Abstract
Insertion sequences (ISs) are mobile elements that are commonly found in bacterial genomes. Here, the structural and functional diversity of these mobile elements in the genome of the cyanobacterium Crocosphaera watsonii WH8501 is analyzed. The number, distribution, and diversity of nucleotide and amino acid stretches with similarity to the transposase gene of this IS family suggested that this genome harbors many functional as well as truncated IS fragments. The selection pressure acting on full-length transposase open reading frames of these ISs suggested (i) the occurrence of positive selection and (ii) the presence of one or more positively selected codons. These results were obtained using three data sets of transposase genes from the same IS family that were collected based on the level of amino acid similarity, the presence of an inverted repeat, and the number of sequences in the data sets. Neither recombination nor ribosomal frameshifting, which may interfere with the selection analyses, appeared to be important forces in the transposase gene family. Some positively selected codons were located in a conserved domain, suggesting that these residues are functionally important. The finding that this type of selection acts on IS-carried genes is intriguing, because although ISs have been associated with the adaptation of the bacterial host to new environments, this has typically been attributed to transposition or transformation, thus involving different genomic locations. Intragenic adaptation of IS-carried genes identified here may constitute a novel mechanism associated with bacterial diversification and adaptation.
Insertion sequences (ISs) are common mobile DNA elements of bacterial genomes that do not require extensive DNA homology for transposition. They are believed to undergo horizontal transfer more frequently than other genes, possibly as a consequence of their transfer between the bacterial genome and plasmids (13, 14, 39). The original view was that ISs exhibit a parasitic lifestyle, because they replicate at the expense of the host without delivering any obvious benefit. If so, the host needs to keep the number of IS copies in check. However, both transposition and horizontal transfer of ISs may lead to the acquisition and mobilization of host genes in addition to those of the IS. Moreover, changes in the expression levels of host genes may occur if ISs integrate in regulatory regions. The combination of their capacity to invade many different regions of a bacterial genome and their impact on host genes suggests that IS elements may occasionally also increase host fitness. As a consequence, ISs may be important for bacterial adaptation and niche differentiation (4, 15, 17, 21, 24, 28), among other ways through their potential for orchestrating regulatory circuits (42).
The intragenomic diversity of IS-carried genes such as transposase genes differs substantially from that of other duplicated gene classes in bacterial genomes in that it is typically much lower. Because this difference is evident both for synonymous and for nonsynonymous mutations, it is unlikely to be a consequence of high levels of constraint (47). Genes carried by ISs frequently also comprise a high number of identical intrachromosomal copies, which is readily observable in similarity searches if transposase genes are used as queries. The most logical explanation of these features is high rates of intragenomic transposition and duplication and frequent horizontal transfer (13) coupled with frequent extinctions and invasions of bacterial genomes by ISs (47).
One opportunity to get a better understanding of IS evolution is through examination of the selection pressures that act on IS-encoded proteins. These analyses are still in their infancy, because the majority of the work on ISs derives from the pregenomics era, which did not allow the identification of all ISs in a genome. In addition, bacterial genomes typically span large evolutionary distances, which simply do not allow accurate assessments of the IS dynamics (47). Another cause for the lack of knowledge of the evolution of ISs is that a considerable number of suitably divergent ISs is required for analyses of selection of protein-encoding genes (1). Only a few bacterial genomes contain divergent IS families of sufficient size. The pattern that emerges from a limited number of studies is that different forces may affect different IS families (15, 39). IS copies may experience purifying selection (inefficient and ongoing selection against deleterious substitutions), while other IS families are under positive or adaptive selection (15). Because of rapid protein evolution, the latter outcome is typically interpreted to result from adaptation to a new host or to new environmental challenges experienced by the host. To gain insight into the role of natural selection in the maintenance and evolution of IS elements, we investigated the selection pressure on a large and slightly divergent IS family in the genome of the cyanobacterium Crocosphaera watsonii WH8501. This strain is a member of a novel genus of marine unicellular diazotrophic cyanobacteria with a diameter of 2.5 to 6 μm that occurs in ocean waters warmer than 24°C. Because of its rapid doubling time, it is believed to contribute significantly to oceanic carbon and nitrogen budgets in the tropical oceans.
MATERIALS AND METHODS
Collection of the transposase gene data sets.
Because the C. watsonii genome has not yet been closed, we concatenated all contigs according to the contig number. Subsequently, we identified a large group of similar nucleotide stretches of ∼1.3 kb, which lacked internal stop codons and which are annotated as IS66 transposases. We first identified the possible functional category of these open reading frames (ORFs) using several types of similarity searches and determined the ORF length distribution throughout the genome, the similarity between the stretches, and their frequency in contigs. We used sequences with the GenBank accession numbers ZP_00515223.1 (Cwatdraft_4681) and ZP_00515197 (Cwatdraft_5058) as queries for BLASTX and for PRODOM and PSI-BLAST searches, respectively. Throughout this study, we have numbered sites starting at amino acid position 1 of the query sequences. The sequences identified through BLASTX searches that comprised ORFs of ∼1.3 kb were collected (data set I).
Because transposase genes are part of mobile elements such as ISs and transposons, we identified inverted repeat (IR) sequences in the vicinity of the ORFs using the EMBOSS package (36). By collecting sequences with a conserved IR in the vicinity of the ORFs (while allowing variable numbers of mismatches to the left copy of the IR [IRL]), we identified a large set of sequences that were similar at the nucleotide level. This data set is referred to as data set IA. We subsequently studied the length of the genome stretches between IRs, the identity of genes flanking the ISs, the nature of the mutations (deletions, insertions, and in-frame stop codons), and the relationship between divergence and the genomic position of these ORFs.
