Abstract
FtsZs from Mycoplasma pulmonis (MpuFtsZ) and Bacillus subtilis (BsFtsZ) are only 46% and 53% identical in amino acid sequence to FtsZ from Escherichia coli (EcFtsZ). In the present study we show that MpuFtsZ and BsFtsZ can function for cell division in E. coli provided we make two modifications. First, we replaced their C-terminal tails with that from E. coli, giving the foreign FtsZ the binding site for E. coli FtsA and ZipA. Second, we selected for mutations in the E. coli genome that facilitated division by the foreign FtsZs. These suppressor strains arose at a relatively high frequency of 10−3 to 10−5, suggesting that they involve loss-of-function mutations in multigene pathways. These pathways may be negative regulators of FtsZ or structural pathways that facilitate division by slightly defective FtsZ. Related suppressor strains were obtained for EcFtsZ containing certain point mutations or insertions of yellow fluorescent protein. The ability of highly divergent FtsZs to function for division in E. coli is consistent with a two-part mechanism. FtsZ assembles the Z ring, and perhaps generates the constriction force, through self interactions; the downstream division proteins remodel the peptidoglycan wall by interacting with each other and the wall. The C-terminal peptide of FtsZ, which binds FtsA, provides the link between FtsZ assembly and peptidoglycan remodeling.
FtsZ assembles into the Z ring at the center of the bacterial cell, and this ring eventually constricts as the cell divides. There is much evidence that the Z ring provides the force for constriction, and although not definitively proven (an alternative for cells with a cell wall is that invagination of the cell wall generates the constriction force), we will assume that this is the case. Other proteins associate with the Z ring and are essential for the eventual constriction of the Z ring. FtsZ is required for the association of all other division proteins. In Escherichia coli, Z-ring assembly requires either FtsA or ZipA, which serve to tether FtsZ to the membrane (39, 40). FtsK is the next protein to associate with the Z ring, and it requires FtsA and perhaps ZipA (17, 40). In E. coli the downstream division proteins assemble in a mostly linear fashion (4, 5). However, recent work suggests that the pathway is not strictly linear and involves cooperative binding of some downstream proteins to two or more partners (18-20, 27, 47). For more detailed discussion of Z-ring assembly and bacterial cell division, see recent reviews (15, 19, 32, 44, 51).
Several proteins are known to bind directly to FtsZ. FtsA and ZipA both bind the same site: a 15- to 17-amino-acid peptide at the very C terminus of FtsZ (23, 31, 49, 52). In Mycobacterium tuberculosis, which has no FtsA or ZipA, this peptide binds FtsW (9), and this binding may occur in other species. MinC binds FtsZ (26), probably on the bottom protofilament interface of FtsZ. MinC is not essential for the assembly and constriction of Z rings, but in its absence extra Z rings assemble near the cell ends, resulting in mini-cells (10). ZapA, SlmA, and ClpX bind FtsZ, but the sites of binding are not known; these are not essential division proteins, although deletion of some pairs (ZapA and EzrA, or SlmA and MinCDE) results in synthetic lethality (3, 21, 50).
We will consider two extreme models for how the Z ring might generate the force for constriction. The first model is via a motor molecule, similar to the known mechanism of eukaryotic cytokinesis. This model would propose that motor molecules bind FtsZ and pull protofilaments together. Although no candidate motor molecules have been identified in bacteria, there could be an unidentified motor, or one of the downstream proteins might bind FtsZ protofilaments and act as a motor. The second model is the Z-centric hypothesis, which postulates that FtsZ provides not only the cytoskeletal scaffold but also generates the constriction force. We have previously proposed one possible mechanism, which involves the change in protofilament conformation from straight to curved (14, 30). An alternative model, in which FtsZ provides the scaffold and one or more of the downstream proteins generates the constriction force, will be addressed in the Discussion, below.
If FtsZ generated the constriction force entirely through self interactions, one might be able to replace E. coli FtsZ with the FtsZ from widely divergent bacterial species. Of course it would be necessary to give the foreign FtsZ the C-terminal peptide from E. coli FtsZ so it could bind FtsA. On the other hand, if constriction involved a motor binding to FtsZ protofilaments, the binding site for the E. coli motor would likely be lost on divergent FtsZs.
FtsZ is highly divergent across prokaryotic species. The amino acid sequence of E. coli FtsZ (EcFtsZ) is only 46% and 53% identical to those of Mycoplasma pulmonis (MpuFtsZ) and Bacillus subtilis (BsFtsZ). In addition, M. pulmonis is missing all of the downstream proteins identified in E. coli. It is therefore unlikely that MpuFtsZ would retain the ability to bind any E. coli downstream division proteins or a hypothetical motor molecule. We therefore decided to investigate whether FtsZ from M. pulmonis and other bacteria could function for division in E. coli.
Before beginning these studies we made a discovery that was essential for their success. While experimenting with mutants of EcFtsZ that could not complement the ftsZ null in our standard assay (38, 43), we found several FtsZ mutants that could generate suppressor strains in which a second-site genomic mutation permitted the defective FtsZ to function for cell division. This same approach generated suppressor strains in which foreign FtsZ could function for division.
