Abstract
Escherichia coli AcrB is a multidrug efflux transporter that recognizes multiple toxic chemicals and expels them from cells. It is a proton antiporter belonging to the resistance-nodulation-division (RND) superfamily. Asp407, Asp408, Lys940, and Arg971 in transmembrane (TM) helices of this transporter have been identified as essential amino acid residues that probably function as components of the proton relay system. In this study, we identified a novel residue in TM helix 11, Thr978, as an essential residue by alanine scanning mutagenesis. Its location close to Asp407 suggests that it is also a component of the proton translocation pathway, a prediction confirmed by the similar conformations adopted by T978A, D407A, D408A, and K940A mutant proteins (see the accompanying paper). Sequence alignment of 566 RND transporters showed that this threonine residue is conserved in about 96% of cases. Our results suggest the hypotheses that Thr978 functions through hydrogen bonding with Asp407 and that protonation of the latter alters the salt bridging and hydrogen bonding pattern in the proton relay network, thus initiating a series of conformational changes that ultimately result in drug extrusion.
Multidrug transporters cause serious problems in the chemotherapy of cancer as well as in the antibiotic treatment of bacterial infections. These membrane proteins recognize many structurally dissimilar toxic compounds and actively extrude them from the cell. The Escherichia coli AcrB transporter (10, 11), which is a member of the resistance-nodulation-division (RND) family of transporters (26), is responsible for most of the intrinsic drug resistance of this organism (14, 15, 22) and is one of the best-studied multidrug pumps. It occurs as a multiprotein complex (24, 25, 32), with an outer membrane channel TolC protein (4, 9) and a periplasmic linker protein, AcrA (10), and this complex structure allows the direct export of drugs to the external medium (14). The structural work of Murakami et al. (12) revealed that unbound AcrB is a homotrimer, where each subunit contains 12 transmembrane (TM) helices and two large periplasmic domains, between TM1 and TM2 and between TM7 and TM8. The top of the periplasmic domain of AcrB is thought to associate with TolC.
AcrB utilizes proton motive force (PMF) as energy for its transport function (10, 30, 31). The molecular mechanism that couples proton translocation to the efflux of drugs has not been elucidated completely. However, the simplest mechanism seems to operate in a small multidrug transporter, EmrE, of E. coli (19, 23, 28). In this transporter, there exists only one membrane-embedded charged residue, Glu14, and this residue is involved in the recognition of both the substrates and the coupling proton. For another well-studied PMF-dependent transporter, the lactose permease LacY from E. coli (1), the proton translocation pathway involves several charged residues in the TM helices and is separate from the substrate transport pathway.
Charged residues embedded in the membrane were also shown to be essential for the activities of RND transporters. In 1999, Goldberg et al. (6) showed for the toxic cation efflux pump CzcA of Ralstonia that Asp402 and Asp408 of TM4 are essential for the pumping of divalent cations. Furthermore, by proteoliposome reconstitution, they showed that mutants in which these aspartate residues were changed to neutral residues were still capable of catalyzing facilitated diffusion of Zn2+, producing a strong case that the residues are involved specifically in proton translocation. Two years later, Guan and Nakae (7) showed for an AcrB homolog, MexB, of Pseudomonas aeruginosa that three charged residues in the transmembrane domain, Asp407, Asp408 (both in TM4), and Lys939 (in TM10), which are also conserved in AcrB, are essential for transport. Asp407 corresponds to Asp408 of CzcA, whereas Asp408 is not conserved in CzcA and other RND pumps of metal cations. Remarkably, the crystallographic structure of AcrB shows that Asp407 is very close to the Lys940 side chain (12). One additional essential charged residue, Arg971 (13), exists close to the cytoplasmic end of TM11. Thus, an Asp407-Lys940-Arg971 proton translocation pathway can be hypothesized (13). However, other amino acids might also be involved in this pathway.