Especially when dealing with closely related sequences, the power to detect positive selection strongly depends on the size of the data set (1). To increase the number of sequences in data sets I and IA, we sequenced the ORFs of a living culture of C. watsonii WH8501. To this end, we extracted total DNA (cf. reference 15) and amplified part of the transposase genes (see Results) using primers 5′ AAAACAGTTCAGTCCCCC 3′ and 5′ AGCCAACATCAACACACAGACC 3′ using the QIAGEN HotStarTaq DNA polymerase. This polymerase has no proofreading, but its error rate is sufficiently low for sequencing short portions of genes. Subsequently, we cloned the PCR product into TOPO vectors (Promega Inc.). After picking and boiling clones in 10 μl of water, we used 1 μl to amplify the transposase gene fragment with T7 and T3. The DNA Clean and Concentrator-5 kit (Baseclear; ZY-D4004) was used to purify PCR products. The PCR insert was then sequenced from both directions using ABI sequence kits that use the Big Dye technology. Subsequent sequencing reactions were performed using the ABI PRISM Big Dye terminator v3.1 (Applied Biosystems) using 1 μl Big Dye. Prior to sequence determinations on a four-capillary ABI3100 automated sequencer using the POP7 polymer, sequence products were purified using Sephadex plates (Sephadex G-50 superfine; Amersham) and multiscreen HV (Millipore; MAHVN4510). After elimination of sequences with frameshifts and internal stop codons, the sequences of data set I were added to the new sequence variants (data set II).
Sequence and structural characteristics of multigene transposase ORFs and the IS family.
The sequences of each of the data sets were aligned using ClustalX (46). For identification of protein coding frames and collection of summary statistics, the program DNASP version 4.0 (37) was used. Using this program, we also determined the diversity of the transposase ORF data sets using standard summary statistics such as the number of segregating sites (S), the pairwise number of differences (K), and the number of haplotypes (H). One important tool for illustration of the data, phylogenetic analysis, was performed using PAUP* (45). Optimal nucleotide substitution models for the transposase data sets were identified using Modeltest version 3.06 (34). Tree construction used the likelihood criterion. For reconstruction of the substitutions onto the tree of the transposase genes, the program BASEML of the PAML package (48) was used.
Analyses of selection pressures on transposase genes.
In contrast to data sets I and II, data set IA has only sequences with IRs. If IRs are required for transposition, this data set might comprise a set of functionally divergent ORF variants relative to the ORFs of data sets I and II, which may lack IRs. Because analysis of functional diversification in these data sets might also differ, selection analyses were carried out for all three data sets.
The selection pressure on protein-encoding genes can be measured by comparing nonsynonymous (dN) and synonymous (dS) substitution rates. Under neutrality (nonsynonymous changes have no associated advantage or disadvantage), the expected ratio of dN/dS (or ω) is 1 and significant deviation from this value can be used to identify genes that are either under purifying selection (dN < dS, nonsynonymous changes are deleterious) or under positive selection (dN > dS, nonsynonymous changes are favored because of a fitness advantage). Due to new methods to detect positive selection, the reports of positive selection are increasing rapidly (also in bacteria, e.g., references 3, 16, and 43). By focusing on genes that change rapidly at the amino acid level, which is typically taken to reflect adaptation at the molecular level, these comparative analyses expand the scope for studies of gene functionality relative to the highly constrained genes typically targeted by functional genomics. We calculated the dN/dS ratio using models in the program package PAML version 3.14 (48). We deleted all sequences with gaps and premature and internal stop codons from data sets IA and II (data set I did not contain these types of mutations). Subsequently, we used neutral (M1 and M7) and selection (M2 and M8) models of codon evolution to establish whether positive selection was at hand and, if so, to identify the codons that are under positive selection (29, 48). Models M1 and M7 assume a different distribution of ω values smaller than 1. These models differ from the selection models M2 and M8 in the presence of a class of codons with ω constrained to be larger than 1 (ω2), thereby distinguishing positive selection from purifying evolution (ω < 1), neutral evolution (ω = 1), and positive selection. The fit of the model pairs M1-M2 and M7-M8 can be compared using a χ2 distribution with 2 degrees of freedom. In these model pairs, 9.21 units of difference is required for a significantly better fit of the selection model relative to the neutral model at the 1% level (M1 versus M2 and M7 versus M8). If the fit of the selection models is significantly better than the corresponding neutral models, the selection models can also be used to identify which codons are under positive selection (49).
Because we know nothing about the selection pressures acting on codons of transposase genes, nor of the distribution of ω in these genes, we used an additional method to examine the occurrence of positively selected codons. We also analyzed the three data sets using a conservative method for detection of positively selected codons that is based on the parsimony method of Suzuki and Gojobori (44) and which is implemented as the single likelihood ancestor counting method (31). The parsimony method uses a binomial distribution of parsimoniously inferred synonymous and nonsynonymous substitutions to assess the significance of their numbers. For the reconstruction, only a single tree is used. For the selection analyses, a tree was constructed based on the nucleotide substitution models as inferred from Modeltest (34), followed by likelihood searches. As noted above, we applied these two tests because of the fully unknown dynamics and the forces acting on the bacterial transposase genes of ISs. We assume that the use of multiple tests, which are based on different assumptions, allows a more robust identification of sites under positive selection than does any method used singly.
