Abstract
Expression of the Streptomyces exfoliatus lipA gene, which encodes an extracellular lipase, depends on LipR, a transcriptional activator that belongs to the STAND class of P-loop nucleoside triphosphatases. LipR is closely related to activators present in some antibiotic biosynthesis clusters of actinomycetes, forming the LipR/TchG family of regulators. In this work we showed that purified LipR protein is essential for activation of lipA transcription in vitro and that this transcription depends on the presence of a conserved inverted repeat, the LipR box, located upstream of the lipA promoter. Mutagenesis of the lipA promoter region indicated that most transcription depends on LipR binding to the proximal half-site of the LipR box in close proximity to the −35 region of the promoter. Our experiments also indicated that LipR establishes contact with the RNA polymerase on both sides of the LipR box, since some activation was observed when only the distal half-site was present or when the entire LipR box was moved further upstream. We also showed that the LipR proteins of S. exfoliatus and Streptomyces coelicolor are functionally interchangeable both in vitro and in vivo, revealing the functional conservation of the regulatory elements in these two species.
Gram-positive bacteria belonging to the genus Streptomyces are sporulating soil bacteria with a mycelial growth habit that undergo complex morphological and physiological differentiation (2, 4). Streptomycetes have a remarkable capacity for synthesis of secondary metabolites, which are usually produced in a growth phase-dependent manner; in liquid batch culture, synthesis of antibiotics and other secondary metabolites usually starts at the onset of the stationary growth phase (2).
Lipases (triacylglycerol acylhydrolases [EC 3.1.1.3]) are an important group of enzymes with many industrial applications (32). A survey of lipase-producing microorganisms from soil showed that members of the genus Streptomyces can be highly lipolytic (27), and different lipase genes have been cloned from members of this genus. We have previously described the lipase genes of Streptomyces exfoliatus (18), Streptomyces albus (5), and Streptomyces coelicolor (30) and have shown that these highly similar lipases are related to a psychrophilic lipase from Moraxella sp. strain TA144 (9) and to human platelet-activating factor acetylhydrolase II (33). More recently, it has been shown that two hydrolases from Acidovorax delafieldii and Thermobifida fusca capable of degrading synthetic polyesters exhibit high levels of similarity to these Streptomyces lipases (13, 29). Other lipase-encoding genes have been cloned from Streptomyces species, including the gene for a Streptomyces cinnamomeus lipase related to Pseudomonas lipases (26) and a Streptomyces rimosus gene encoding an enzyme representing a third lipase family of Streptomyces (31).
In previous work we showed that transcription of the S. exfoliatus lipase gene, lipA, occurred from a single promoter, which was preceded by an inverted repeat (18); transcription was completely dependent on the product of a downstream gene, which we designated lipR as it appeared to encode a transcriptional activator (24). Similar organization was observed in S. coelicolor and S. albus, including the presence of homologous lipR genes, well-conserved −10 and −35 regions, and a well-conserved inverted repeat; this prompted us to suggest that the conserved inverted repeat could function as the target site for the LipR regulators (30).
The observed similarity of the LipR proteins to other bacterial transcriptional regulators led to a proposal for a novel protein family, described as the MalT or LAL family of bacterial regulators (7, 30). The features distinguishing these proteins were a large size (typically around 900 amino acids), a conserved C-terminal LuxR-type DNA-binding domain, and the presence of conserved Walker A and Walker B motifs in the N-terminal region (7). More recently, these proteins have been shown to be part of the much larger STAND class of P-loop nucleoside triphosphatases (15). Detailed phylogenetic analysis has shown that the bacterial regulators previously described as members of the LAL family are members of separate families of the STAND class. The LipR proteins are more closely related to activators encoded in some polyketide biosynthetic gene clusters of actinomycetes than to MalT, constituting the LipR/TchG family of actinobacterial proteins; MalT-related proteins, on the other hand, are prevalent in the γ-proteobacteria, although some are also present in other bacterial groups, including the actinomycetes (15). Even though there is experimental evidence indicating that members of the LipR/TchG family are transcriptional activators, so far there have been no studies in which the mechanism by which these proteins activate transcription has been analyzed.
In this paper we examine the importance of the conserved inverted repeat upstream of the S. exfoliatus lipA promoter for transcriptional activation by LipR, a member of the LipR/TchG family of actinobacterial regulators. We show that the half-site close to the −35 region is essential for activation but that full activation also requires the upstream half-site, strongly suggesting that LipR establishes specific contact with the RNA polymerase molecule on both sides of the inverted repeat. We also show that the S. exfoliatus and S. coelicolor lipA genes can be activated by the LipR protein from either species, which indicates the functional conservation of elements required for regulation of the lipase genes in these two species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and microbiological procedures.
The Escherichia coli strains used were JM101, XL1-Blue, and BL21(DE3) Rosetta 2 (Novagen). These strains were grown and transformed by standard methods (22). Wild-type Streptomyces lividans 66 (strain 1326 from the John Innes Centre collection) was used throughout this study. Transformation of Streptomyces strains with plasmid DNA was carried out as described previously (12), except that protoplasts were plated in a hypertonic soft agar overlay (16). The plasmids used are listed in Table 1.
TABLE 1.