In this study, we identified Thr978 in TM11 of AcrB as another key residue in the function of this transporter. Thr978 is highly conserved in RND transporter sequences and is located at a hydrogen-bonding distance from Asp407. Our hypothesis that Thr978 is a component of the proton relay network was confirmed by the accompanying X-ray crystallographic studies of Thr978 mutant proteins, which are shown to be very similar in conformation to proteins with mutations of other residues of this putative network, i.e., Asp407, Asp408, and Lys940 (21).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli DH5α was used for the construction and propagation of various plasmid constructs. Cells were grown in Luria-Bertani (LB) broth supplemented with ampicillin (100 μg/ml) when necessary.
TABLE 1.
E. coli strains and plasmids used for this study
| Strain or plasmid | Description or genotype | Reference or source |
|---|---|---|
| Strains | ||
| DH5α | Standard host strain for cloning | 17 |
| HNCE1a | marR1 acrB::kan ΔacrD | 3 |
| Plasmids | ||
| pSportI | High-copy-number cloning and expression vector with amp marker and lac-inducible expression | Gibco BRL |
| pUCK151A | A 6.5-kb BglII fragment containing the entire acrAB operon cloned into the pUC19 vector | 10 |
| pSAcrBHis | acrB gene with His4 tag sequence at its 3′ end cloned into pSportI | This study |
Construction of plasmid pSAcrBHis.
pSAcrBHis, which was used to express hexahistidine-tagged AcrB, was constructed from pUCK151A (10) by the overlap extension method (8), using the primers NSFw (5′-ATGCATGTCGACTCAGCCTGAACAGTCCA-3′), HisRv (5′-TCAGTGGTGGTGGTGATGATGATCGACAGTATGG-3′), HisFw (5′-CATCACCACCACCACTGATACAACGTGTAATCACTA-3′), and CB104Rv (5′-GCGGGATCCTTATCCGTGGTTAATACTG-3′) (singly underlined and italic sequences represent SalI and BamHI restriction sites, respectively, and doubly underlined sequences encode four histidine residues and were introduced at the 3′ end of the acrB gene). The amplified fragment was digested with SalI and BamHI, and the 3.3-kb fragment was inserted into the corresponding site of pSportI, from which acrB was expressed under the control of the lac promoter. Since the intrinsic C-terminal sequence of AcrB is His-His, the resulting His-tagged AcrB protein has a His6 sequence at its C terminus.
Site-specific mutagenesis.
Site-directed mutagenesis was carried out by using the QuickChange site-directed mutagenesis system from Stratagene. Point mutations were introduced into the plasmid pSAcrBHis by PCR, using Pfu Ultra high-fidelity DNA polymerase (Stratagene) according to the manufacturer's instructions. The coding region for acrB in each plasmid was sequenced to confirm that there were no unintended nucleotide sequence alterations.
Drug susceptibility assays.
E. coli HNCE1a (3) cells harboring pSportI-derived plasmids were tested, without IPTG (isopropyl-β-d-thiogalactopyranoside) induction, for drug susceptibility by the gradient plate method (2). This method was used because conventional MIC determinations tended to give nonreproducible results for strains expressing AcrB alone from multicopy plasmids. A linear concentration gradient of cholic acid (Sigma) was prepared in square LB agar plates containing 8,000 μg/ml of cholic acid in the lower layer. Mid-exponential-phase cultures were diluted to an optical density at 660 nm of 0.1 with LB broth and streaked as a line across the plate, in parallel with the drug gradient. Bacterial growth across the plates, from low to high drug concentrations, was measured after 24 h at 37°C. All mutants were assayed at least four times, with strains harboring wild-type pSAcrBHis and the vector pSportI as controls. The relative activity of each mutated AcrB protein was calculated by dividing the length of growth of each mutant strain, minus that of the strain carrying the vector alone, by the length of growth of the strain containing pSAcrBHis, minus that of the vector-containing strain, and multiplying the result by 100. Thus, full efflux activity and no activity should produce values of 100 and 0%, respectively.