MATERIALS AND METHODS
Strains and plasmids used are listed in Table 1. E. coli strain JKD7-1/pKD3 (7) was used for complementation assays and for generating suppressor strains. pJSB2 was used to express the mutant or foreign FtsZ as previously described (43, 45) (Fig. 1). pJSB2 was derived from the pBAD plasmid, which has an arabinose-inducible promoter. ftsZ genes from Yersinia pestis KIM5 and Pseudomonas aeruginosa PAO1 were cloned by colony PCR. The ftsZ genes from B. subtilis, M. pulmonis, and Mycoplasma pneumoniae were cloned by PCR from genomic DNA, from plasmid pCX66 (generously provided by Joe Lutkenhaus, University of Kansas [48]), and from a plasmid provided by Jörg T. Regula, Adolf Butenandt Institut, LMU München. Azotobacter vinelandii ftsZ was cloned from our pET11 plasmid (29). All ftsZ genes were blunt cloned into the SmaI site in pJSB2 (45).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| JKD7-1 | ftsZ::Kanr (genomic ftsZ null) recA | 7 |
| JSN2 | JKD7-1/pKD3/pJSB2 | 45 |
| wt | JKD7-1/pJSB2-EcFtsZ | 45 |
| SUP{D96A}-D96A | JKD7-1/pJSB2-D96A (mut?) | This study |
| SUP{D96A}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{Q47K}-Q47K | JKD7-1/pJSB2-Q47K (mut?) | This study |
| SUP{Q47K}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{D212G1}-D212G | JKD7-1/pJSB2-D212G (mut?) | This study |
| SUP{D212G1}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{D212G2}-D212G | JKD7-1/pJSB2-D212G (mut?) | This study |
| SUP{D212G2}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{CtYFP1}-CtYFP1 | JKD7-1/pJSB2-CtYFP (mut?) | This study |
| SUP{CtYFP1}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{CtYFP2}-CtYFP1 | JKD7-1/pJSB2-CtYFP (mut?) | This study |
| SUP{CtYFP2}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{326YFP}-326YFP1 | JKD7-1/pJSB2-326YFP (mut?) | This study |
| SUP{326YFP}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{CtYFP1//D96A}-D96A | JKD7-1/pJSB2-D96A (mut? mut?) | This study |
| SUP{CtYFP1//D96A}-wt | JKD7-1/pKD3/pBAD18Spcr (mut? mut?) | This study |
| SUP{CtYFP1//Q47K}-Q47K | JKD7-1/pJSB2-D96A (mut? mut?) | This study |
| SUP{CtYFP1//Q47K}-wt | JKD7-1/pKD3/pBAD18Spcr (mut? mut?) | This study |
| SUP{D212G1//CtYFP}-CtYFP | JKD7-1/pJSB2-CtYFP (mut? mut?) | This study |
| SUP{D212G1//CtYFP}-wt | JKD7-1/pKD3/pBAD18Spcr (mut? mut?) | This study |
| SUP{Av}-Av | JKD7-1/pJSB2-AvftsZ (mut?) | This study |
| SUP{Pa}-Pa | JKD7-1/pJSB2-PaftsZ (mut?) | This study |
| SUP{Bs/EcCt}-Bs/EcCt | JKD7-1/pJSB2-Bs/EcCt ftsZ (mut?) | This study |
| SUP{Mpu/EcCt11}-Mpu/EcCt | JKD7-1/pJSB2-Mpu/EcCt ftsZ (mut?) | This study |
| SUP{Mpu/EcCt11}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{Mpu/EcCt13}-Mpu/EcCt | JKD7-1/pJSB2-Mpu/EcCt ftsZ (mut?) | This study |
| SUP{Mpu/EcCt13}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| SUP{Mpu/EcCt28}-Mpu/EcCt | JKD7-1/pJSB2-Mpu/EcCt ftsZ (mut?) | This study |
| SUP{Mpu/EcCt28}-wt | JKD7-1/pKD3/pBAD18Spcr (mut?) | This study |
| Plasmids | ||
| pKD3 | pSC101 origin; repA(Ts) ftsZ Ampr | 7 |
| pBAD18 | pBR322 origin; arabinose regulation; Ampr | 22 |
| pBAD18Spcr | pBAD18-bla::aadA (Amps Spcr) | This study |
| pJSB1 | pBAD18 with MCSa for blunt insertion | 45 |
| pJSB2 | pJSB1-bla::cat (Amps Cmr) | 45 |
| pJSB2-wt | pJSB2-EcftsZ | 45 |
| pJSB2-Q47K | pJSB2-EcftsZ-Q47K | 43 |
| pJSB2-D96A | pJSB2-EcftsZ-D96A | 45 |
| pJSB2-D212G | pJSB2-EcftsZ-D212G | 45 |
| pJSB2-248-20aa | pJSB2-EcftsZ-248-20aa | 38 |
| pJSB2-326YFP | pJSB2-EcftsZ-326-YFP | 38 |
| pJSB2-348-20aa | pJSB2-EcftsZ-348-20aa | 38 |
| pJSB2-348YFP | pJSB2-EcftsZ-348YFP | 38 |
| pJSB2-CtYFP | pJSB2-EcftsZ-CtYFP | This study |
| pJSB2-Yp | pJSB2-YpftsZ | This study |
| pJSB2-Pa | pJSB2-PaftsZ | This study |
| pJSB2-Av | pJSB2-AvftsZ | This study |
| pJSB2-Bs | pJSB2-BsftsZ | This study |
| pJSB2-Bs/EcCt | pJSB2-Bs/EcCtftsZ | This study |
| pJSB2-Mpu | pJSB2-MpuftsZ | This study |
| pJSB2-Mpu/EcCt | pJSB2-Mpu/EcCtftsZ | This study |
| pJSB2-Mpn | pJSB2-MpnftsZ | This study |
| pJSB2-Mpn/EcCt | pJSB2-Mpn/EcCtftsZ | This study |
mut? indicates an unknown suppressor mutation in the genome; mut? mut? indicates unknown double suppressor mutations in the genome.
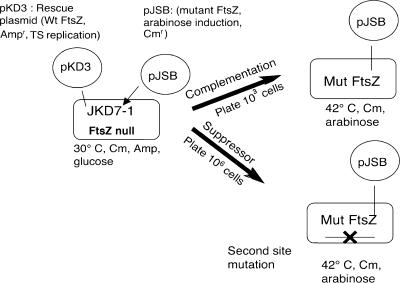
FIG. 1.
Schematic illustration of the complementation system and production of suppressor strains. See Results for details.
The yellow fluorescent protein (YFP) was the variant Venus (37), which we have found superior to other green fluorescent proteins for fusion to FtsZ (38).