Plasmids and oligonucleotides used in this study
| Plasmid or oligonucleotide | Description and/or sequence | Reference or source |
|---|---|---|
| Plasmids | ||
| pB13 | Original clone carrying the S. exfoliatus lipA and lipR genes in pIJ486 | 18 |
| pB17 | Original clone carrying the S. exfoliatus lipA and lipR genes; independent from pB13 | 18 |
| pB48 | pJV1-derived, Hygr cloning vector | 12 |
| pB92 | HindIII-NcoI fragment from pB13 cloned in pUCBM21 (Boehringer); carries lipA and first half of lipR | This study |
| pB94 | pIJ486 derivative carrying S. exfoliatus lipA and lipR genes | 24 |
| pB99 | pIJ6021 carrying S. exfoliatus lipR modified by introduction of an NdeI site at the start codon | 24 |
| pB100 | S. exfoliatus lipA gene and promoter region cloned in Hygr pJV1-derived vector | 24 |
| pB150 | pIJ6021 carrying S. coelicolor lipR modified by introduction of an NdeI site at the start codon | This study |
| pB151 | S. coelicolor lipA gene and promoter region cloned in Hygr pJV1-derived vector | This study |
| pB606 | S. exfoliatus lipR gene from pB99 cloned in the expression vector pET28a (Novagen) | This study |
| pBZ619 | Derivative of pB94 with deletion of insert DNA upstream of position −83; carries the inverted repeat of the lipA promoter region | This study |
| pBZ621 | Derivative of pB94 with deletion of insert DNA upstream of position −52; carries only the proximal half-site of the inverted repeat of the lipA promoter region | This study |
| pBZ623 | Derivative of pB94 with deletion of insert DNA upstream of −35 region of the promoter; lacks the inverted repeat of the lipA promoter region | This study |
| pBZ662 | Derivative of pB94 in which the sequence of the proximal half-site of the repeat has been modified | This study |
| pBZ663 | Derivative of pBZ619 with a 5-bp insertion between the two half-sites of the inverted repeat | This study |
| pBZ667 | Derivative of pBZ621 with an 11-bp insertion between the −35 region of the lipA promoter and the inverted repeat | This study |
| pBZ670 | S. coelicolor lipR gene from pB150 cloned in the expression vector pET28a (Novagen) | |
| pBZ687 | Derivative of pBZ621 with a 5-bp insertion between the −35 region of the lipA promoter and the inverted repeat | This study |
| pBZ688 | Derivative of pBZ621 with a 10-bp insertion between the −35 region of the lipA promoter and the inverted repeat | This study |
| pIJ6021 | pIJ101-derived Kanr expression vector carrying inducible tipA promoter | 28 |
| Oligonucleotides | ||
| DUA1050PLIPA | CCGGTCCGCTGAGGATCTAGATCCCTTTCGGCG; used for construction of pBZ619a | This study |
| DUB2050PLIPA | CGCCGAAAGGGATCTAGATCCTCAGCGGACCGG; used for construction of pBZ619 | This study |
| DPDA1090PLIPA | CGCCGGTCCGTCTAGACCGGTGATACCGC; used for construction of pBZ621 | This study |
| DPDB2090PLIPA | GCGGTATCACCGGTCTAGACGGACCGGCG; used for construction of pBZ621 | This study |
| DP1A137PLIPA | GACCGGTGATACTCTAGATGCGCGTCCACGGG; used for construction of pBZ623 | This study |
| DP2B137PLIPA | CCCGTGGACGCGCATCTAGAGTATCACCGGTC; used for construction of pBZ623 | This study |
| MUTREPEDER1 | CCGGTCCGTCAAGGAATTCCGTACTCTAGATGCG; used for construction of pBZ662 | This study |
| MUTREPEDER2 | CGCATCTAGAGTACGGAATTCCTTGACGGACCGG; used for construction of pBZ662 | This study |
| INSER5BLIP1 | GGTATCGCCGGTC(GATAT)CGTCAAGACCGGTGATAC; used for construction of pBZ663b | This study |
| INSER5BLIP2 | GTATCACCGGTCTTGACG(ATATC)GACCGGCGATACC; used for construction of pBZ663 | This study |
| INS5PBRI35A | GGTGATACCGCCGA(TATCC)TGCGCGTCCACGGG; used for construction of pBZ687 | This study |
| INS5PBRI35B | CCCGTGGACGCGCA(GGATA)TCGGCGGTATCACC; used for construction of pBZ687 | This study |
| INS10PBRI35A | GGTGATACCGCCGA(ACGATATCCT)TGCGCGTCCACGGG; used for construction of pBZ688 | This study |
| INS10PBRI35B | CCCGTGGACGCGCA(AGGATATCGT)TCGGCGGTATCACC; used for construction of pBZ688 | This study |
| INS11pbPLIPA1a | CGGTGATACCGCCGA(ATCGATATCCT)TGCGCGTCCACGGGC; used for construction of pBZ667 | This study |
| INS11pbPLIPA2b | GCCCGTGGACGCGCA(AGGATATCGAT)TCGGCGGTATCACCG; used for construction of pBZ667 | This study |
| PCRS1PLIPSE | GGTCAAGGGCGCGCTGC | This study |
| PCRS1PLIPA2SE | CCCGTCAGCGTACGGGGG | This study |
| LIPAP1EX | TCAAGGGCGCGCTGC | This study |
| LIPAP2EX | TCGGCGGTCGCCACT | This study |
Underlining indicates the mutagenic base pair changes compared to the wild-type sequence.
Bases in parentheses indicate the base pair insertions in the wild-type sequence.
Media and growth conditions.
Strains of E. coli were grown in LB or 2XYT medium (22). When needed, carbenicillin was added at a final concentration of 200 μg/ml, kanamycin was added at a final concentration of 60 μg/ml, and chloramphenicol was added at a final concentration of 50 μg/ml. Media and conditions for growth of Streptomyces strains have been described previously (12).
Mutagenesis and plasmid construction.