Analysis of AcrB expression levels in the inner membrane fraction.
Exponential-phase cells grown in LB broth were harvested, resuspended in buffer A (10 mM HEPES-KOH [pH 7.0], 1 mM dithiothreitol), and broken by sonication. Unbroken cells were removed by low-speed centrifugation, and the total membrane fraction was collected by ultracentrifugation at 150,000 × g for 1 h at 4°C. Inner membrane proteins were solubilized in buffer A containing 1.5% (wt/vol) N-lauroylsarcosine. The extract was ultracentrifuged again, and proteins in the supernatant were resolved by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to a nitrocellulose membrane (Bio-Rad) for Western blot analysis. Binding of polyclonal rabbit anti-AcrB (31) was detected with an alkaline phosphatase-conjugated anti-rabbit secondary antibody (Sigma). Protein-antibody conjugates were visualized with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate.
RESULTS
Alanine scanning mutagenesis of the TM domains of AcrB.
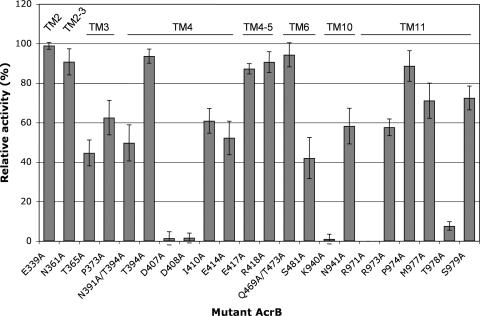
To identify an amino acid(s) which might be involved in proton translocation, we mutagenized some amino acid residues in TM helices whose side chains might work as proton donors or acceptors in hydrogen bonds. We tried to include most charged residues (Asp, Glu, Lys, and Arg), except for those residues located on the outer surface of the molecule in the three-dimensional structure of Murakami et al. (12). These included Lys334, Lys342, Glu346, Arg363, Arg432, Lys433, Arg468, Glu947, Lys950, Asp951, Asp954, Lys955, Glu956, Lys958, Glu962, Asp966, and Arg969. We also included some residues containing hydroxyl or amide groups (Ser, Thr, Asn, and Gln) as well as Pro373 and Met977. The transport activity of each mutant was evaluated by its drug susceptibility on gradient plates containing 8,000 μg/ml cholic acid in the lower layer, as described in Materials and Methods. Since overexpression of AcrB alone tended to produce nonreproducible drug susceptibility patterns (29), acrB was expressed without IPTG induction. The lengths of growth across the gradient were reproducibly 60 to 70 and 24 to 31 mm for the acrB::kan ΔacrD host strain HNCE1a expressing wild-type AcrB and the strain containing vector only, respectively. As shown in Fig. 1, conversion to Ala of Asp407, Asp408, and Lys940, which were previously shown to be essential in the AcrB homolog MexB of P. aeruginosa (7), totally abolished activity, as did that of Arg971, again shown earlier to be essential for AcrB activity (13). On the other hand, the conversion of other charged amino acid residues, such as Glu339, Glu414, Glu417, Arg418, and Arg973, did not severely affect the transport function (Fig. 1).
FIG. 1.
Alanine scanning mutagenesis of AcrB residues that might be involved in proton translocation. The mutant AcrB proteins were expressed in the acrB acrD host strain HNCE1a, and their efflux activities were estimated from their levels of resistance to cholate. The positions of the mutations in the AcrB protein are indicated at the top. For details, see Materials and Methods.
A significant outcome of this mutagenesis study was the identification of Thr978, which is located in the TM11 helix, as an essential residue, as conversion of this residue to alanine nearly completely abolished the activity of the pump (Fig. 1). The transport defect of the T978A mutant, as well as those of the D407A, D408A, and K940A mutants, was not limited to the transport of cholate, as host strains expressing these mutant proteins were found to be fully susceptible to other substrates of the AcrB pump, including tetracycline, chloramphenicol, ciprofloxacin, erythromycin, dequalinium, ethidium bromide, rhodamine 6G, and acriflavine (results not shown).