To create chimeric molecules with the C-terminal tail (amino acids [aa] 325 to 383) from E. coli and the N-terminal domains from foreign bacteria, a silent SpeI site was introduced at aa 326 in EcFtsZ in pJSB2. C-terminal (Ct)-deleted BsFtsZ (aa 1 to 323), MpuFtsZ (aa 1 to 320), and MpnFtsZ (aa 1 to 336) were amplified by PCR from their pJSB2 plasmids. The sequence went from a BamHI site located in the vector about 30 bp 5′ of the start codon and added a SpeI site on the C terminus. The EcFtsZ-SpeI vector was cut with BamHI and SpeI, and the foreign ftsZ was ligated to replace the N-terminal segment of EcFtsZ.
In order to study the suppressor strains with wild-type FtsZ, we removed the pJSB2 plasmid expressing the mutant or foreign FtsZ. We prepared a new pBAD plasmid in which the ampicillin resistance was replaced with spectinomycin resistance. This plasmid is incompatible with pJSB2 (which is a pBAD-based plasmid). We transformed the suppressor strain with pKD3, to provide wild-type FtsZ, and with the new pBAD plasmid and grew the cells at 30°C in glucose, ampicillin, and spectinomycin. Clones that were chloramphenicol sensitive and spectinomycin resistant were identified as suppressor strains that were now using wild-type FtsZ from pKD3. Suppressor strains were later retransformed with pJSB2 expressing the original mutant FtsZ or a new one.
For light microscopy cells were grown in Luria broth (LB)-based medium containing 34 μg/ml chloramphenicol and 0.2% arabinose until the optical density at 600 nm reached ∼0.8 (except as noted). Cells were visualized with differential interference contrast and fluorescence microscopy using a Zeiss Axiophot with a 100× objective, 1.3 numerical aperture lens. Filter cubes optimized for YFP and green fluorescent protein were used for fluorescence microscopy. Images were acquired with a Coolsnap HQ charge-coupled device camera (Roper Scientific).
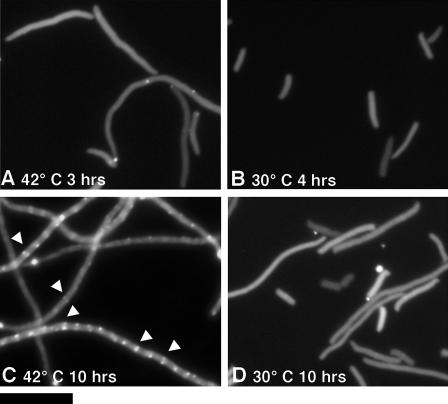
To determine the localization of (Bs/EcCt)CtYFP in its suppressor strain, a stationary culture was diluted 200-fold and grown for several hours (this strain grew very slowly). To observe the localization in the original JKD7-1 background, without the suppressor mutation, JKD7-1/pKD3 was transformed with pJSB2 expressing (Bs/EcCt)CtYFP. An overnight culture was diluted 200-fold at 30°C or at 42°C plus 0.2% arabinose. Specimens were made at 3 to 4 h, when the cells were just starting to elongate, and at 10 h, when they were in stationary phase and starting to die.
RESULTS
Production of suppressor strains.
Complementation assays were performed as previously described (43, 45) and as diagrammed in Fig. 1. The bacterial strain JKD7-1 has a chromosomal ftsZ null mutation and is supported by a rescue plasmid pKD3, which expresses wild-type FtsZ (7). pKD3 is temperature sensitive for replication, and wild-type FtsZ is eliminated at 42°C. The mutant FtsZ to be tested is expressed from plasmid pJSB2, which has an arabinose-inducible promoter. When cells are plated on LB agar containing 0.2% arabinose and grown at 42°C, the wild-type FtsZ is lost and is replaced by the mutant FtsZ. For our standard complementation assay, we plated about 1,000 cells. We found that some mutants complemented and produced 1,000 colonies and some mutants did not complement and produced no colonies (43, 45). There were no intermediate results.
We used a modification of this complementation system to produce suppressor strains for FtsZ mutants and for FtsZ from foreign species. Instead of plating 1,000 cells, we plated 106 to 107 cells. A substantial background of colonies appeared at a frequency of 10−5 to 10−6. These background colonies were large and generally retained resistance to ampicillin, which suggested that the pKD3 plasmid had lost its temperature sensitivity. Some FtsZ mutants generated additional colonies at frequencies of 10−4 to 10−6. These colonies were smaller and were sensitive to ampicillin and dependent on arabinose for growth, indicating that they were depending on the mutant FtsZ expressed from pJSB2. We provisionally identified them as suppressor strains and confirmed this with additional tests described below. The mutants that generated suppressor strains and the frequencies at which they arose are given in Table 2.
TABLE 2.
| FtsZ strain | Complementation in suppressor strain (at indicated frequency)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ct YFP (10−4 to 10−5) | 326 YFP (10−4 to 10−5) | 348 YFP (10−4 to 10−5) | 348 20 aa (10−4 to 10−5) | 248 20 aa (10−4 to 10−5) | D212G (10−4 to 10−5) | D212A (10−4 to 10−5) | D96A (10−5 to 10−6) | Q47K (10−4 to 10−5) | |
| SUP{CtYFP1} | C | C | C | C | NC | NC | NC | NC | |
| SUP{326YFP} | C | C | C | NC | NC | NC | |||
| SUP{D96A} | NC | NC | NC | NC | NC | C | C | ||
| SUP{Q47K} | NC | NC | NC | NC | NC | C | C | ||
| SUP{D212G1} | NC | NC | C | NC | C | C | C | C | |
| SUP{Mpu/EcCt11} | NC | NC | NC | NC | C | NC | C | ||
| SUP{Mpu/EcCt13} | NC | C | C | C | C | NC | C | ||
CtYFP indicates FtsZ with a C-terminal YFP; 326 YFP indicates YFP was inserted at aa 326 of FtsZ; 348 20 aa means a 20-amino-acid peptide was inserted at aa 348 (38); D212G indicates a point mutant. None of these constructs could complement in our standard assay, but each gave rise to suppressor strains when tested at high plating density. The value in parentheses following each suppressor strain is the frequency at which suppressor strains arose. Each mutant construct was tested in each of the suppressor strain backgrounds. SUP{CtYFP1} designates one of two suppressor strains generated originally by FtsZ with a C-terminal YFP (SUP{CtYFP2} is tested in Table 3). Likewise, SUP{D212G1} and SUP{D212G2} indicate suppressor strains from two different colonies from the original generation. SUP{Mpu/EcCt11} and -13 indicate strains from two colonies generated by M. pulmonis FtsZ. C, the construct complemented the ftsZ null in that suppressor strain; NC, the construct did not complement. Point and insertion mutations were from references 43 and 38, respectively.