Mutagenesis was carried out using a QuikChange site-directed mutagenesis kit (Stratagene) with plasmid pB92 as the template and the complementary oligonucleotides listed in Table 1. To generate deletion plasmids pBZ619, pBZ621, and pBZ623 complementary oligonucleotides were designed that introduced an XbaI site at different locations in the wild-type lipA promoter region; XbaI-BstXI fragments were transferred to pB94 cut with the same enzymes, thus eliminating the sequence upstream of the newly introduced XbaI site (Table 1 and Fig. 1). The other mutants were obtained by further mutagenesis of the pB92 derivatives using the oligonucleotides listed in Table 1 and transferring the mutant fragments to pB94 as described above or as a HindIII-BstXI fragment in the case of pBZ662. All mutations were verified by nucleotide sequence determination.
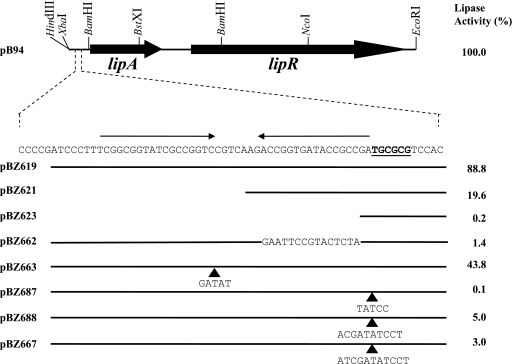
FIG. 1.
Effects of mutations in the lipA promoter region on lipase activity. The insert carried by plasmid pB94, including relevant restriction sites used for plasmid construction, is shown. The −35 region of the lipA promoter is indicated by boldface type and underlined; the two arrows above the sequence indicate the inverted repeat constituting the LipR box. Plasmids pB94 and pBZ662 carry an additional 55 bp upstream of the sequence shown; for the other plasmids the horizontal lines indicate the amounts of DNA retained in the different mutants. The sequence changes in pBZ662 are indicated. The arrowheads indicate the points where the bases shown below them were inserted. The values on the right indicate the lipase activities obtained for S. lividans cultures carrying the plasmids, expressed as percentages of the activity obtained with pB94, which was 4.751 lipase units per ml of supernatant.
To construct plasmid pB150, an NdeI site was introduced by site-directed mutagenesis at the start codon of the S. coelicolor lipR gene, which was then cloned under the control of the tipA promoter in pIJ6021 (28). Plasmid pB151 was constructed by transferring the insert of plasmid pB110 (30) to the pJV1-derived cloning vector pB48, which is compatible with pIJ6021 (12).
Growth of cultures and lipase activity assay.
Growth of S. lividans and an assay for lipase activity in culture supernatants were carried out as described previously (18, 24). The lipase activities in supernatants were determined after 72 h of growth.
S1 nuclease protection assays.
A 309-nucleotide probe for S1 mapping was prepared by PCR amplification using Platinum Taq DNA polymerase (Invitrogen) and oligonucleotides PCRS1PLIPSE and PCRS1PLIP2SE (Table 1). Oligonucleotide PCRS1PLIP2SE was labeled with [γ-32P]ATP using T4 polynucleotide kinase (Invitrogen) before the PCR, resulting in a PCR product uniquely labeled at one end, which was purified from a low-melting-point agarose gel. For each S1 nuclease protection assay 105 Cerenkov counts per minute of probe was hybridized to 50 μg of total RNA, which was purified by standard protocols (12). Hybridizations were performed in 20 μl sodium trichloroacetate buffer (17) for 5 h at 45°C, after brief denaturation at 65°C for 10 min. Further processing of the samples and denaturing gel electrophoresis of the protected fragments were done as previously described (25). End-labeled pBR322 HpaII fragments were used as size markers. The expected size of fragments protected by lipA transcripts was 177 nucleotides.
Overproduction and purification of LipR proteins.
Recombinant E. coli BL21(DE3) Rosetta 2 cells harboring plasmid pBZ606 or pBZ670 (Table 1) were grown overnight in LB medium with 60 μg/ml kanamycin and 50 μg/ml chloramphenicol. Two milliliters of each overnight culture was used to inoculate 250 ml of the same medium. When the optical density at 600 nm reached 0.6, 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce LipR synthesis. Cells were harvested by centrifugation 4 h after induction, resuspended in 20 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 0.4 M NaCl, 2 mM EDTA 2, 1 mM phenylmethylsulfonyl fluoride), and lysed in a French press. The solution was centrifuged at 12,000 × g for 20 min at 4°C. The supernatant was discarded, and the pellet was resuspended in 20 mM Tris-HCl (pH 8.0), 2 M urea, 2 mM EDTA, 1% Triton X-100 and centrifuged again as described above. This step was repeated twice. The LipR inclusion bodies were washed twice with buffer without urea and stored at −20°C.
The inclusion bodies were solubilized by incubation at room temperature for at least 1 h at 30°C in solubilization buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 6 M urea; pH 8.0) and clarified by centrifugation at 12,000 × g for 20 min at 4°C, and the supernatant was retained for analysis. Under these conditions approximately 85 to 90% of the recombinant LipR protein was solubilized.
LipR was purified by affinity chromatography using a fast protein liquid chromatography Hi-Trap chelating column (Amersham) equilibrated in solubilization buffer. The affinity-purified LipR protein was dialyzed against buffer A (50 mM NaH2PO4 [pH 7.0], 6 M urea, 10 mM NaCl) and loaded on a fast protein liquid chromatography Hi-Trap SP column (Amersham). After the column was washed with the same buffer, the protein was eluted with an NaCl gradient; LipR eluted around 0.3 M NaCl.
Refolding of the LipR proteins.