Asn361 (TM2-3 loop), Thr365 (TM3), Asn391, Thr394 (both in TM4), Gln469, and Thr473 (both in TM6) are located between the Asp407-Lys940-Asp408 ion-pair/hydrogen-bonding structure and the central cavity in the crystal structure (12). Conversion of these residues to alanine did not abolish transport, although the conversion of Thr365 or Asn391 significantly reduced the relative activity (Fig. 1).
Notably, all of our mutant proteins were produced in the inner membrane at levels that are not very different from that of wild-type AcrB (Fig. 2). Although the expression levels of some mutant proteins appeared to be somewhat different, this seems unlikely as the explanation for the reduced pumping activity of some mutants, since there was no correlation between the apparent expression level and drug resistance. Furthermore, the small variation in the observed expression level showed good correlation with the cell density at the time of harvest, and examination of the cells of the same strains harvested at different optical densities at 600 nm confirmed this correlation (results not shown).
FIG. 2.
Expression levels of mutant AcrB proteins. Inner membrane proteins were extracted from exponential-phase cells of the HNCE1a host strain containing plasmids with the mutant acrB genes, as described in Materials and Methods. A fixed amount (4.5 μg protein) was applied to each lane, and proteins were separated by sodium dodecyl sulfate-PAGE. AcrB was detected by a rabbit polyclonal antibody, as described in Materials and Methods. Scanning of the Western blots showed that the expression levels were similar, with a standard deviation of 27% of the average. The small variation in expression levels had no correlation with the levels of drug resistance. M, molecular size marker.
Purified T978A protein migrated as a trimer by blue native PAGE, suggesting that trimerization was not affected by the mutation (results not shown).
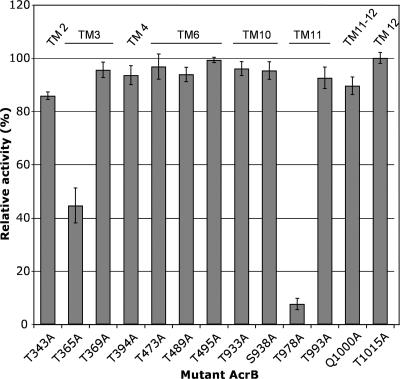
Thr978 is the only essential and conserved threonine residue in the TM segments.
In the transmembrane region of AcrB, there are 15 Thr residues, and 10 of these are conserved as Thr or Ser in seven homologous RND multidrug transporters, including AcrB, AcrF, AcrD, and YhiV of E. coli, MexB and MexD of P. aeruginosa, and MtrD of Neisseria gonorrhoeae (not shown). We converted all of these conserved Thr residues to alanine and evaluated their transport functions (Fig. 3). Thr993, which is present in the same TM helix as Thr978, was also included. Because the crystal structure (12) suggests that Gln1000 may work as a bridge between Thr343 (TM2) and Thr993 (TM11), Gln1000 was also tested by substitution. Although the T365A mutant had a 50% decreased activity, as shown in Fig. 1, no other substitution except for T978A affected the transporter function significantly, showing that Thr978 is uniquely important.
FIG. 3.
Activities of AcrB mutants in which one of the conserved Thr residues in the transmembrane domain was changed to Ala. All 10 conserved Thr residues plus Thr993, located in the same TM helix as Thr978, and Asn1000 were examined by alanine scanning mutagenesis, and the activity of each protein was assessed by the extent of resistance to cholate of the host strain expressing the protein, as described in the legend to Fig. 1.
Ser residues at positions 481, 938, and 979 are conserved in the seven homologues mentioned above. Conversion of these serine residues to alanine did not inactivate the transport function of AcrB (Fig. 1 and 3).