The following point mutations did not produce suppressor strains: N24K, D45A, G103K, E138K, R142D, F182D, N207C, D209C, D209K, F210A, D212C, RLD271-3AKK, and D209A. At insertion site 1 (N-terminal fusion), the following 20-aa insertion mutations did not produce suppressor strains: 8, 98, 134, 141, 158, 173, 195, 214, 234, 239, 288, and 377.
To test the possibility that the second site mutation for suppression might be in the pJSB2 encoding the mutant ftsZ, we removed pJSB2, replacing it with pKD3. We then retransformed with the original mutant ftsZ in pJSB2 (or eventually with other mutants or foreign ftsZ) and performed a second complementation assay, plating 1,000 cells on chloramphenicol and 0.2% arabinose plates at 42°C. For all suppressor strains the reintroduced mutant ftsZ on pJSB2 fully complemented the chromosomal ftsZ::kan null allele. From this we conclude that the suppressor mutations were on the E. coli genome.
We will use the abbreviation SUP{D96A} to refer to the suppressor strain generated by ftsZ mutant D96A. In some cases we characterized two separate colonies from the original suppressor generation, and we have designated these by appending a number, e.g., SUP{D212A1} and SUP{D212A2} (Table 2).
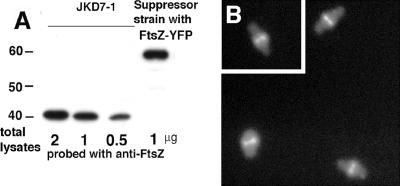
We confirmed by immunoblotting the absence of wild-type FtsZ in strain SUP{CtYFP1} (the suppressor strain produced by FtsZ with YFP fused at the C terminus, described below). Wild-type FtsZ was completely absent from the suppressor strain, and FtsZ-CtYFP was expressed at about a 2 to 3 times higher level than the wild-type FtsZ expressed from pKD3 in JKD7-1 (Fig. 2A). This result is in good agreement with other reports that pKD3 supplies FtsZ at the same level as genomic FtsZ (8) and that pJSB2 can express ∼3 to 5 times higher levels than genomic FtsZ (43, 45).
FIG. 2.
Expression of FtsZ-CtYFP in the suppressor strain. (A) The original strain JKD7-1 and the suppressor strain generated from FtsZ-CtYFP were compared by immunoblotting, as described previously (45). One microgram of total protein from the suppressor strain and 2, 1, and 0.5 μg from JKD7-1 were loaded. The cells were cultured with 0.2% arabinose at 42°C for the suppressor strain and at 30°C for JKD7-1. (B) Fluorescence microscopy localizing FtsZ-CtYFP in the suppressor strain SUP{CtYFP1}, where it is the sole source of FtsZ.
The colonies generated by any given mutant typically showed a variety of cellular phenotypes. For example, some colonies of SUP{CtYFP} had cells with normal length and Z rings (Fig. 2B), while others were variably elongated (not shown). The different phenotypes suggest that there are multiple mutation sites for suppression, some more effective than others.
Two suppressor strains were previously described for the temperature-sensitive allele ftsZ84 (G105S), permitting growth at 42°C. One of these was due to a twofold overexpression of ZipA, and the other was not characterized (42). To see if something similar was occurring in our results, we checked several suppressor strains generated by FtsZ-CtYFP for alterations in FtsA and ZipA. In one strain, SUP{CtYFP1}, the level of FtsA was reduced to about half. Six others strains had normal levels of FtsA. All strains had normal levels of ZipA, and there were no sequence changes in either gene.
Suppressor strains for insertional mutants in EcFtsZ.
We next extended the study to mutant FtsZs that had insertions of either YFP or a shorter 20-amino-acid insert (38). The YFP insertions that generated suppressor strains were either at the very C terminus (FtsZ-CtYFP) (Fig. 2) or within the ∼50-amino-acid “spacer” between the C-terminal globular domain and the C-terminal peptide that binds FtsA and ZipA (38). Interestingly, one 20-amino-acid insertion at aa 248 also produced a suppressor strain. The functionality of the FtsZ protein with this insertion is surprising, because it is in a highly conserved sequence, SPL248LE.
Testing for cross-suppression.
We then addressed the question of whether a suppressor strain generated by one mutant FtsZ could suppress the defects of other FtsZ mutants or insertions. For example, suppressor strain SUP{D96A} carrying wild-type ftsZ on pKD3 was transformed in succession by pJSB2 carrying each of the other ftsZ mutant alleles. We then plated ∼1,000 cells and compared colony formation under control (30°C, glucose) and complementation (42°C, 0.2% arabinose) conditions. For pJSB2-Q47K, we observed full complementation in SUP{D96A}, but none of the other FtsZ mutants produced colonies in this strain at 42°C. We will say that SUP{D96A} suppresses Q47K. We also found that SUP{Q47K} suppressed D96K, but neither of these strains suppressed any of the other mutants. SUP{D212G} was of broader potency, as it could suppress D96A and Q47K, as well as one 20-amino-acid insertion. The suppressor strains generated by YFP insertions at the C terminus and at aa 326 could suppress most of the other insertions but none of the point mutants. These suppressor strains seem specialized for insertions in the C-terminal region. Table 2 summarizes the tests of each FtsZ mutant in each suppressor strain.
In some cases, a second round of suppressor strains was generated using a new mutant. For example, the suppressor strain SUP{CtYFP1}, originally produced by FtsZ with YFP fused to its C terminus, was retransformed with pJSB2 carrying the D96A mutation. When tested under complementation conditions (1,000 cells plated), no colonies were obtained. However, when plated at high levels (106 to 107 cells), new suppressor strains developed. These strains presumably retained the original suppressor mutation and had an additional genomic mutation and are designated as SUP{CtYFP1//D96A} (the double slash indicates a double suppressor).
