | TetR family transcriptional regulator |
| CD0630-CD2599 | Putative general stress protein |
| CD0630-CD3140 | Putative membrane protein |
| CD0630-CD3144 | Putative transcriptional regulator |
| CD0630-CD3145 | Putative serine-aspartate-rich surface-anchored fibrinogen binding protein |
Close scrutiny of the gene content in this clade suggests some microheterogeneity that may be chronologically significant. It appears that two subgroups were isolated after 2001 (BI-6 onwards). For example, specific fragments of conjugative transposons CTn2 (CD0404-CD0437) and CTn5 (CD1864-CD1868) are present in the U.S. strains BI-6, -6p, -6p2, -7, -8, -10, -12, and -15 as well as R20291 (Stoke Mandeville 027), R20352 (Canadian 027), and R20928 (USA 027) but are absent in BI-1, -2, -3, -4, -5, -9, -11, -14, and -16 and R12087 (Popoff 027). BI-1, -2, -3, -4, and -5 are nonepidemic older U.S. isolates from 1984 to 1993. The role(s) of CTn2 and CTn5 is unproven, but these loci have genes that may encode ABC transporter proteins and CD0434 within CTn2 encodes a protein that has amino acid similarity to MatE, a drug/antiporter protein (12). The potential impact of the presence of these conjugative transposons in hypervirulent strains upon clinical management of patients is unknown. However, prescription of proton pump inhibitors and fluoroquinolone antibiotics has been suggested to exacerbate the CDAD and contribute to the emergence of the hypervirulent strains (25).
Toxin-defective clade.
All 14 A− B+ strains grouped in a tight subclade that was part of a larger clade that included seven other strains, with a subclade of A− B− strains (M3, M13, and M23) and four animal isolates that were more distantly related. The A− B+ strains were from outbreaks of CDAD in Ireland, the United Kingdom, and the United States, again confirming the wide geographical distribution of an epidemic C. difficile clone. Similar observations have been made when other collections of A− B+ strains have been examined by independent typing methods, such as MLST (22). The hypervirulent and A− B+ isolates cluster into two independent highly homogeneous phylogenetic lineages. Taken together, these results suggest a low genetic diversity of the hypervirulent and A− B+ variant strains and of the wide geographical spread of these lineages. Also, all 14 A− B+ strains have a version of CTn5 that lacks CD1864. Table 3 shows a list of genes absent from all A− B+ strains except strain CF5.
TABLE 3.
A− B+-specific deletions (except strain CF5)
| Loci | Deletion(s)/divergent genes |
|---|---|
| CD0630-CD3575 | Putative sodium:solute symporter |
| CD0630-CD3574 | Putative membrane protein |
| CD0630-CD3573 | Hypothetical protein |
| CD0630-CD0654a | Putative ABC transporter, permease protein |
| CD0630-CD0590a | Hypothetical protein |
Absent in CF5.
Human and animal clades.
Two further clades distinct from the hypervirulent and toxin A− B+ strains could be distinguished. One clade designated HA1 had mainly (14/20) human isolates, with a single porcine strain, two equine strains, and a tight subclade of the three murine strains. Human isolates in clade HA1 include toxinotype 0, REA types B1, J9, and K14 that have caused CDAD outbreaks in U.S. hospitals in Minnesota, Illinois, New York, Arizona, Massachusetts, and Florida, and reference strain VPI 10463, a toxinotype 0 hyperproducer of toxins A and B (18; D. N. Gerding, personal communication). The other clade designated HA2 had predominantly (10/14) pig and bovine isolates and four human isolates and a preponderance of ribotype A strains (9/9 with known ribotypes were ribotype A). Analysis of animal isolates showed that bovine (six of seven in HA2 clade) and murine (three of three tightly grouped in HA1 clade) strains were clustered (Fig. 1). By contrast, porcine and equine strains were distributed across three different clades, in many cases mixing with clusters of human isolates. Since the comparative phylogenomic analysis in this study failed to define any host specificity, it could be presumed that animals constitute a source for human infection. CDAD occurs in various forms in domestic animals, but only in the pig has its widespread occurrence been documented; porcine CDAD affects neonates, with significant economic losses (29). Given that C. difficile is not part of the normal human flora, one could argue that strains causing CDAD must ultimately come from some outside source and pork consumption remains a possibility.
Genes/genomic islands that relate to niche adaptation and potential virulence.