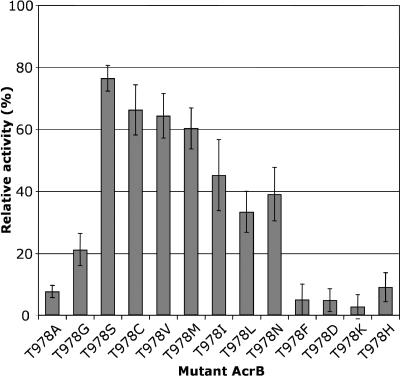
Replacement of Thr978 with other amino acid residues.
Based on the crystal structure of AcrB (12), the side chain oxygen of Thr978 and the carboxylate oxygen of Asp407 are close enough (3.2 Å) to form a hydrogen bond. Thus, the easiest interpretation of the inactivity of the T978A mutant was that the hydrogen bond between Thr978 and Asp407 is functionally important. When Thr978 was replaced with serine, which could form a hydrogen bond with Asp407, the T978S mutant retained 77% ± 4% of the wild-type activity (Fig. 4). Similarly, substitution with cysteine, in which the thiol group might work as a hydrogen bond donor, also resulted in retention of 66% of the activity (Fig. 4). A change to asparagine, with its potential for hydrogen bond formation, also produced a moderately active transporter (Fig. 4).
FIG. 4.
Activities of AcrB mutants in which Thr978 was replaced by various other amino acids. The activities of the mutant AcrB proteins were determined as described in the legend to Fig. 1.
However, when Thr978 was replaced by valine, the activity was retained almost at the level of the T978C mutant, even though there is no hydrogen bond-forming group in the valine side chain (Fig. 4). Similarly, significant activity was found when the residue was replaced with other residues with aliphatic side chains, such as isoleucine or leucine, although the bulky side chain of phenylalanine was not tolerated (Fig. 4).
Double mutants containing the T978V mutation.
The retention of activity by the T978V mutant suggested that if hydrogen bonding between Asp407 and Thr978 is essential, an alternative hydrogen bond might be formed between another residue and Asp407. Since the Ser979 and Pro974 side chains in TM11 appeared to be close to that of Thr978 in the crystal structure (12), these two residues were examined by combining their alteration with the T978V mutation (Table 2).
TABLE 2.
Activities of AcrB proteins with mutations in TM11
| Mutation(s) | Residue at positiona:
|
Relative activity (%)b | ||
|---|---|---|---|---|
| 974 | 978 | 979 | ||
| Wild type | Pro | Thr | Ser | 100 |
| T978A | Pro | Ala | Ser | 8 ± 2 |
| T978V | Pro | Val | Ser | 64 ± 7 |
| S979A | Pro | Thr | Ala | 73 ± 5 |
| T978V S979A | Pro | Val | Ala | 6 ± 3 |
| P974A | Ala | Thr | Ser | 88 ± 7 |
| T978V P974A | Ala | Val | Ser | 65 ± 7 |
| T978S | Pro | Ser | Ser | 77 ± 4 |
| T978S S979A | Pro | Ser | Ala | 7 ± 4 |
Mutated residues are shown in bold.
Data are means ± standard deviations.
Single alterations, such as S979A or P974A, did not affect the transport activity (as deduced from drug resistance levels). Introducing the S979A mutation into the T978V background, however, destroyed the activity nearly completely, suggesting that the hydroxyl moiety of S979 may substitute for that of Thr978 in the T978V mutant. However, adding the S979A mutation to the T978S background also led to the loss of function (Table 2, bottom row), possibly because even substitution with serine at position 978 creates a situation that requires assistance from the neighboring serine residue. The P974A/T978V double mutant also retained 65% of the wild-type activity, suggesting that the Pro974 residue is not important in the formation of this network.
Thr978 is highly conserved among RND family homologues.