Refolding of LipR was performed as described by De Bernardez Clark et al. (6). The protein was diluted to obtain a concentration of 50 μg/ml in solubilization buffer. The denaturing agent was removed after dilution by successive dialysis against refolding buffer, which reduced the urea concentration in 0.5 M intervals from 6 M to 0 M at 4°C (2 h per interval). After renaturation the protein solution was filtered using 0.22-μm Millipore filters to remove protein aggregates. The solubilized protein was concentrated in Centriprep 10 concentrators (Amicon). Finally, the protein solution was dialyzed against 20 mM Tris-HCl (pH 8.0), 5 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol, 50% glycerol and stored at −20°C. Proteins were visualized by electrophoresing the samples on 10% denaturing sodium dodecyl sulfate-polyacrylamide gels and staining the gels with Coomassie brilliant blue R-250.
Electrophoretic mobility shift assays.
Gel retardation assays were performed either with the same 309-bp fragment used for S1 nuclease mapping or with a 140-bp probe extending from position −133 to position 7 relative to the S. exfoliatus lipA transcriptional start. To detect binding, the labeled fragment was incubated in a 20-μl reaction mixture containing 10 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 100 mM KCl, 1 mM dithiothreitol, 1 μg poly(dI-dC), and 5% glycerol. LipR protein was added at the appropriate concentration, and the mixture was incubated for 30 min at room temperature. After incubation, samples were electrophoresed in 5% nondenaturing polyacrylamide gels with a running buffer containing 25 mM Tris-HCl (pH 8) and 190 mM glycine at 100 V for 2 h. For the small probe gels were run in a Hoefer Mighty Small II electrophoresis apparatus, whereas for the larger probe gels were run in a Hoefer Sturdier SE400 apparatus. After electrophoresis the gels were dried and subjected to autoradiography.
In vitro transcription.
RNA polymerase holoenzyme purified from S. coelicolor M145 (a kind gift from Mark Buttner, John Innes Centre, United Kingdom) was used. Runoff transcripts were synthesized in vitro using the protocol described by Kieser et al. (12). Where appropriate, the template fragments were incubated with LipR protein before addition of the RNA polymerase. The synthesized RNA fragments were resolved on 6% denaturing polyacrylamide gels, using a 32P-end-labeled 25-bp ladder (Invitrogen) as the size standard.
RESULTS
Mutagenesis of the lipA promoter region.
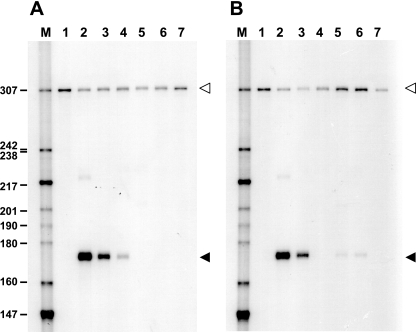
To examine the role of the inverted repeat upstream of the lipA promoter in transcription, we constructed mutations that removed different parts of the promoter region. Cultures containing a construct that retained the inverted repeat but lacked DNA sequences upstream of position −83 exhibited wild-type lipase activity (pBZ619). Cultures containing constructs with longer deletions with an endpoint at position −52 (with the distal half-site removed; pBZ621) and with an endpoint at position −35 (with the complete inverted repeat removed; pBZ623) exhibited lipase activities that were 19% and <1% of the wild-type activity, respectively (Fig. 1). Replacement of the proximal half-site of the putative LipR binding sequence with a different nonhomologous sequence (pBZ662) while the distal half-site was retained again resulted in reduced lipase activity (1.4% of the wild-type activity). Cultures of S. lividans carrying the vector alone did not show significant lipase activity (data not shown). S1 nuclease protection experiments were carried out to determine the levels of lipA transcripts and to confirm that the observed lipA expression was due to transcription from the single lipA promoter in the mutants (Fig. 2A); these experiments confirmed the results obtained for the lipase activities at the transcriptional level. Taken together, these results show that the conserved inverted repeat upstream of the lipA promoter is essential for activation of lipA transcription by LipR, and we therefore refer to it as the “LipR box.”
FIG. 2.
S1 nuclease mapping of lipA transcripts from S. lividans RNA obtained from cultures carrying different plasmids. The size markers used (lane M) were a 32P-end-labeled pBR322 HpaII digest; their sizes (in nucleotides) are indicated on the left. The solid arrowheads indicate the band corresponding to the lipA transcripts, while the open arrowheads indicate the position of undigested probe. (A) Lane 1, no RNA; lane 2, pB94; lane 3, pBZ619; lane 4, pBZ621; lane 5, pBZ662; lane 6, pBZ623; lane 7, pIJ486. (B) Lane 1, no RNA; lane 2, pB94; lane 3, pBZ663; lane 4, pBZ687; lane 5, pBZ688; lane 6, pBZ667; lane 7, pIJ486.
Importance of the location of the LipR box for activation.
To examine the effects of spacing between the lipA promoter elements, we constructed four derivatives with altered spacing. When the two half-sites of the LipR box were separated from each other by one half-turn of the DNA helix (5 bp), keeping normal spacing between the proximal half-site and the −35 region, the lipase activity was about 43% of the wild-type activity (pBZ663) (Fig. 1). On the other hand, when the entire LipR box was separated from the −35 region by a 5-bp insertion placing it on the opposite face of the DNA helix, no lipase activity was detected (pBZ687) (Fig. 1), indicating that there was no transcription from the lipA promoter. Two derivatives were constructed that changed the spacing between the LipR box and the −35 region by approximately one turn of the DNA helix (10 bp, pBZ688; 11 bp, pBZ667), placing the LipR box on the same face of the molecule but further upstream; the lipase activities of cultures carrying pBZ688 and pBZ667 were 5% and 3% of the wild-type level, respectively.
S1 nuclease mapping of RNA isolated from cultures carrying these derivatives (Fig. 2B) confirmed that the lipase activities obtained corresponded to the transcript levels. These results show that the spacing between the promoter and the LipR box and the spacing between the two half-sites of the LipR box are crucial for proper activation.
Confirmation of the S. exfoliatus LipR sequence.