Can FtsZ from foreign bacteria achieve cell division in E. coli?
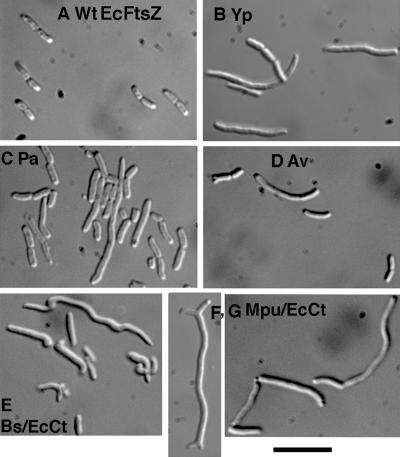
We used the complementation and suppressor assays to test whether FtsZ from divergent bacterial species could function for division in E. coli. FtsZs from Yersinia pestis, Pseudomonas aeruginosa, Azotobacter vinelandii, Bacillus subtilis, Mycoplasma pulmonis, and Mycoplasma pneumoniae are 97%, 67%, 68%, 53%, 46%, and 23% identical, respectively, to E. coli FtsZ (these percent identities have been determined for the conserved core domain, aa 1 to 316 in EcFtsZ). Only YpFtsZ was functional for division in our standard complementation assay. YpFtsZ was not fully functional, since colonies grew slower on the agar plates and cells were somewhat elongated (Fig. 3B).
FIG. 3.
Morphology of strains using foreign FtsZ for division. (A and B) The original JKD7-1 strain complemented with EcFtsZ (A) and YpFtsZ (B). (C to G) Other foreign FtsZs are shown in the suppressor strains they generated. Those strains were cultured with 0.2% arabinose at 37°C. Bar, 10 μm.
PaFtsZ and AvFtsZ did not produce colonies under complementation conditions; however, both of them produced suppressor strains. The frequency of generating suppressor strains was 10−3 for PaFtsZ and 10−4 to 10−5 for AvFtsZ (Table 3). The morphology of both PaFtsZ and AvFtsZ suppressor strains showed variably elongated cells and mini-cells (Fig. 3C and D). The presence of mini-cells suggests that these FtsZs do not interact with MinC, which is not surprising given the sequence divergence. The AvFtsZ suppressor strain also exhibited some cell swelling.
TABLE 3.
Foreign FtsZs tested for complementation and production of suppressor strainsa
| FtsZ strain | Complementation with foreign FtsZ from:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | Y. pestis | P. aeruginosa | A. vinlandii | B. subtilis | Bs/EcCt | M. pulmonis | Mpu/EcCt | M. pneumoniae | Mpn/EcCt | |
| Wild type | C | C | S (∼10−3) | S (10−4-10−5) | NS | S (10−4-10−5) | NS | S (10−4-10−5) | NS | NS |
| SUP{D96A} | C | C | NC | NC | NC | NC | NC | NS | ||
| SUP{Q47K} | C | C | NC | NC | NC | NC | NC | NC | ||
| SUP{D212G1} | C | C | C | C? (0.1) | C? (0.1) | C | NC | |||
| SUP{D212G2} | C | C | C | C? (0.1) | NC | C | NC | |||
| SUP{CtYFP1} | C | C | C | C | NS | C | NS | NC, NS | NS | NS |
| SUP{CtYFP2} | C | C | C | NC | C | NC | NC | |||
| SUP{CtYFP1//D96A} | C | C | C? (0.1) | NC | C | C | NC | |||
| SUP{CtYFP1//Q47K} | C | C | C | C | C | C? (0.1) | NC | |||
| SUP{D212G1//CtYFP} | C | C | C | C? (0.4) | C? (0.4) | C? (0.3) | NC | |||
| SUP{Mpu/EcCt13} | C | C | C | NC | C | C | NC | |||
Bs/EcCt, B. subtilis FtsZ with the C-terminal peptide from E. coli; Mpu/EcCt, M. pulmonis FtsZ with the C-terminal peptide from E. coli; Mpn/EcCt, M. pneumoniae FtsZ with the C-terminal peptide from E. coli. Wild-type results show the results when each foreign FtsZ was tested in the wild-type E. coli background. C, the foreign FtsZ complemented in our standard assay; S, it did not complement, but it produced suppressor strains at the indicated frequency; NS, the construct was tested under suppressor generation conditions, but none were obtained; C? (0.1), colonies were obtained under complementing conditions but at 1/10 the normal frequency.
FtsZ from B. subtilis did not generate suppressor strains. However, as shown in Fig. 4, the 17-amino-acid C-terminal peptide that binds FtsA and ZipA has several amino acid changes from that of E. coli. It has been demonstrated that several point mutations in this sequence are lethal (23, 31). We therefore replaced the C-terminal segment of BsFtsZ with the E. coli C-terminal segment, aa 326 to 383. This segment contains most of the nonconserved spacer in addition to the FtsA/ZipA-binding peptide. The chimeric protein, which we call (Bs/EcCt)FtsZ, comprised BsFtsZ amino acids 1 to 323 and EcFtsZ amino acids 326 to 383. This chimeric FtsZ produced suppressor strains. These suppressor strains showed more severe defects than the previous ones, including elongation, swelling, twisted shape, and mini-cells (Fig. 3E). Despite the defects, cell division succeeded often enough to avoid extensive filamentation.
FIG. 4.
Sequence alignment of the 17-amino-acid C-terminal peptide that binds FtsA and ZipA.
FtsZ from M. pulmonis gave results similar to BsFtsZ. It produced suppressor strains when given the E. coli C-terminal tail. These strains also showed unusual morphology, including some branched shapes (Fig. 3F and G). Some colonies gave cells with lots of branching, and others were more normal in appearance.