Ribosomal frameshifting in transposase genes.
In ISs, whose most important component is transposases, ribosomal frameshifting is common (9). This phenomenon, which occurs during translational elongation and entails the shift of a reading frame by mostly a single nucleotide by a ribosome, results in drastically different, mostly shorter, protein sequences. Slippery nucleotide stretches, which typically comprise mononucleotide stretches, may cause high rates of ribosomal frameshifting. For example, approximately 50% of the ribosomes shift frames when encountering the heptamer A AAA AAG in the dnaX gene of Escherichia coli (12). Most other IS-carried genes, however, have a much lower frameshifting efficiency. Because ribosomal frameshifts may dramatically affect the amino acid sequence of a protein and because they are common in bacterial ISs, this mechanism may also affect analysis of codon evolution, in which typically a single reading frame is assumed. As a consequence, it is imperative to identify overlapping ORFs in different frames, to identify sequence stretches which are liable to ribosomal frameshifting during translation, and to assess the presence of secondary structures such as pseudoknots and hairpins that promote frameshifting (27). In the transposase gene family, the localization of these stretches and that of positively selected sites were compared.
Intragenic recombination and gene conversion in the transposase gene family.
Because signatures of positive selection and recombination may be confused (1, 2), we assessed the importance of recombination and attempted to identify the stretches involved in recombination. Two methods were used to evaluate the evidence for recombination in the transposase gene family. First, evidence for gene conversion was assessed using Geneconv version 1.81 (40), which detects whether pairs of sequences share unusually long stretches of similarity (in Geneconv called fragments) given their overall similarity. In this program, two methods are used to assess the significance of putative stretches of gene conversion, a BLAST-like scoring method and a permutation test. Second, we used the program Recombination Detection Program version 2 (RDP2), which in contrast to the sequence similarity criterion which underlies Geneconv employs a criterion for detection of recombination based on phylogenetic incongruence. Specifically, it searches every possible combination of groups of three sequences in the alignment for evidence of recombination based on the similarity between pairs of sequences. Shifts in the affiliation of two sequences relative to a reference sequence are then taken as an indication of recombination. Stretches of nucleotide that may be involved in recombination may be identified using sliding window analyses (25).
Nucleotide sequence accession numbers.
Forty-three partial transposase gene sequences were deposited in GenBank under accession numbers DQ518778 to DQ518820. Of these, 21 were novel gene variants compared to the genomic sequence in the C. watsonii genome.
RESULTS
Affiliations of the duplicated ORFs.
We used BLASTX, PSI-BLAST, and protein domain searches to identify the functional category of the similar genome stretches in the C. watsonii genome. Disregarding self-hits, BLASTX searches suggested links to transposase genes of IS66 of Anabaena variabilis ATCC 29413 and Deinococcus geothermalis DSM 11300 (see file 1 in the supplemental material). Further, among the best hits in PRODOM searches were additional links to the IS66 family. The Position-Specific Iterative (PSI)-BLAST searches, designed to detect weak but biologically relevant sequence similarities, indicated the highest levels of similarity to hypothetical proteins of E. coli. However, this search also found many hits to transposase-like genes from a variety of bacteria (E-values e−105 or higher; not shown). Overall, these findings suggest that the gene family studied here comprises transposase gene copies. It should be noted, however, that all of these relationships are only distant (see file 1 in the supplemental material).
Characteristics of the three transposase data sets. (i) Data set I.
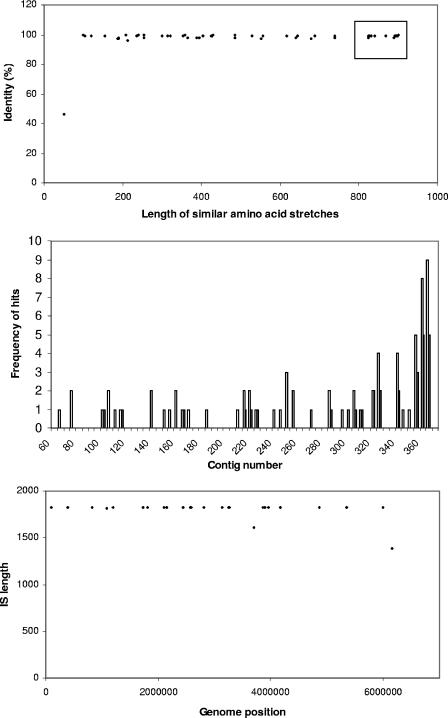
For BLASTX searches of the “nr” database, we used cutoff E-values (3e-07) and scores (50.4 bits) to include short and similar amino acid stretches. Seventy-two amino acid stretches with similarity to the query in the C. watsonii genome were identified. The large number of structural variants indicates that truncation, which occurred at both gene ends, is a major process in the structural diversification of this transposase gene family (Fig. 1, top panel). These genome stretches were strikingly similar as judged from the contrast of the score, which is sensitive to the length of the region of similarity, and the amino acid identity, which is insensitive to hit length. Although most hits were found in the contigs 358 through 362, these contigs are also much longer than the other contigs (totaling 2.20 Mb), and overall the hits were distributed across contigs (Fig. 1, middle panel). The 28 hits that corresponded to long ORFs of ∼1.3 kb are also distributed across contigs (Fig. 1, middle panel). The nucleotide and the protein alignments are available as files 1 and 2, respectively, in the supplemental material.