Even though the three Streptomyces LipR proteins described so far are homologous members of the same protein family, the S. exfoliatus LipR protein appears to be different since it lacks a Walker A motif. This observation prompted the suggestion that the sequence previously reported might contain a sequencing error (7). Analysis of the sequence failed to reveal a clear Walker A motif in the two remaining reading frames, suggesting that the absence cannot be explained by a single frameshift error in the reported sequence. In addition, FRAME analysis (carried out at http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl) did not show any discontinuities in the region where the Walker A motif would be expected. In order to definitely rule out the possibility of a sequencing error, this region of the lipR gene was resequenced. The sequence was also determined for an independent clone of the S. exfoliatus lipase gene (18). The sequence obtained was identical to the sequence previously reported, confirming the absence of a Walker A motif in the S. exfoliatus LipR protein.
Overexpression and purification of LipR.
The S. exfoliatus lipR gene was cloned in the pET28a expression vector and used to overexpress and purify the six-His-tagged LipR protein from E. coli BL21(DE3) Rosetta 2 cells (Novagen). Induction of the construct resulted in high-level expression of a 97-kDa polypeptide, which was not seen in uninduced cells or in cells carrying the vector alone. The six-His-tagged LipR protein expressed under these conditions was found in the insoluble fraction corresponding to inclusion bodies. We decided to purify the protein under denaturing conditions, followed by renaturation as described in Materials and Methods. Confirmation that the 97-kDa purified protein was indeed LipR was provided by staining with the InVision His tag in-gel stain (Invitrogen) (data not shown).
In vitro analysis of LipR activity.
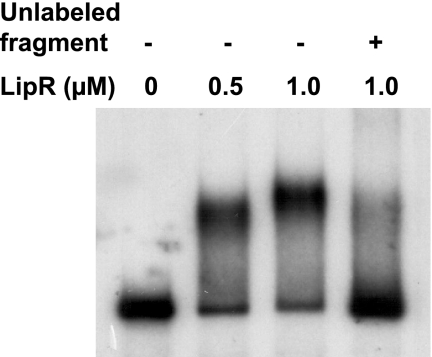
In order to determine whether the purified LipR protein is functional and able to bind to the lipA promoter region, electrophoretic mobility shift assays were performed with this protein and a DNA fragment carrying the lipA promoter region. Stable DNA-protein complexes were observed at a LipR concentration of 1 μM (Fig. 3); binding of LipR to the probe was specifically inhibited by the addition of unlabeled DNA fragment. However, attempts to determine if LipR binds specifically to the LipR box in this fragment were unsuccessful, and no clear DNA footprint could be obtained despite repeated efforts (data not shown).
FIG. 3.
Binding of purified S. exfoliatus LipR protein to the lipA promoter region. Electrophoretic mobility shift assays were performed using a 32P-end-labeled fragment containing the lipA promoter and inverted repeat (extending from position −133 to position 7 relative to the lipA transcriptional start site) in the absence (−) or presence (+) of 200 ng of the same unlabeled fragment. The concentrations of LipR protein in the samples are indicated above the lanes.
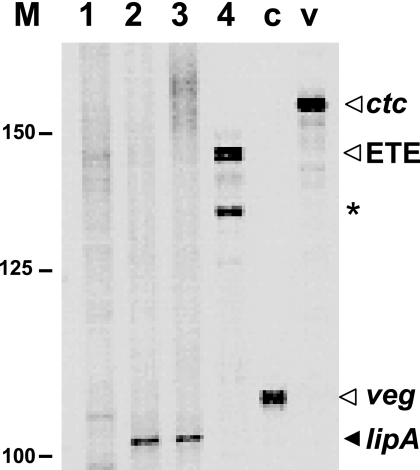
We therefore asked whether purified LipR protein could activate transcription from the lipA promoter, and we used in vitro transcription experiments to obtain evidence that the LipR box is indeed the site for LipR action. Figure 4 shows the results of runoff transcription assays performed with purified S. coelicolor RNA polymerase. When a fragment containing the wild-type lipA promoter region (derived from pB94) was used, the RNA polymerase was unable to synthesize a transcript (Fig. 4, lane 1); however, a band corresponding to the expected size of runoff transcripts originating at the lipA promoter (104 nucleotides) was clearly apparent when LipR protein was included in the reaction mixture (Fig. 4, lane 2). An identical transcript was obtained when a shorter DNA fragment (derived from pBZ619 and therefore lacking any DNA upstream of the inverted repeat) was used as the template, thus confirming that the observed band corresponded to transcripts originating at the lipA promoter (Fig. 4, lane 3). No transcripts were synthesized by the RNA polymerase when the template (derived from pBZ623) lacked the LipR box upstream of the −35 region of the promoter (Fig. 4, lane 4). Therefore, RNA polymerase can initiate transcription from the lipA promoter in vitro only when the LipR box is included in the template fragment; this transcription is dependent on the presence of the LipR protein, confirming that the LipR box is its binding site. A transcript that was about 10 nucleotides shorter than the transcripts obtained by end-to-end transcription of all the linear templates used was also observed (shown for the shorter template used in Fig. 4). Therefore, this transcript originated in the strand opposite that carrying the lipA promoter and has no relationship to the lipA transcripts.
FIG. 4.
Runoff in vitro transcription of lipA promoter fragments. The numbers on the left indicate the positions of a 25-bp ladder of DNA fragments (Invitrogen). The integrity of the RNA polymerase preparation was confirmed by carrying out two control reactions with fragments containing the veg and ctc promoters (lanes c and v, respectively), which have been described previously (34). Lane 1, in vitro transcription of the 249-bp XbaI-BamHI fragment from pB94 in the absence of LipR protein; lane 2, transcription of the same fragment with 1 μM LipR added; lane 3, transcription of the 187-bp XbaI-BamHI fragment from pBZ619 with 1 μM LipR added; lane 4, transcription of the 139-bp XbaI-BamHI fragment from pBZ623 with 1 μM LipR added. The expected size of runoff lipA transcripts was 104 nucleotides. The arrowhead labeled ETE indicates the position of bands corresponding to end-to-end transcription of the linear fragment, and the asterisk indicates the position of a transcript which originated in the strand opposite the lipA promoter.