Finally, we tested FtsZ from M. pneumoniae, which has an extremely divergent sequence. This FtsZ did not produce suppressors even when given the E. coli C-terminal tail. MpnFtsZ has a pI of 7.5, versus pIs of 4.5, 4.9, and 5.1 for EcFtsZ, BsFtsZ, and MpuFtsZ. It is likely that MpnFtsZ is designed to operate in a higher pH environment and may have defective assembly in the E. coli cytoplasm. One other FtsZ, that from Mycobacterium tuberculosis, also failed to produce a suppressor strain, even with the E. coli C-terminal peptide (data not shown).
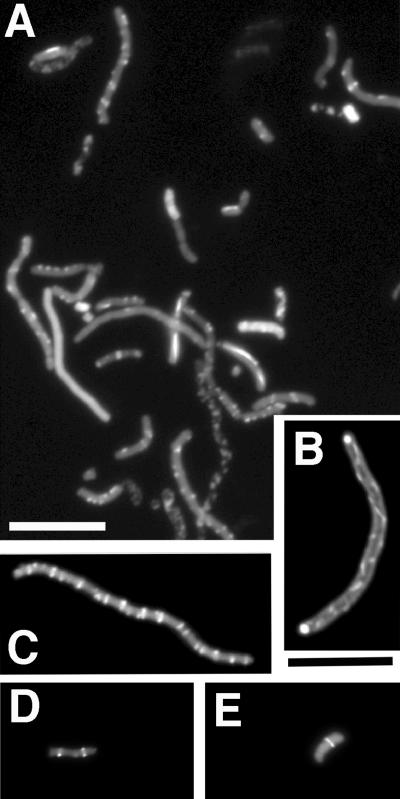
To determine the localization of (Bs/EcCt)FtsZ and the morphology of Z rings, we constructed (Bs/EcCt)CtYFP, where YFP was fused to the C terminus. We had already characterized the suppressor strain SUP{CtYFP1}, which was originally generated from E. coli FtsZ-CtYFP, and demonstrated that it suppressed Bs/EcCt. We therefore tested (Bs/EcCt)CtYFP in this strain and found that it produced colonies. The frequency of colony formation was only 10% under the complementing conditions. The cells examined by light microscopy were variably elongated and bent, similar to Bs/EcCt in its original suppressor strain (data not shown). Fluorescence microscopy showed a highly variable localization of (Bs/EcCt)CtYFP (Fig. 5A). Many cells had spiral filaments (Fig. 5B), multiple Z rings (Fig. 5C), or straight filaments running longitudinally or with a patchy distribution (Fig. 5A). A frequent structure was a small spot on one side of the cell, which looked like half of a Z ring. Some normal-looking Z rings appeared in short cells, but these were rare (Fig. 5D and E). Immunofluorescence was also used to localize (Bs/EcCt)FtsZ in its original suppressor strain, SUP{Bs/EcCt}. The immunofluorescence was more diffuse than YFP but in general agreement (data not shown).
FIG. 5.
The Z-ring structures of B. subtilis FtsZ (Bs/EcCt)CtYFP visualized in SUP{CtYFP1}, a suppressor strain generated with E. coli FtsZ-YFP. The cells were observed with YFP fluorescence. (A) Field with various types of localization. (B and C) Spiral and multiple Z rings, respectively. (D and E) Sharp Z rings in short cells, which were observed only occasionally. Bars, 10 μm.
We then tested the localization of (Bs/EcCt)CtYFP under complementation and dominant-negative conditions in wild-type E. coli to determine whether it could form Z rings in the absence of the suppressor mutation. (Bs/EcCt)CtYFP was expressed in JKD7-1(pKD3) at two different temperatures. At 30°C the mutant FtsZ is expressed along with wild-type EcFtsZ, whereas at 42°C the expression of EcFtsZ should be gradually reduced (7). The distribution was mostly diffuse at 3 to 4 h at both 42° and 30°C, although at 42°C there were some bright spots that may have been nascent or partial Z rings (Fig. 6A and B). After 10 h at 30°C, where EcFtsZ remains present, the distribution remained diffuse. However, after 10 h at 42°C, where EcFtsZ should be largely depleted, (Bs/EcCt)CtYFP was able to form Z-ring-like structures in the filamented cells. These cultures had reached stationary phase, and this may be a significant factor, since we did not find convincing Z-ring structures in log phase. Overall, these results show that (Bs/EcCt)CtYFP is able to assemble into protofilaments and further assemble into Z rings, but only in the absence of EcFtsZ. We conclude that the suppressor mutation is not required for (Bs/EcCt)FtsZ to assemble into Z rings, although we do not understand why this assembly is inhibited by EcFtsZ.
FIG. 6.
Distribution of (Bs/EcCt)CtYFP under complementation and dominant-negative conditions. JKD7-1 transformed with pJSB2 expressing (Bs/EcCt)CtYFP was cultured overnight at 30°C without arabinose and then the cells were diluted 200-fold into LB containing 0.2% arabinose. The cells were cultured at 42°C (A and C, complementation) or 30°C (B and D, dominant negative). The fluorescence of (Bs/EcCt)CtYFP was observed at 3, 4, or 10 h after starting the culture, as indicated. Arrowheads show Z-ring-like structures in panel C. Bar, 10 μm.
Overlap of suppressor strains for EcFtsZ mutants and foreign FtsZ.
We next tested whether the suppressor strains for foreign FtsZs were related to those already isolated for EcFtsZ point mutants and insertions. As shown in Table 3, PaFtsZ, AvFtsZ, and Bs(EcCt)FtsZ were all functional in SUP{CtYFP1}, the suppressor strain originally generated by EcFtsZ with a C-terminal YFP. The suppressor strains generated by point mutant D212G could also suppress the defects of almost all of the foreign FtsZs; however, some showed a 10-fold reduction in colony number. We prepared several double suppressor strains from our original collection of suppressors of point mutations and YFP insertions. The single suppressor strain SUP{CtYFP1} already suppressed the defects of most foreign FtsZs, and the double suppressor strain SUP{CtYFP1//D96A} extended its range to include (Mpu/EcCt)FtsZ.
We identified two distinct suppressor strains produced by (Mpu/EcCt)FtsZ, designated 11 and 13, and we tested them against all of the mutants in Table 2. Strain 13 was one of the most powerful suppressor strains we have generated, and it appears to be equivalent to the double suppressor strain SUP{CtYFP1//D96A}.