FIG. 1.
Characteristics of stretches with similarity to the IS family in the C. watsonii genome. The length and similarity of short amino acid stretches (top panel), the frequency of hits of similar amino acid stretches across contigs (middle panel), and the length and distribution across the genome of nucleotide stretches along the genome (bottom panel) are shown.
(ii) Data set IA.
A class of highly conserved IRs of 24 bases was found in the vicinity of many of the transposase genes. The sequence of IRL was 5′ GTAACTGCTCACCGCAACAAAAAA 3′, mostly with three mismatches to the IRR (5′ TTTTTTGTTGCGGTGAGCAGTTAC 3′). Genome sequences between IRL and IRR that varied between 200 and 3,000 bases and that had variable numbers of mismatches to the consensus IRL sequence were retrieved. Using three and four and using five and six mismatches gave 25 and 26 hits, respectively. Most of these matches ranged between 1,821 and 1,825 bases, and also these were distributed across the C. watsonii genome (Fig. 1, bottom panel). Alignment of the sequences between IRL and IRR identified two copies with length mutations, one copy with a 1-bp frame-disrupting insertion and one copy with an internal stop codon, leaving 21 full-length ORF sequences with an IR (data set IA). Examination of the nucleotide stretches next to the IRL and IRR indicated that the insertion sites of these ISs were not conserved (not shown).
(iii) Data set II.
By using a portion of the IS66 gene (nucleotide positions 41 through 657 of data set I), 21 additional sequence variants of the IS66 transposase gene were collected. These were added to data set I (resulting in 49 transposase gene variants; data set II). The positions of the variable nucleotide positions in the C. watsonii genome correspond to the mutations seen in the additional sequences. This, in combination with the low error rate of the DNA polymerase, suggests that the mutations seen in the additional sequence are genuine.
Analyses of selection pressure on the transposase family.
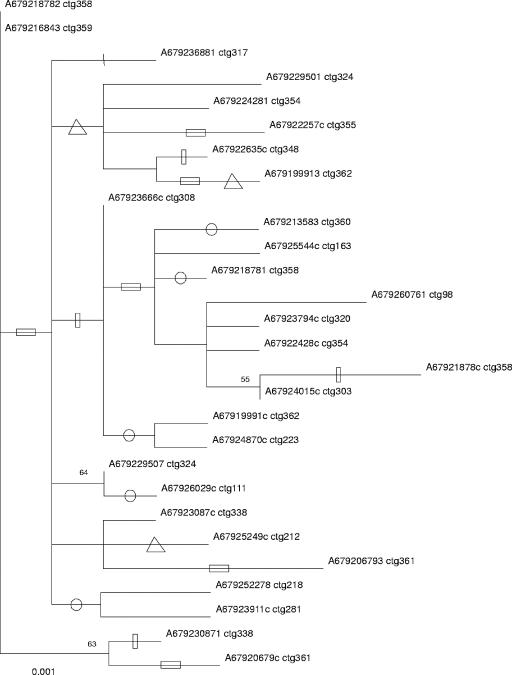
As noted before, if the IR is required for the functioning of the IS or the transposase gene, the results of selection analyses of data sets I and II may differ from those of data set IA. Therefore, we conducted selection analyses using the PAML package using all data sets. All data sets comprised contiguous ORFs, without insertions or deletions of codons. The data sets had very similar levels of diversity (Table 1). Phylogenetic trees reconstructed from closely related sequence data sets lacked strongly supported internodes in that bootstrap support was always lower than 90% (tree shown only for data set I; Fig. 2).
TABLE 1.
Summary of statistics of the three IS66 data sets of C. watsonii
| Data set | Content | No. of sequences | Sequence length (bases) | No. of segregating sites | Avg pairwise no. of differences | No. of haplotypes |
|---|---|---|---|---|---|---|
| I | Transposase ORFs based on BLAST searches | 28 | 1,305 | 43 | 5.06 | 27 |
| IA | Transposase ORFs between IRL and IRR | 21 | 1,299 | 28 | 4.82 | 21 |
| II | Transposase ORFs expanded | 49 | 606 | 37 | 3.36 | 49 |
FIG. 2.
Maximum likelihood phylogram based on 28 IS66 ORF sequences. A single tree was found (ln = −2,211.39). The accession numbers and contig numbers are indicated. The four positive selected codons are plotted onto the tree using the Baseml program. A vertical line (nucleotide position 70) and an open vertical bar (position 71) mark codon 24. An open circle marks changes in codon 62. An open horizontal bar marks changes of codon 128. An open triangle marks changes at codon 184.