The amino acid sequence around Thr978 was reported as the highly conserved region motif C in the 14 members of the RND family and was suggested to play an essential structural or functional role in these proteins (16). The multiple alignment was expanded to include nearly 600 RND transporter homologues by using the Pfam database (http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF00873 [accessed on 25 February 2005]) (Table 3).
TABLE 3.
Conservation of Thr978 and related residues in 566 RND transporter sequencesa
| Residues at positions 407-408-978-979b-981-982 | No. of transporter sequences |
|---|---|
| Asp-Asp-Thr-Ser/Thr/Ala-(Ala)c-(Phe/Ala)c | 437 |
| Asp-Asn-Thr-Ala-Thr-Thr | 1 |
| Asp-various-Thr-various-Thr-various | 27 |
| Asp-various-Thr-various-various-various | 77 |
| Asp-Ser/Cys-Ser-Thr/Ser-Thr-Ser | 2 |
| Asp-Asn/Thr-Ala/Gly/Asn-Ala/Ser/Asn-Thr-Thr | 19 |
| Asp-Gly/Ala-Ala-Ala/Gly-Leu/Ala-Ala/Asn | 3 |
The Pfam database (http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF00873) was accessed on 25 February 2005. Partial sequences were excluded from tabulation.
Ser, Thr, or Ala occupies this position in 418 of 437 cases.
These positions are occupied predominantly, but not always, by Ala and Phe (or Ala), respectively.
Asp407 of AcrB was completely conserved in all 566 sequences examined, after the removal of partial sequences. Asp408 was conserved in 437 sequences (77%). Lys940 was conserved in 366 sequences and was replaced by Arg in 61 homologues.
Thr978, which was found to be essential in this study, was conserved in 542 sequences and was replaced by Ser in two cases. Although the Thr residue was replaced by other residues in 22 sequences (Ala in 17 cases, Gly in 3 cases, and Asn in 2 cases), in most of these proteins (19 sequences) positions 981 and 982, which face the same side of TM11 as Thr978, contain two consecutive Thr residues (Table 3), which presumably are involved in a similar hydrogen-bonding scheme.
DISCUSSION
The molecular mechanism of drug translocation by the PMF-dependent drug transporter AcrB is not known. One needs to know how H+ transport and substrate transport are coupled and how the protein changes its conformation during this process. In this work, we identified Thr978, which is close to the essential Asp-Lys-Asp ion-pair/hydrogen-bonding network in the TM segments, as an important residue for the function of AcrB. The T978A mutant almost completely lost its function, even though the mutant protein was properly expressed, localized to the inner membrane of the cells, and detected as a trimer by native PAGE. It was also crystallized as a trimer, as shown in the accompanying paper (21). Thr978 was conserved in 542 RND sequences among 566 examined, and in 21 of 24 nonconserved cases, it was either replaced by Ser or had the Thr-Thr sequence one turn downstream on the same helix. Although AcrB contains 15 threonine residues in TM helices, Thr978 appeared to be the only residue whose conversion to alanine resulted in negligible transport function.
Based on the crystal structure of wild-type AcrB (12), the side chains of Thr978 and Asp407 are close enough (3.2 Å) to form a hydrogen bond (see Fig. 1 in reference 21), suggesting that Thr978 is part of the putative proton relay network involving Asp407, Lys940, and Asp408. The accompanying crystallographic study of proteins with mutations in these residues indeed showed that there is a wide-ranging, characteristic conformational alteration in the T978A mutant protein which is very similar to that seen in D407A as well as D408A and K940A mutant AcrB proteins (21), giving strong support to this hypothesis.
Replacement of Thr978 with cysteine in the T978C mutant retained 66% of the wild-type activity. Cysteine scanning mutagenesis is a commonly used technique for structural and functional studies of membrane transporters. However, it is possible that the importance of Thr or Ser as a hydrogen-bonding partner might be missed by this approach. The T978V mutant retained 64% of the wild-type activity, but the T978V/S979A double mutant was inactive, possibly because the side chain hydroxyl group of S979 may contribute to the hydrogen-bonding scheme in this background.