DISCUSSION
Several previous studies have examined whether FtsZ from one bacterial species can function for cell division in another. One successful example was the FtsZ of M. tuberculosis functioning in Mycobacterium smegmatis (12). This is not surprising, since there are only eight amino acid changes apart from the spacer domain. Two reports claimed that very divergent ftsZ could rescue the temperature-sensitive mutant ftsZ84 in E. coli (16, 34). These reports seem questionable, because the sequences are very divergent from EcFtsZ and are even missing the FtsA-binding peptide. ftsZ genes from Caulobacter crescentus (41) and from Buchnera aphidicola (2) failed to complement ftsZ84. This is understandable, since these have several changes in the FtsA-binding peptide which would probably prevent their binding to E. coli FtsA and ZipA.
The C-terminal FtsA/ZipA-binding peptide has been described as the last 17 amino acids in the E. coli sequence (13, 35, 36, 46). However, AvFtsZ and PaFtsZ have substantial variation in the first three amino acids, from KEP to DDL (Fig. 4). It is likely that these three amino acids are not part of the FtsA/ZipA-binding segment, because AvFtsZ and PaFtsZ can generate suppressor strains without changing their peptides.
FtsZ binds directly to MinC (26) and probably to SlmA (3) and ClpX (50), all of which are negative regulators of FtsZ. It also probably binds ZapA, a positive regulator (21). It is likely that divergent FtsZs have lost the ability to bind these E. coli proteins. The observation of mini-cells in all strains containing a divergent FtsZ is consistent with a loss of MinC binding. Although deletion of SlmA alone had no effect on E. coli, deletion of SlmA in a Min minus background was lethal for cells growing in rich medium (3). However, Bernhardt and de Boer did obtain colonies when cells were plated at a 104- to 105- higher density (see Fig. 1F in reference 3). We suggest that these colonies may be generated by second-site mutations equivalent to those in our suppressor strains.
A recent study found that deletion of mreB, -C, or -D was lethal in E. coli (28). Cells lacking any of the three mre genes were round and swollen. The lethality was apparently due to a defect in cell division, because it could be rescued by overexpression of FtsQAZ. Remarkably, the lethality could also be rescued by unknown mutations that occurred at a frequency of 10−5. These suppressor strains kept the round phenotype but were able to achieve division. We suggest that these mutations may be related to those producing our suppressor strains.
Our second-site mutations arose at frequencies of 10−3 to 10−5. This high frequency suggests that they are loss-of-function mutations, perhaps in a multigene pathway. A multigene pathway is also suggested by (i) our observation that a given foreign or mutant FtsZ can generate suppressor strains with different phenotypes and efficiency, (ii) the partial overlap of suppressor strains generated by different mutants, and (iii) the ability to generate double suppressor strains with a wider range of suppression. The pathways may involve negative regulation of FtsZ activity or, alternatively, a mechanical resistance of the cell to constriction that, when relaxed, permits division by a partly defective FtsZ.
Implications for the role of FtsZ in division.
How divergent might a protein be and still function in a foreign species? This probably depends on its interactions with other proteins. A soluble enzyme that operated alone could probably be extremely divergent, as it would only need to be able to fold and function in the new ionic environment. However, a cytoskeletal protein generally has multiple protein-binding partners. To operate in a foreign host it would have to preserve the binding to the foreign binding partners. One example of a cytoskeletal protein that is functional in a foreign host is gamma-tubulin. Gamma-tubulins from human and arabidopsis are both functional in fission yeast (24, 25). This means that the binding interfaces for alpha/beta-tubulin and for the gamma-tubulin accessory proteins must have been preserved. These gamma-tubulins share 70% sequence identity.
MpuFtsZ and BsFtsZ share only 46% and 53% sequence identity to EcFtsZ and 49% identity to each other, substantially less than the 70% identity of the gamma-tubulins. It seems unlikely that these divergent FtsZs would have maintained an interface for a hypothetical motor protein in E. coli. Binding of MpuFtsZ to the downstream proteins in E. coli is even more unlikely, since M. pulmonis does not have homologs of any of these proteins. An example of loss of binding from our own study is the FtsA-binding peptide. The 15-amino-acid peptides of BsFtsZ and MpuFtsZ each have several amino acid changes from EcFtsZ, and these were apparently sufficient to block binding of EcFtsA. We would expect similar loss of binding of any other E. coli proteins to these diverse FtsZs. We note, however, that this is still a speculation, not a definitive conclusion.
We have suggested that FtsZ may generate the constriction force through self-interactions, not requiring interaction with downstream cell division proteins. We should consider one alternative possibility, that FtsA or one of the downstream proteins is the motor that generates the constriction force. In this scenario, FtsZ provides the framework for the Z ring and the attachment sites for FtsA and the downstream proteins. From the arguments above we would suggest that this hypothetical downstream motor would not bind directly to FtsZ protofilaments but would operate by itself or with another downstream protein to generate the constriction force. This model would be consistent with our observations and interpretation, but it does have a philosophical objection. FtsZ is found in almost all bacteria and the euryarchaeota, but each of the other cell division proteins identified in E. coli and B. subtilis are missing in various other species (14, 33). Indeed, Mycoplasma and euryarchaeota have FtsZ but no recognizable homolog of any other known cell division protein. If we suggest that one of the downstream proteins in E. coli is the motor, we have to postulate a novel motor in the species that lack it. This is certainly possible, but we prefer to think that the motor activity is associated with the one protein that is universally present, FtsZ.
The ability of the divergent FtsZs to function in E. coli is consistent with a two-part mechanism for the division machine. One part involves self-interactions of FtsZ to assemble protofilaments and the Z ring. MpuFtsZ can apparently assemble protofilaments in the E. coli cytoplasm as well as in its own, which is expected, since protofilament assembly involves only self-interaction of FtsZ subunits. When provided with the FtsA-binding E. coli-binding peptide, it can assemble into a Z ring. Any force generation mechanism that involved only FtsZ self-interactions would presumably also function in E. coli. The second part of the division machine involves the downstream proteins remodeling the cell wall. The downstream proteins can apparently function by interacting with themselves but do not need to interact directly with FtsZ. The link between the two parts is provided by the E. coli C-terminal peptide, which permits FtsA, ZipA, and perhaps FtsW to bind and localize to the Z ring.