Analyses of the selection pressures of the three data sets of transposase gene variants in the IS provided strong evidence for positive selection based on the comparison of neutral and selection models and Bayesian site identification in PAML (Table 2). All PAML analyses indicated significantly better fits for the selection models M2 and M8 than for the neutral models M1 and M7, respectively (Table 2). Furthermore, the codons under positive selection were largely identical across model comparisons M2 and M8, albeit that overlapping subsets of positively selected codons were identified in the different data sets. In data set I, evidence for positive selection is found for four codons under M2 (24, 62, 128, and 184) and the same four sites under M8. Although data sets IA and II gave overlapping sets of positively selected sites, all three data sets agreed on two positively selected sites (codons 24 and 128). Each of these codons had strong evidence for positive selection according to the Bayes empirical Bayes estimates (ω2 estimates in Table 2). Interestingly, codon 128 (and also codon 184, which was indicated as positively selected in a substantial number of analyses) is located in a conserved domain typical of transposase genes (Fig. 3; the transposase_25 domain). Because the conserved domain database contains “building blocks” that are believed to modulate protein function, the presence of positively selected codons in these domains suggests that (mutations at) these codons are functionally important. The conserved domain transposase_25 is found predominantly in proteobacteria (e.g., the entries in Fig. 3 are from Agrobacterium tumefaciens, Rhizobium sp. strain NGR234, E. coli L0015, Pseudomonas putida TF4-IL, and Rhodobacter capsulatus).
TABLE 2.
Tests of positive selection and positively selected codons in the three transposase data sets of C. watsonii according to neutral models (M1 and M7) and selection models (M2 and M8) of PAML
| Model | Data set | Likelihooda | Tree lengthb | Kappa | Parametersc,d | Codon | Amino acide | Probability for positive selection (BEB)f | dN/dS codon and SE |
|---|---|---|---|---|---|---|---|---|---|
| M1 | I | 2,230.86 | 0.13 | 1.27 | ω0 = 0.00, ω1 = 1.00, p0 = 0.67, p1 = 0.33 | ||||
| IA | 2,129.05 | 0.11 | 1.1 | ω0 = 0.00, ω1 = 1.00, p0 = 0.69, p1 = 0.31 | |||||
| II | 1,315.87 | 0.32 | 2.32 | ω0 = 0.00, ω1 = 1.00, p0 = 0.54, p1 = 0.46 | |||||
| M2 | I | 2,196.17 | 0.14 | 1.91 | ω0 = 0.67, ω1 = 1.00, ω2 = 53.90, p0 = 0.98, p1 = 0.00, p2 = 0.02 | 24 | P | 1.00 | 10.13 ± 0.68 |
| 62 | P | 1.00 | 10.12 ± 0.73 | ||||||
| 128 | S | 1.00 | 10.13 ± 0.67 | ||||||
| 184 | K | 0.99 | 10.02 ± 1.20 | ||||||
| IA | 2,092.65 | 0.12 | 1.75 | ω0 = 0.75, ω1 = 1.00, ω2 = 71.43, p0 = 0.97, p1 = 0.00, p2 = 0.03 | 24 | P | 1.00 | 10.21 ± 0.58 | |
| 47 | P | 0.97 | 9.96 ± 1.61 | ||||||
| 62 | T | 1.00 | 10.18 ± 0.73 | ||||||
| 128 | R | 1.00 | 10.21 ± 0.58 | ||||||
| 136 | V | 0.98 | 9.99 ± 1.53 | ||||||
| 259 | M | 1.00 | 10.18 ± 0.75 | ||||||
| 305 | A | 0.97 | 9.95 ± 1.62 | ||||||
| 421 | F | 1.00 | 10.18 ± 0.75 | ||||||
| II | 1,289.29 | 0.33 | 3.14 | ω0 = 0.77, ω1 = 1.00, ω2 = 29.41, p0 = 0.98, p1 = 0.00, p2 = 0.02 | 24 | P | 1.00 | 9.98 ± 0.81 | |
| 128 | R | 1.00 | 9.98 ± 0.82 | ||||||
| 137 | V | 0.99 | 9.72 ± 1.70 | ||||||
| M7 | I | 2,230.93 | 0.13 | 1.23 | B(p = 0.01, q = 0.01) | ||||
| IA | 2,130.19 | 0.11 | 0.99 | B(p = 0.005, q = 0.02) | |||||
| II | 1,316.12 | 0.32 | 2.27 | B(p = 0.02, q = 0.02) | |||||
| M8 | I | 2,196.17 | 0.14 | 1.91 | B(p = 99, q = 47.61), p0 = 0.98, p1 = 0.02, ω2 = 53.92 | 24 | P | 1.00 | 10.12 ± 0.70 |
| 62 | P | 1.00 | 10.12 ± 0.73 | ||||||
| 128 | S | 1.00 | 10.12 ± 0.70 | ||||||
| 184 | K | 0.99 | 10.04 ± 1.11 | ||||||
| IA | 2,092.66 | 0.12 | 1.75 | B(p = 0.02, q = 0.01), p0 = 0.97, p1 = 0.03, ω2 = 71.96 | 24 | P | 1.00 | 10.21 ± 0.57 | |
| 47 | P | 0.99 | 10.09 ± 1.22 | ||||||
| 62 | T | 1.00 | 10.20 ± 0.64 | ||||||
| 128 | R | 1.00 | 10.21 ± 0.57 | ||||||
| 136 | V | 0.99 | 10.10 ± 1.16 | ||||||
| 259 | M | 1.00 | 10.20 ± 0.65 | ||||||
| 305 | A | 0.99 | 10.09 ± 1.23 | ||||||
| 421 | F | 1.00 | 10.20 ± 0.64 | ||||||
| II | 1,289.33 | 0.33 | 3.11 | B(p = 0.02, q = 0.02), p0 = 0.98, p1 = 0.02, ω2 = 20.83 | 24 | P | 1.00 | 9.96 ± 0.83 | |
| 128 | R | 1.00 | 9.96 ± 0.83 | ||||||
| 137 | V | 0.98 | 9.80 ± 1.47 | ||||||
| 184 | Q | 0.95 | 9.55 ± 2.07 |
The likelihood indicates the fit of the data to the different models.