Whole-genome comparisons of all 75 strains revealed several loci that are deleted or are highly divergent in several strains that could be important in niche adaptation and potential virulence (Fig. 3). Among these are flagellin-related genes that are likely to be important in motility (Fig. 4). In the 630 genome, two loci encode potential flagellum-associated proteins (CD0226-CD0240 and CD0245-CD0271), between which lies a third interflagellar locus of four genes of unknown function (CD0241-CD0244). All A− B+ strains have retained the three flagellum-associated loci (excluding CD0252-CD0255). The full gene complement was retained in only 7 of the other 62 strains, including three murine (JGS355, JGS356, and JGS360), two equine (JGS692 and JGS6047), and two human (CD1 and T7) strains (Fig. 4). All other strains have lost the first locus (CD0226-CD0240) and interflagellar locus (CD0241-CD0244). All three loci relating to potential flagellin biosynthesis are absent in HA2 strains. These observations on the flagellin gene complements in C. difficile suggest that motility and chemotaxis are unlikely to be essential in the survival and virulence of the organism in the human host.
FIG. 3.
Whole-genome analysis of all 75 strains. A vertical color bar indicates array competitive genomic hybridization of a single strain, and a horizontal line represents the presence (yellow lines) or absence/high divergence (blue lines) of each gene from CD0001 (top) to CD3679 (bottom). Selected genomic islands of interest are labeled at the sides. In the clade blocks, dark blue represents strains in the A− B+ clade, light blue represents strains in the HY clade, yellow represents strains in the HA1 clade, and red represents strains in the HA2 clade.
FIG. 4.
Selected gene map on flagellin-associated genes. A vertical color bar indicates array competitive genomic hybridization of a single strain, and a horizontal line represents the presence (yellow lines) or absence/high divergence (blue lines) of each gene from CD0226 (top) to CD0271 (bottom). F1 indicates flagellar loci CD0226-CD0240, F2 indicates interflagellar loci CD0241-CD0244, and F3 indicates flagellar loci CD0245-CD0271. In the clade blocks, dark blue represents strains in the A− B+ clade, light blue represents strains in the HY clade, yellow represents strains in the HA1 clade, and red represents strains in the HA2 clade.
A 19-gene cluster in C. difficile 630 (CD1906-CD1926) is likely to be involved in ethanolamine degradation and bears several hallmarks of a laterally acquired genomic island; the entire cluster is inserted into and disrupts CD1927, and the last CDS in the cluster encodes a site-specific recombinase (CD1905). This ethanolamine degradation protein is highly similar to those of other enterics, including Salmonella enterica serovar Typhimurium, Yersinia enterocolitica, and Enterobacter faecalis. In this study, the ethanolamine degradation island was completely intact in all strains except the three murine strains and strains AA1 and AA2. It has been suggested that the use of ethanolamine by C. difficile may be important for its anaerobic gastrointestinal lifestyle, since ethanolamine is a carbon and nitrogen source provided by the host's dietary intake. It is unclear why this island appears to have specifically been deselected in the murine and AA strains, but it may reflect niche adaptation.
Antibiotic resistance-related genes.
C. difficile 630 contains 36 potential drug resistance-associated genes, the majority of which are common to all strains tested. However, gene absence generally falls into specific clades. Lantibiotic resistance loci CD0643-CD0646 and CD1349-CD1352 were absent exclusively from all HA2 strains. The putative antibiotic resistance ABC transporter gene that encodes daunorubicin resistance (CD0456) was absent from all HA2 strains and the majority of HA1 strains (except B-one, K14, and JGS692). However, it was present in all hypervirulent strains, all A− B+ strains, and four A− B− strains (CD1, M3, M13, and M23). A candidate streptogramin A acetyltransferase (CD2226) that may encode streptogramin resistance A was present in all strains except BI-9, the outlier in the hypervirulent clade.
Toxin-related genes.
Surprisingly, DNA from all A− B+ strains failed to hybridize with the first two tcdB reporters but did hybridize with all eight tcdA reporters (Fig. 2). Analysis of published tcdB sequence from CF2 (A− B+) (29) identifies numerous point mutations in the region of the first two tcdB reporters. Therefore, the tcdB apparent deletions may be due to sequence divergence beyond the specificity of the microarray. Interestingly, CF2 tcdB was virtually identical to tcdB from C. difficile strain 8864, which has been described as having a 5′ end similar to that of the toxin gene of Clostridium sordellii (3). The explanation for why the A− B+ strains have apparent intact toxin A genes is unknown. However, all eight strains that were classified as A− B− clearly lacked evidence for toxin B (all reporters nonhybridizing) and toxin A (first five reporters nonhybridizing) (Fig. 2). A− B− strains are represented in three of the four clades, suggesting that the absence of toxins is not a feature of clonality and that the PaLoc can readily be lost.