This two-part mechanism is consistent with several studies showing that downstream proteins interact with FtsA and each other, but not with FtsZ (6, 11, 27) (a weak interaction indicated between FtsZ and FtsK [11] may use the FtsA-binding peptide or is nonessential). It is also consistent with a recent study demonstrating that the Z ring assembles in two phases: FtsZ and FtsA assemble first into the Z ring, and most of the downstream proteins associate later (1).
Acknowledgments
This work was supported by NIH grant GM66041.
REFERENCES
- 1.Aarsman, M. E., A. Piette, C. Fraipont, T. M. Vinkenvleugel, M. Nguyen-Disteche, and T. den Blaauwen. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55:1631-1645. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, L., and P. Baumann. 1998. Characterization of ftsZ, the cell division gene of Buchnera aphidicola (endosymbiont of aphids) and detection of the product. Curr. Microbiol. 36:85-89. [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt, T. G., and P. A. de Boer. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 5.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315-1327. [DOI] [PubMed] [Google Scholar]
- 6.Corbin, B. D., B. Geissler, M. Sadasivam, and W. Margolin. 2004. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186:7736-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, K., and J. Lutkenhaus. 1991. ftsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 173:3500-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta, P., A. Dasgupta, S. Bhakta, and J. Basu. 2002. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J. Biol. Chem. 277:24983-24987. [DOI] [PubMed] [Google Scholar]
- 10.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1990. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl. Acad. Sci. USA 87:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 12.Dziadek, J., S. A. Rutherford, M. V. Madiraju, M. A. Atkinson, and M. Rajagopalan. 2003. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology 149:1593-1603. [DOI] [PubMed] [Google Scholar]
- 13.Erickson, H. P. 2001. The FtsZ protofilament and attachment of ZipA-structural constraints on the FtsZ power stroke. Curr. Opin. Cell Biol. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, H. P. 1997. FtsZ, a tubulin homolog, in prokaryote cell division. Trends Cell Biol. 7:362-367. [DOI] [PubMed] [Google Scholar]
- 15.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaikwad, A., V. Babbarwal, V. Pant, and S. K. Mukherjee. 2000. Pea chloroplast FtsZ can form multimers and correct the thermosensitive defect of an Escherichia coli ftsZ mutant. Mol. Gen. Genet. 263:213-221. [DOI] [PubMed] [Google Scholar]
- 17.Geissler, B., D. Elraheb, and W. Margolin. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geissler, B., and W. Margolin. 2005. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol. 58:596-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 20.Goehring, N. W., M. D. Gonzalez, and J. Beckwith. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 61:33-45. [DOI] [PubMed] [Google Scholar]
- 21.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. A. de Boer. 2001. Genetic analysis of the E. coli FtsZ-ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 24.Horio, T., and B. R. Oakley. 2003. Expression of Arabidopsis gamma-tubulin in fission yeast reveals conserved and novel functions of gamma-tubulin. Plant Physiol. 133:1926-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horio, T., and B. R. Oakley. 1994. Human gamma-tubulin functions in fission yeast. J. Cell Biol. 126:1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, Z. L., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 29.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima: quantitation, GTP hydrolysis, and assembly. Cell Motility Cytoskel. 40:71-86. [DOI] [PubMed] [Google Scholar]
- 30.Lu, C. L., M. Reedy, and H. P. Erickson. 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6:862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 34.Momynaliev, K. T., O. V. Smirnova, V. N. Lazyrev, T. A. Akopian, V. V. Chelysheva, J. A. Ayala, A. N. Simankova, S. N. Borchsenius, and V. M. Govorun. 2002. Characterization of the Mycoplasma hominis ftsZ gene and its sequence variability in mycoplasma clinical isolates. Biochem. Biophys. Res. Commun. 293:155-162. [DOI] [PubMed] [Google Scholar]
- 35.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moy, F. J., E. Glasfeld, L. Mosyak, and R. Powers. 2000. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry 39:9146-9156. [DOI] [PubMed] [Google Scholar]
- 37.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90.11753368 [Google Scholar]
- 38.Osawa, M., and H. P. Erickson. 2005. Probing the domain structure of FtsZ by random truncation and insertion of GFP. Microbiology 151:4033-4043. [DOI] [PubMed] [Google Scholar]
- 39.Pichoff, S., and J. Lutkenhaus. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722-1734. [DOI] [PubMed] [Google Scholar]
- 40.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quardokus, E., N. Din, and Y. V. Brun. 1996. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc. Natl. Acad. Sci. USA 93:6314-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.RayChaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redick, S. D., J. Stricker, G. Briscoe, and H. P. Erickson. 2005. Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J. Bacteriol. 187:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romberg, L., and P. A. Levin. 2003. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57:125-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stricker, J., and H. P. Erickson. 2003. In vivo characterization of Escherichia coli ftsZ mutants: effects on Z-ring structure and function. J. Bacteriol. 185:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughan, S., B. Wickstead, K. Gull, and S. G. Addinall. 2004. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58:19-29. [DOI] [PubMed] [Google Scholar]
- 47.Vicente, M., and A. I. Rico. 2006. The order of the ring: assembly of Escherichia coli cell division components. Mol. Microbiol. 61:5-8. [DOI] [PubMed] [Google Scholar]
- 48.Wang, X., and J. Lutkenhaus. 1996. Characterization of FtsZ from Mycoplasma pulmonis, an organism lacking a cell wall. J. Bacteriol. 178:2314-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, X. D., J. A. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weart, R. B., S. Nakano, B. E. Lane, P. Zuber, and P. A. Levin. 2005. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol. Microbiol. 57:238-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 52.Yan, K., K. H. Pearce, and D. J. Payne. 2000. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 270:387-392. [DOI] [PubMed] [Google Scholar]