S. exfoliatus and S. coelicolor LipR proteins are functionally interchangeable.
The lipA promoter regions of S. exfoliatus and S. coelicolor are similar, showing conservation of the promoter and of the LipR box, and the LipR proteins of these two organisms exhibit an end-to-end level of sequence identity of 47% (30). This raises the question whether they are functionally interchangeable. We therefore purified the S. coelicolor LipR protein using the same expression and purification system used previously for the S. exfoliatus protein.
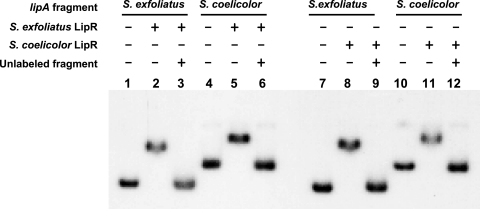
Electrophoretic mobility shift assays were performed to analyze whether the S. coelicolor LipR protein was able to bind the S. exfoliatus lipA promoter region and vice versa. Figure 5, lanes 4 to 6, show that S. exfoliatus LipR protein was able to bind to a fragment carrying the S. coelicolor lipA promoter region at the same concentration (1 μM) used to bind to its cognate promoter region (Fig. 5, lanes 1 to 3). Stable DNA-protein complexes were also observed at an S. coelicolor LipR protein concentration of 1 μM using fragments carrying lipA promoter regions from either strain (Fig. 5, lanes 7 to 12). This experiment showed that both LipR proteins are indeed able to bind the homologous promoter sequence of either species.
FIG. 5.
Electrophoretic mobility shift assays of S. exfoliatus and S. coelicolor lipA promoter regions bound to the LipR proteins. Assays were performed with a 309-bp fragment containing the S. exfoliatus lipA promoter region or a 526-bp fragment containing the S. coelicolor lipA promoter region, with or without 200 ng of unlabeled fragment, as indicated at the top. The S. exfoliatus and S. coelicolor LipR proteins were added at a concentration of 1 μM. The gel was run in a Hoefer SE400 electrophoresis apparatus.
Given the results described above, we decided to analyze whether the LipR activators were also functionally interchangeable in vivo. To do this, plasmid pB150 was constructed, in which the S. coelicolor lipR gene was placed under the control of the tipA promoter in the cloning vector pIJ6021 (28) after introduction of an NdeI site at the lipR start codon (Table 1). This plasmid was therefore equivalent to plasmid pB99 (Table 1) that carried the S. exfoliatus lipR gene under control of the tipA promoter in the same vector and that has been described previously (24). Both of these plasmids and pIJ6021 were introduced into S. lividans carrying the S. exfoliatus lipA gene cloned in a compatible pJV1-derived vector (pB100) (Table 1) (24). In this experiment thiostrepton (5 μg/ml) was added after 12 h of growth to induce the tipA promoter, and the lipase activities of S. lividans cultures carrying pB100 with lipR provided in trans were as follows (averages ± standard deviations for three independent cultures): culture with no lipR gene, 0.019 ± 0.003 U/ml of supernatant; culture with lipR gene from S. exfoliatus (pB99), 5.782 ± 1.562 U/ml of supernatant; and culture with lipR gene from S. coelicolor (pB150), 3.911 ± 1.515 U/ml of supernatant. The lipase activity obtained in the absence of either lipR gene was very low, as expected. On the other hand, high levels of activity were observed in the presence of either the S. exfoliatus or S. coelicolor lipR gene; the levels of activity obtained with the S. coelicolor lipR gene were about 70% of the levels obtained with S. exfoliatus lipR, indicating that in vivo the S. coelicolor lipR product might be slightly less efficient for activation of the S. exfoliatus lipA than its cognate lipR product. In spite of the small difference, these results clearly show that the two LipR activators are functionally interchangeable in vivo. A similar experiment was done to analyze activation of the S. coelicolor lipA gene, cloned in pB151 (Table 1), by both lipR products. Again, the lipase activity was dependent on the presence of lipR, irrespective of which lipR gene was used to activate transcription of the S. coelicolor lipA gene. In this case, however, activity was highly variable between cultures, which can be attributed to the extreme instability of the S. coelicolor lipase (30; data not shown).