Tree length is measured as the number of mutations per codon.
Kappa is the transversion/transition ratio. Pi denotes the proportion of sites falling in site class ωi.
Parameters p and q are the shape parameters of the beta distribution which underlies M7 and M8.
The reference sequence for the amino acid designation is ZP_00519072.
The probability that codons were under positive selection was determined using Bayes empirical Bayes (BEB), with the ω and its standard error indicated per codon.
FIG. 3.
Conserved domain in the IS66 transposase ORFs. Asterisks mark positively selected amino acids corresponding to codons 128 and 184. The other two positively selected codons are not within the conserved domain. Numbers in brackets indicate the number of amino acids in length-variable regions. The sequence marked 679213 is an IS66 transposase gene copy from contig 360; the other sequences are from the conserved domain transposase_25.
The conservative parsimony method of Suzuki and Gojobori (44) found one codon under positive selection (codon 24 in data set II; P = 0.04), and this codon was also found to be under positive selection by the maximum likelihood method. No positively selected sites were found in data sets I and IA using this method (not shown). This is not surprising, because the parsimony method is generally considered to be conservative and unlikely to detect sites under positive selection in relatively small data sets of a few dozen sequences (1). This holds true especially when dealing with closely related sequences (Table 1) (32). In fact, the finding of a positively selected codon using the parsimony method of Suzuki and Gojobori in this closely related IS data set suggests that the results of the PAML analyses (Table 2) are not methodological artifacts. The 27 haplotypes in data set I are due disproportionately to the four positively selected codons, which give rise to 13 haplotypes. The other 14 haplotypes are due to the remaining 31 variable codons.
Recombination in the transposase genes of C. watsonii.
Because recombination can cause signatures reminiscent of those of selection (1, 2), we assessed the importance of recombination in the transposase data sets of C. watsonii. Geneconv detected no significant pairwise fragments, nor significant inner or outer global fragments (not shown). Recombination leads to conflicting phylogenetic relationships among gene stretches. However, there was no evidence of incongruent phylogenetic relationships among groups of three sequences in the transposase data set, and gene regions indicative of recombination also were missing according to this criterion (not shown). These results suggest that there is little evidence for a role of recombination or gene conversion in the IS66 gene family.
Ribosomal frameshifting.
There are two reasons for examining ribosomal frameshifting in the transposase ORFs. The first is due to the potential impact of frameshifting on selection analyses of protein-encoding genes. Ribosomal frameshifts impact the analyses of selection pressures, because a recoding of synonymous and nonsynonymous mutations is required. The alternative transposase reading frames in the C. watsonii genome are devoid of internal stop codons (34 in −1 frame, 32 in +1 frame), and as a consequence, there is little room for generating large proteins using overlapping reading frames. Second, the location of positively selected sites may be associated with slippery sequence stretches or with stabilizing secondary structures such as hairpin-loop stems that are common in ISs. Although frameshifting mechanisms identified in ISs are notoriously heterogeneous, we therefore attempted to identify the slippery sequence stretches and secondary structures in the transposase ORFs. The number of slippery sequences in the transposase ORFs is extremely high as judged from the distribution of these stretches in random sequences of the same length as the full-length ORF (27). Typically, none or a single slippery stretch is present in random sequences (not shown), whereas six were found in the ORF variants studied (Fig. 4). Consistent with the literature on retroviruses, animal and plant viruses, retrotransposons, bacterial genes, bacteriophage genes, and bacterial insertion sequences the frameshifting stretches were all of the most common −1 type (7). The secondary structure of hairpin-loop stems involves a hairpin whose single-stranded looping region pairs with upstream DNA stretches. This additional stem is separated from the first hairpin by a spacer region of variable length. Of these six slippery stretches, four had stabilizing hairpin-loop stems (their putative location involves the underlined residues in Fig. 4). In the hairpin-loop stem structures of the C. watsonii transposase genes, the spacer regions range in length from 4 to 10 bases. The stem of the hairpins was 4 or 5 nucleotides long. In the C. watsonii genome, the slippery heptamers are distributed along the IS66 genes and only one slippery sequence stretch and one hairpin stem-loop were associated with a positively selected codon (Fig. 3; slippery stretch starting from nucleotide 183, codon 62; boxed in Fig. 4). Thus, the overlap of the location of the positively selected and slippery stretches and secondary structural elements was only marginal. In addition, there are no obvious overlapping ORFs that could generate proteins of substantial length. Although this may be taken to suggest that ribosomal frameshifting is unlikely to affect the selection analyses, different parts of transposases may serve different functions (8). For most ISs identified in bacterial genomes, it is currently unknown what function truncated protein variants could serve.
FIG. 4.
Overview of the IS66 transposase gene of C. watsonii WH8 using sequence A679218782 (contig 358) as a reference. Indicated are the locations of slippery sequences (bold and underlined), secondary downstream structures (underlined), synonymous substitutions (above the nucleotide sequence), nonsynonymous substitutions (below the amino acid sequence), and the four positively selected codons according to the PAML analysis (boxed codons) of Table 2. The location of synonymous substitutions is marked by an asterisk, and the frequency of alternative nucleotides is indicated. The frequency of the least frequent amino acid is indicated, together with the number of changes on the gene tree in parentheses (Fig. 2). Alternative amino acids at a single position are indicated by a slash. One overlapping slippery sequence and a positively selected codon were found (codon 62).