Core gene set.
Using only genes designated present (EPP of 100%), an unusually low core gene content of 19.7% was derived for all 75 strains. Table 4 gives estimates of the core gene set for all of the strains represented in the respective clades. These core genes encode mainly metabolic, biosynthetic, cellular, and regulatory processes. However, many potential virulence determinants are also conserved, indicating that they are indispensable for C. difficile to cause disease in humans. These included genes that are likely to encode a capsule (CD2769-CD2780), a type IV pilus (CD3294-CD3297 and CD3503-CD3513), and fibronectin binding proteins (CD1304 and CD2592).
TABLE 4.
Conserved reportersa
| Clade | No. (%) of genes in core gene set |
|---|---|
| HY | 1,826 (49.0) |
| A− B+ | 1,348 (36.2) |
| HA1 | 1,561 (41.9) |
| HA2 | 1,785 (47.9) |
| All strains | 734 (19.7) |
Of 3,723 microarray reporters.
Comparative phylogenomics.
The comparative phylogenomic method has previously proven to be useful for highlighting potential infection sources and identifying potential virulence determinants in other enteric pathogens (6, 16). In this study, the method confirmed the clonal nature of the hypervirulent and A− B+ strains; it largely validates and complements existing typing methods used for C. difficile, such as toxinotyping, PFGE, REA, PCR ribotyping, and MLST (22). Comparative phylogenomics has a higher discriminatory power than traditional typing methods, and features relevant to strain groupings can be related to gene content.
Given the emergence of hypervirulent strains, the continued use of broad-spectrum antibiotics (including fluoroquinolones), and the rising numbers of immunocompromised and elderly patients, the incidence of CDAD is unlikely to recede. This study is the first genomic microarray comparison of multiple C. difficile strains and, through Bayesian-based algorithms, was able to group strains into four independent clades. This method has also identified many genetic loci that contribute to the formation of each clade, thereby identifying several potential determinants that may help to explain niche adaptation and the differences in pathogenicity observed.
Acknowledgments
We acknowledge BμG@S (the Bacterial Microarray Group at St. George's, University of London) for supplying the 630 microarray and the Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative. We acknowledge Adam Roberts for supplying C. difficile 630 and Michael Gaunt for phylogenetic analysis.
The U.S. Department of Veterans Affairs Research Service provided support for D.N.G. This work was supported by a Medical Research Council grant to B.W.W.
REFERENCES
- 1.Brazier, J. S. 2001. Typing of Clostridium difficile. Clin. Microbiol. Infect. 7:428-431. [DOI] [PubMed] [Google Scholar]
- 2.Champion, O. L., M. W. Gaunt, O. Gundogdu, A. Elmi, A. A. Witney, J. Hinds, N. Dorrell, and B. W. Wren. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. USA 102:16043-16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaves-Olarte, E., P. Low, E. Freer, T. Norlin, M. Weidmann, C. von Eichel-Streiber, and M. Thelestam. 1999. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J. Biol. Chem. 274:11046-11052. [DOI] [PubMed] [Google Scholar]
- 4.Clabots, C. R., S. Johnson, K. M. Bettin, P. A. Mathie, M. E. Mulligan, D. R. Schaberg, L. R. Peterson, and D. N. Gerding. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J. Clin. Microbiol. 31:1870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day, W. A., Jr., R. E. Fernández, and A. T. Maurelli. 2001. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 69:7471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorrell, N., S. J. Hinchliffe, and B. W. Wren. 2005. Comparative phylogenomics of pathogenic bacteria by microarray analysis. Curr. Opin. Microbiol. 8:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fawley, W. N., J. Freeman, and M. H. Wilcox. 2003. Evidence to support the existence of subgroups within the UK epidemic Clostridium difficile strain (PCR ribotype 1). J. Hosp. Infect. 54:74-77. [DOI] [PubMed] [Google Scholar]
- 9.Gal, M., G. Northey, and J. S. Brazier. 2005. A modified pulsed-field gel electrophoresis (PFGE) protocol for subtyping previously non-PFGE typeable isolates of Clostridium difficile polymerase chain reaction ribotype 001. J. Hosp. Infect 61:231-236. [DOI] [PubMed] [Google Scholar]
- 10.Geric, B., R. J. Carman, M. Rupnik, C. W. Genheimer, S. P. Sambol, D. M. Lyerly, D. N. Gerding, and S. Johnson. 2006. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J. Infect. Dis. 193:1143-1150. [DOI] [PubMed] [Google Scholar]
- 11.Geric, B., S. Johnson, D. N. Gerding, M. Grabnar, and M. Rupnik. 2003. Frequency of binary toxin genes among Clostridium difficile strains that do not produce large clostridial toxins. J. Clin. Microbiol. 41:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, G.-X., T. Kuroda, T. Mima, Y. Morita, T. Mizushima, and T. Tsuchiya. 2004. An H+-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 186:262-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinds, J., K. G. Laing, J. A. Mangan, and P. D. Butcher. 2002. Glass slide microarrays for bacterial genomes, p. 83-99. In B. W. Wren and N. Dorrell (ed.), Methods in microbiology: functional microbial genomics. Academic Press, London, United Kingdom.