DISCUSSION
In this study we examined the activation of the S. exfoliatus lipase gene, lipA, by its transcriptional activator, LipR, a member of the LipR/TchG family of actinobacterial proteins. It was previously suggested that an inverted repeat present upstream of the −35 region of the S. exfoliatus lipA promoter (which is conserved in the lipA promoter regions of S. coelicolor and S. albus) functions as the site of LipR binding and is therefore necessary for promoter activation to occur (30). This idea was consistent with the fact that most bacterial activators bind inverted repeat sequences (11), even though this has not been sufficiently documented for streptomycete regulatory proteins. The experiments described here clearly show that the inverted repeat, which we named the LipR box, is the only element upstream of the −35 region of the lipA promoter necessary for transcription activation. This was shown in vivo by eliminating the entire LipR box, which resulted in a lack of transcription activation, and in vitro by showing that LipR-dependent initiation of lipA transcription by purified RNA polymerase depends on the presence of the LipR box, confirming its role as the site of LipR binding. The results presented in this paper indicate that LipR must be bound to the proximal half-site in close proximity to the −35 region of the promoter for significant activation to occur, suggesting that LipR acts primarily as a type II transcriptional activator (19). The fact that moving the LipR box further upstream resulted in transcription levels similar to those obtained in the absence of the proximal half-site suggests that, like most type II activators, LipR bound to the proximal half-site establishes specific contacts with the sigma factor of the RNA polymerase holoenzyme (3, 19). Even though this interaction is critical for activation by LipR, our results show that the upstream half-site of the LipR box also plays a role in activation of the lipA promoter. This is shown by the fact that full transcription levels can be achieved only when both half-sites are present and by the low levels of transcription obtained in the absence of the proximal half-site. These levels suggest that LipR bound to the upstream half-site is able to establish contact with the RNA polymerase different from the contact established by LipR bound to the proximal half-site; this interaction most likely involves the C-terminal domain of the alpha subunit, which is known to interact with transcription factors that bind further upstream of the promoter (19). This is consistent with the low levels of transcription observed when the entire LipR box is moved further upstream on the same face of the DNA helix, since in this case the contacts established by LipR bound to the proximal half-site would be lost, while those established with the alpha subunit would be retained due to the well-known flexibility of the C-terminal domain, which would allow these contacts to be maintained when the activator is bound further upstream (3, 19). Therefore, LipR appears to act like LuxR, an ambidextrous activator that binds an inverted repeat (the LuxR box) and establishes contact with the RNA polymerase on both sides for full promoter activation (8, 10). This is in contrast to the mechanism of activation used by MalT, which also belongs to the STAND class and activates transcription by a different mechanism, which involves oligomeric assembly of the protein and its cooperative binding to several copies of the recognition motif (14, 23), in response to maltotriose and ATP (20, 21). The difference between the mechanisms of activation used by LipR and MalT is relevant, as it has been suggested previously that comparisons with MalT might yield clues about the mechanism by which LipR and related proteins activate transcription (1, 35). It will be interesting to explore whether LipR-related activators present in actinomycete antibiotic biosynthetic gene clusters activate transcription by a mechanism similar to the one described here for LipR, especially since these proteins exhibit end-to-end sequence similarity to the Streptomyces LipR proteins and have a conserved domain organization (15).
One interesting aspect of the entire STAND family is the presence of an N-terminal nucleotide binding domain consisting of well-conserved Walker A and Walker B motifs (15). In the case of PikD it has been shown that elimination of these motifs by site-directed mutagenesis results in a nonfunctional protein unable to activate transcription of the pikromycin gene cluster (35). Previous analysis of the S. exfoliatus LipR protein sequence revealed that it lacks a Walker A motif, an observation that prompted doubts about the reported sequence (7). In this work, however, we confirmed that the sequence is correct. This implies either that the S. exfoliatus LipR protein does not require nucleotide binding in order to activate transcription or that the Walker B motif is capable of coordinating nucleotide binding with some other structure not clearly apparent from the S. exfoliatus LipR sequence. Since the S. coelicolor and S. albus LipR protein sequences contain clear Walker A and Walker B motifs, we favor the idea that the S. exfoliatus LipR protein is a member of the LipR/TchG family that has lost the ability for nucleotide binding while retaining its activation properties. It is interesting that although the LipR proteins were fully interchangeable in vitro, there was a small difference in the ability of the S. coelicolor protein to recognize the S. exfoliatus lipA promoter, which was not apparent in the in vitro experiments, most likely because these experiments were carried out at a high, saturating protein concentration. In spite of the small differences observed between the S. exfoliatus and the S. coelicolor LipR proteins, our results show that they are similar enough that they are functionally interchangeable in vivo and in vitro, revealing conservation of the regulatory elements in the two species, which is evident from the similar locations and conservation of the LipR box. It is likely that this functional conservation also extends to S. albus, which has similar regulatory elements (30).
We have previously shown that S. exfoliatus and S. coelicolor lipase expression is regulated by the growth phase, with most lipase synthesis occurring at the onset and during the stationary phase (24). Attempts to identify molecules that might specifically induce lipase synthesis, such as triglycerides or fatty acids having different chain lengths, have not produced any significant results; this is consistent with our observation that in vitro the LipR proteins do not appear to require a coactivator to bind their target sequence or to activate transcription. The fact that lipase expression is regulated by proteins closely related to activators involved in the regulation of antibiotic synthesis, which is also developmentally regulated, raises the interesting possibility that it responds to the same developmental signals, which are still poorly understood (2). Clearly, more experimentation is required to answer this important question.
Acknowledgments
We are grateful to Mark Buttner for the gift of S. coelicolor RNA polymerase and to Laura Camarena for comments on the manuscript.
Z.E.-M. was supported by Ph.D. scholarship 138467 from the Consejo Nacional de Ciencia y Tecnología (CONACyT) and received additional support from the Programa de Apoyo a Estudiantes de Posgrado (grant 203318) of the Universidad Nacional Autónoma de México. This work was supported by grant 32558N from CONACyT.