DISCUSSION
Summary of results.
Multiple methods were used to support the inference of positive selection and the location of positively selected codons in the IS66 transposase genes of C. watsonii. Both conservative and more powerful methods detected one or more codons under positive selection (Table 2). Also other methods (26, 33) consistently found several positively selected codons including codons 24 and 128 (not shown). These latter methods are not so prone to false positives as the PAML methods but are more powerful than the parsimony method (32). Therefore, the selection analyses support the occurrence of positive selection on these transposase genes. Tests of recombination and gene conversion indicate that these processes do not likely affect the inference of positive selection in the IS66 family. This is because of the high ω2 estimates in the IS66 data sets (Table 2) (1). In addition, if recombination is present, it most likely involves only low rates. Under these conditions, the PAML analyses are still trustworthy (1). Apart from these direct tests, other attributes also deny a major role for recombination and gene conversion in the evolution of the IS66 family. First, the genes are spread over the C. watsonii genome and this conformation is less likely to undergo gene conversion relative to tandemly duplicated genes, at least in eukaryotes (7). If gene conversion is mechanistically distinct in prokaryotes, which is possible (38), then highly expressed genes such as ribosomal protein-encoding genes could use gene conversion to slow down the accumulation of deleterious mutations in their gene products (47). However, expression levels of transposase genes are notoriously low (19) and an adaptive explanation for the occurrence of gene conversion in transposase genes is lacking. In sum, there are few grounds to invoke a role for recombination or gene conversion in the evolution of this IS66 transposase gene family.
IS function and host adaptation.
The numbers of transposase ORFs and intact ISs in the C. watsonii genome suggest that it is one of the larger IS families with divergent gene variants. Does this imply that this genome is saturated with ISs or that mutational mechanisms may differ from those in smaller IS families? The finding that conserved target sequences of the IS66 family are lacking (see Results) and the fact that ISs are generally not very selective in their target sites (23) suggest that a saturation of integration sites is unlikely. Distinct target site specificities and transposition mechanisms have been invoked to explain the different selective forces acting on different IS families (15). Our results support the finding that at least some IS families are under adaptive forms of natural selection (see above; 15). This type of selection requires an explanation either in terms of increased host fitness associated with IS activity or in terms of interactions among IS copies within the same genome. Typically, positive selection is invoked when ISs adapt to the host through the rapid evolution of new divergent alleles or gene variants. This may be mediated by the host proteins required for transposition, which were shown to differ among IS classes (5). However, it should be stressed that if IS evolution is related to host fitness, one would not necessarily expect positively selected codons among closely related ISs from a single genome. This is because typically only a single mutant IS copy is involved in host adaptation (cf. reference 41). As such, an increase of copy number of the beneficial IS variant and an increase of divergence among IS copies subsequent to transposition also do not require positive selection per se. An alternative explanation is that the rate of transposition is controlled, for example, by adjustment of the number of different transposase gene variants that differ in transposition activities. Although this is not a sufficient explanation for the diversity of IS families either (see below), it agrees, however, with the observation that single amino acid substitutions can increase transposition activities of a number of ISs (5, 6, 35), with the possibility that mRNA structure may influence expression of transposase genes (18, 24) and with the observation that adapting codons may be located in conserved, and thus functionally important, domains of the transposase genes (Fig. 3). In addition, truncated portions of transposases may serve different functions (e.g., reference 8). Each of these processes may underlie the positive selection detected here.
Although knowledge of the interplay of these regulatory mechanisms and of the age of different IS families is lacking, these features probably differ substantially among IS families. It is important, however, to realize that these aspects of IS evolution are probably not sufficient to explain the diversity patterns of IS families as found in bacterial chromosomes. If transposase gene families have been large for prolonged periods of time, higher levels of divergence than typically observed for IS genes are expected, even if down-regulation of transposition activity occurs (47). Instead, frequent extinctions and invasions of bacterial genomes are an essential component to explain the low levels of intragenomic diversity of ISs in bacterial genomes (47).
In spite of their generally rapid turnover and low levels of diversity, some IS-carried gene families diversify and adapt at the codon level. Examination of transposase gene families will allow us to explore the functional divergence of prokaryotic genes associated with gene duplications. Because duplicated gene classes other than transposases and integrases are generally rare in bacterial genomes (10, 22, 30), their use as targets for studies of functional gene differentiation may complement similar studies in eukaryotes (11). Because of their distinctiveness in terms of diversity and dynamics, transposase genes are frequently treated as a distinct gene class and are commonly excluded from whole-genome analyses (e.g., reference 10). Nevertheless, their role in bacterial adaptation and ecological specialization (20) and the development of new theories that stress the adaptive potential of duplicated genes (11) suggest that these gene families are suitable targets for comparative and functional genomics.
Supplementary Material
Acknowledgments
We thank Beate Averhoff (University of Frankfurt) and Lucas J. Stal (NIOO-CEME) for comments on a previous draft of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Publication 3896 of NIOO-KNAW.