- 15.Hinds, J., A. A. Witney, and J. K. Vass. 2002. Microarray design for bacterial genomes, p. 67-82. In B. W. Wren and N. Dorrell (ed.), Methods in microbiology: functional microbial genomics. Academic Press, London, United Kingdom.
- 16.Howard, S. L., M. W. Gaunt, J. Hinds, A. A. Witney, R. Stabler, and B. W. Wren. 2006. Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 188:3645-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, S., and D. N. Gerding. 1998. Clostridium difficile-associated diarrhea. Clin. Infect. Dis. 26:1027-1034. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, S., M. H. Samore, K. A. Farrow, G. E. Killgore, F. C. Tenover, D. Lyras, J. I. Rood, P. DeGirolami, A. L. Baltch, M. E. Rafferty, S. M. Pear, and D. N. Gerding. 1999. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 341:1645-1651. [DOI] [PubMed] [Google Scholar]
- 19.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:RESEARCH0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuijper, E. J., S. B. Debast, E. Van Kregten, N. Vaessen, D. W. Notermans, and P. J. van den Broek. 2005. [Clostridium difficile ribotype 027, toxinotype III in The Netherlands]. Ned. Tijdschr. Geneeskd. 149:2087-2089. (In Dutch.) [PubMed] [Google Scholar]
- 21.Kyne, L., M. B. Hamel, R. Polavaram, and C. P. Kelly. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin. Infect. Dis. 34:346-353. [DOI] [PubMed] [Google Scholar]
- 22.Lemee, L., A. Dhalluin, M. Pestel-Caron, J. F. Lemeland, and J. L. Pons. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 42:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442-2449. [DOI] [PubMed] [Google Scholar]
- 24.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433-2441. [DOI] [PubMed] [Google Scholar]
- 25.Pepin, J., N. Saheb, M. A. Coulombe, M. E. Alary, M. P. Corriveau, S. Authier, M. Leblanc, G. Rivard, M. Bettez, V. Primeau, M. Nguyen, C. E. Jacob, and L. Lanthier. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254-1260. [DOI] [PubMed] [Google Scholar]
- 26.Rahmati, A., M. Gal, G. Northey, and J. S. Brazier. 2005. Subtyping of Clostridium difficile polymerase chain reaction (PCR) ribotype 001 by repetitive extragenic palindromic PCR genomic fingerprinting. J. Hosp. Infect. 60:56-60. [DOI] [PubMed] [Google Scholar]
- 27.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 28.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmee. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Songer, J. G. 2004. The emergence of Clostridium difficile as a pathogen of food animals. Anim. Health Res. Rev. 5:321-326. [DOI] [PubMed] [Google Scholar]
- 30.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyokawa, M., A. Ueda, H. Tsukamoto, I. Nishi, M. Horikawa, A. Sunada, and S. Asari. 2003. Pseudomembranous colitis caused by toxin A-negative/toxin B-positive variant strain of Clostridium difficile. J. Infect. Chemother. 9:351-354. [DOI] [PubMed] [Google Scholar]
- 32.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
- 33.Wust, J., N. M. Sullivan, U. Hardegger, and T. D. Wilkins. 1982. Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J. Clin. Microbiol. 16:1096-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]