REFERENCES
- 1.Beckett, D. 2001. Regulated assembly of transcription factors and control of transcription initiation. J. Mol. Biol. 314:335-352. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. [DOI] [PubMed] [Google Scholar]
- 3.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 4.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 5.Cruz, H., C. Pérez, E. Wellington, C. Castro, and L. Servín-González. 1994. Sequence of the Streptomyces albus G lipase-encoding gene reveals the presence of a prokaryotic lipase family. Gene 144:141-142. [DOI] [PubMed] [Google Scholar]
- 6.De Bernardez Clark, E., F. Schwarz, and R. Rudolph. 1999. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol 309:217-236. [DOI] [PubMed] [Google Scholar]
- 7.De Schrijver, A., and R. De Mot. 1999. A subafamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology 145:1287-1288. [DOI] [PubMed] [Google Scholar]
- 8.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 9.Feller, G., M. Thiry, and C. Gerday. 1990. Sequence of a lipase gene from the Antarctic psychrophile Moraxella TA144. Nucleic Acids Res. 18:6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell. Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 11.Huffman, J. L., and R. Brennan. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12:98-106. [DOI] [PubMed] [Google Scholar]
- 12.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 13.Kleeberg, I., K. Welzel, J. VendenHeuvel, R.-J. Müller, and W.-D. Deckwer. 2005. Characterization of a new extracellular hydrolase from Thermobifida fusca degrading aliphatic-aromatic copolyesters. Biomacromolecules 6:262-270. [DOI] [PubMed] [Google Scholar]
- 14.Larquet, E., V. Schreiber, N. Boisset, and E. Richet. 2004. Oligomeric assemblies of the Escherichia coli MalT transcriptional activator revealed by cryo-electron microscopy and image processing. J. Mol. Biol. 343:1159-1169. [DOI] [PubMed] [Google Scholar]
- 15.Leipe, D. D., E. V. Koonin, and L. Aravind. 2004. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343:1-28. [DOI] [PubMed] [Google Scholar]
- 16.Matshusima, P., and R. H. Baltz. 1986. Protoplast fusion, p. 170-183. In A. L. Demain and N. A. Solomon (ed.), Manual of industrial microbiology and biotechnology. American Society for Microbiology, Washington, D.C.
- 17.Murray, M. G. 1986. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal. Biochem. 158:165-170. [DOI] [PubMed] [Google Scholar]
- 18.Pérez, C., K. Juárez, E. García-Castells, G. Soberón, and L. Servín-González. 1993. Cloning, characterization and expression in Streptomyces lividans 66 of an extracellular lipase-encoding gene from Streptomyces sp. M11. Gene 123:109-114. [DOI] [PubMed] [Google Scholar]
- 19.Rhodius, V. A., and S. J. W. Busby. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152-159. [DOI] [PubMed] [Google Scholar]
- 20.Richet, E., and O. Raibaud. 1989. MalT, the regulatory protein of the Escherichia coli maltose system, is an ATP-dependent transcriptional activator. EMBO J. 8:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richet, E., D. Vidal-Ingigliardi, and O. Raibaud. 1991. A new mechanism for coactivation of transcription initiation: repositioning of an activator triggered by the binding of a second activator. Cell 66:1185-1195. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Schreiber, V., and E. Richet. 1999. Self-association of the Escherichia coli transcription activator MalT in the presence of maltotriose and ATP. J. Biol. Chem. 274:33220-33226. [DOI] [PubMed] [Google Scholar]
- 24.Servín-González, L., C. Castro, C. Pérez, M. Rubio, and F. Valdez. 1997. bldA-dependent expression of the Streptomyces exfoliatus M11 lipase gene (lipA) is mediated by the product of a contiguous gene, lipR, encoding a putative transcriptional activator. J. Bacteriol. 179:7816-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Servín-González, L., M. R. Jensen, J. White, and M. Bibb. 1994. Transcriptional regulation of the four promoters of the agarase gene (dagA) of Streptomyces coelicolor A3(2). Microbiology 140:2555-2565. [DOI] [PubMed] [Google Scholar]
- 26.Sommer, P., C. Bormann, and F. Gotz. 1997. Genetic and biochemical characterization of a new extracellular lipase from Streptomyces cinnamoneus. Appl. Environ. Microbiol. 63:3553-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sztajer, H., I. Maliszewska, and J. Wieczorek. 1988. Production of exogenous lipases by bacteria, fungi and actinomycetes. Enzyme Microb. Technol. 10:492-497. [Google Scholar]
- 28.Takano, E., J. White, C. J. Thompson, and M. J. Bibb. 1995. Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene 166:133-137. [DOI] [PubMed] [Google Scholar]
- 29.Uchida, H., Y. Shigeno-Akutsu, N. Nomura, T. Nakahara, and T. Nakajima-Kambe. 2002. Cloning and sequence analysis of poly(tetramethylene succinate) depolymerase from Acidovorax delafieldii strain BS-3. J. Biosci. Bioeng. 93:245-247. [DOI] [PubMed] [Google Scholar]
- 30.Valdez, F., G. González-Cerón, H. Kieser, and L. Servín-González. 1999. The Streptomyces coelicolor A3(2) lipAR operon encodes an extracellular lipase and a new type of transcriptional regulator. Microbiology 145:2365-2374. [DOI] [PubMed] [Google Scholar]
- 31.Vujaklija, D., W. Schröder, M. Abramic, P. Zou, I. Lescic, P. Franke, and J. Pigac. 2002. A novel streotomycete lipase: cloning, sequencing and high-level expression of the Streptomyces rimosus GDS(L)-lipase gene. Arch. Microbiol. 178:124-130. [DOI] [PubMed] [Google Scholar]
- 32.Vulfson, E. N. 1994. Industrial applications of lipases, p. 271-288. In P. Wooley and S. B. Petersen (ed.), Lipases—their structure, biochemistry and application. Cambridge University Press, Cambridge, England.
- 33.Wei, Y., L. Swenson, C. Castro, U. Derewenda, W. Minor, H. Arai, J. Aoki, K. Inoue, L. Servín-González, and Z. S. Derewenda. 1998. Structure of a microbial homologue of mammalian platelet-activating factor acetylhydrolases: Streptomyces exfoliatus lipase at 1.9 A resolution. Structure 6:511-519. [DOI] [PubMed] [Google Scholar]
- 34.Westpheling, J., M. Ranes, and R. Losick. 1985. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature 313:22-27. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, D. J., Y. Xue, K. A. Reynolds, and D. H. Sherman. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183:3468-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]