REFERENCES
- 1.Anisimova, M., R. Nielsen, and Z. H. Yang. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164:1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anisimova, M., and Z. H. Yang. 2004. Molecular evolution of the hepatitis delta virus antigen gene: Recombination or positive selection? J. Mol. Evol. 59:815-826. [DOI] [PubMed] [Google Scholar]
- 3.Ansaldi, M., and D. Dubnau. 2004. Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the comX pheromone. J. Bacteriol. 186:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, D. E., and C. M. Berg. 1983. The prokaryotic transposable element Tn5. Bio/Technology 1:417-435. [Google Scholar]
- 5.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 6.Derbyshire, K. M., and N. D. Grindley. 1996. Cis preference of the IS903 transposase is mediated by a combination of transposase instability and inefficient translation. Mol. Microbiol. 21:1261-1272. [DOI] [PubMed] [Google Scholar]
- 7.Drouin, G. 2002. Characterization of the gene conversions between the multigene family members of the yeast genome. J. Mol. Evol. 55:14-23. [DOI] [PubMed] [Google Scholar]
- 8.Duval-Valentin, G., C. Normand, V. Khemici, B. Marty, and M. Chandler. 2001. Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J. 20:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farabaugh, P. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60:103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevers, D., K. Vandepoele, C. Simillon, and Y. van de Peer. 2004. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 12:148-154. [DOI] [PubMed] [Google Scholar]
- 11.Gu, Z. L., L. M. Steinmetz, X. Gu, C. Scharfe, R. W. Davis, and W.-H. Li. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421:63-66. [DOI] [PubMed] [Google Scholar]
- 12.Gurvich, O. L., P. V. Baranov, J. Zhou, A. W. Hammer, R. F. Gesteland, and J. F. Atkins. 2003. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 22:5941-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartl, D. L., and S. A. Sawyer. 1988. Multiple correlations among insertion sequences in the genome of natural isolates of Escherichia coli, p. 91-106. In A. J. Kingsman, S. M. Kingsman, and K. F. Chater (ed.), Transposition. Cambridge University Press, Cambridge, United Kingdom.
- 14.Hartl, D. L., and S. A. Sawyer. 1988. Why do unrelated insertion sequences occur together in the genome of Escherichia coli? Genetics 118:537-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalia, A., A. K. Mukhopadhyay, G. Dailide, Y. Ito, T. Azuma, B. C. Y. Wong, and D. E. Berg. 2004. Evolutionary dynamics of insertion sequences in Helicobacter pylori. J. Bacteriol. 186:7508-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalia, A., and D. E. Bessen. 2004. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J. Bacteriol. 186:110-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidwell, M. G., and D. R. Lisch. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1-24. [DOI] [PubMed] [Google Scholar]
- 18.Kleckner, N. 1989. Transposon Tn10, p. 211-226. In D. Berg and M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 19.Kleckner, N. 1990. Regulating Tn10 and IS10 transposition. Genetics 124:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konstantinidis, K. T., and J. M. Tiedje. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 101:3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence, J. G., H. Ochmann, and D. Hartl. 1992. The evolution of insertion sequences within enteric bacteria. Genetics 131:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerat, E., V. Daubin, H. Ochman, and N. A. Moran. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3:807-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahillon, J., C. Léonard, and M. Chandler. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. 150:675-687. [DOI] [PubMed] [Google Scholar]
- 25.Martin, D., and E. Rybicki. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562-563. [DOI] [PubMed] [Google Scholar]
- 26.Massingham, T., and N. Goldman. 2005. Detecting amino acid sites under positive selection and purifying selection. Genetics 169:1753-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon, S., Y. Byun, H.-J. Kim, S. Jeong, and K. Han. 2004. Predicting genes expressed via −1 and +1 frameshifts. Nucleic Acids Res. 32:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy, Z., and M. Chandler. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387-398. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen, R., and Z. H. Yang. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal, C., B. Papp, and M. J. Lercher. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 37:1372-1375. [DOI] [PubMed] [Google Scholar]
- 31.Pond, S. L. K., and S. D. W. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 32.Pond, S. L. K., and S. D. W. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 33.Pond, S. L. K., and S. V. Muse. 2005. Site-to-site variation of synonymous substitution rates. Mol. Biol. Evol. 22:2375-2385. [DOI] [PubMed] [Google Scholar]
- 34.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 35.Reznikoff, W. S. 2003. Tn5 as a model for understanding DNA transposition. Mol. Microbiol. 47:1199-1206. [DOI] [PubMed] [Google Scholar]
- 36.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: The European molecular biology open software suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 37.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 38.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed] [Google Scholar]
- 39.Sawyer, S., D. Dykhuizen, R. DuBose, L. Green, T. Mutangadura-Mhlanga, D. Wolczyk, and D. Hartl. 1987. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics 115:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawyer, S. A. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 41.Schlenke, T. A., and D. J. Begun. 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101:1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro, J. A. 1999. Transposable elements as the key to a 21st century view of evolution. Genetica 107:171-179. [PubMed] [Google Scholar]
- 43.Smith, E. E., E. H. Sims, D. H. Spencer, R. Kaul, and M. V. Olson. 2005. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeroginosa. J. Bacteriol. 187:2138-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, Y., and T. Gojobori. 1999. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 16:1315-1328. [DOI] [PubMed] [Google Scholar]
- 45.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Mass.
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, A. 2006. Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variability in multiple genomes. Mol. Biol. Evol. 23:723-733. [DOI] [PubMed] [Google Scholar]
- 48.Yang, Z., R. Nielsen, N. Goldman, and A.-M. K. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, Z. H., W. S. W. Wong, and R. Nielsen. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107-1118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.