Assuming that Thr978 is part of the proton relay network involving Asp407, Lys940, and Asp408, we can speculate on how the Thr978 side chain may function in proton translocation. In secondary transporters coupled to proton motive force, a crucial event appears to be the protonation of one of the acidic residues in the transmembrane domain. Examples include Glu14 in the small multidrug resistance family transporter EmrE (19) and Glu269 (and subsequently Glu325) in LacY (1). In the crystal structure of wild-type AcrB (12), it is most likely that the ɛ-amino group of Lys940 is protonated and the carboxyl group of Asp407 is deprotonated because of their close proximity to each other. The protonation of the Asp407 carboxyl group will destroy the salt bridge interaction between these groups and will also affect the hydrogen-bonding interaction between Asp407 and Thr978. The resultant distortion of this region may trigger a series of conformational alterations that would ultimately result in the pumping of drugs. The wide-ranging conformational changes in the T978A mutant, as well as in D407A, D408A, and K940A mutant AcrB proteins, as described in the accompanying paper (21), probably give us glimpses of this process.
There are a few earlier reports of the importance of Thr residues in transporters. In the lactose permease LacY, Thr348, located near the end of TM helix XI, was identified as being required for transport activity by cysteine scanning mutagenesis (5), and Thr393 in TM helix XII was identified as essential by random mutagenesis of the residues in this helix (20). However, the crystal structure (1) shows that these residues are far away from the proton relay region, and the roles played by these Thr residues are not clear at present. In the multidrug transporter Cdr1p of Candida albicans, Thr1351 of TM helix 11 was found to be essential (18). Although Cdr1p is an ATP-binding cassette (ABC) transporter, a recent paper reported that the integral membrane domain of an ABC transporter can function as a PMF-driven transporter (27), and it would be interesting to know the detailed structure surrounding this essential threonine residue.
Acknowledgments
This study was supported in part by USPHS research grant AI-09644. Y.T. was supported by a postdoctoral fellowship from the Japan Society for Promotion of Science during the early stages of this study.
REFERENCES
- 1.Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610-615. [DOI] [PubMed] [Google Scholar]
- 2.Bryson, V., and W. Szybalzski. 1952. Microbial selection. Science 116:45-51. [PubMed] [Google Scholar]
- 3.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frillingos, S., M. Sahin-Tóth, J. Wu, and H. R. Kaback. 1998. Cys-scanning mutagenesis: a novel approach to structure-function relationships in polytopic membrane proteins. FASEB J. 12:1281-1299. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg, M., T. Pribyl, S. Juhnke, and D. H. Nies. 1999. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-Nodulation-cell division protein family. J. Biol. Chem. 274:26065-26070. [DOI] [PubMed] [Google Scholar]
- 7.Guan, L., and T. Nakae. 2001. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 183:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 9.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 10.Ma, D., D. A. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, D., D. A. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 12.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 13.Murakami, S., and A. Yamaguchi. 2003. Multidrug-exporting secondary transporters. Curr. Opin. Struct. Biol. 13:443-452. [DOI] [PubMed] [Google Scholar]
- 14.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):S32-S41. [DOI] [PubMed] [Google Scholar]
- 15.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Shukla, S., S. V. Ambudkar, and R. Prasad. 2004. Substitution of threonine-1351 in the multidrug transporter Cdr1p of Candida albicans results in hypersusceptibility to antifungal agents and threonine-1351 is essential for synergic effects of calcineurin inhibitor FK520. J. Antimicrob. Chemother. 54:38-45. [DOI] [PubMed] [Google Scholar]
- 19.Soskine, M., Y. Adam, and S. Schuldiner. 2004. Direct evidence for substrate-induced proton release in detergent-solubilized EmrE, a multidrug transporter. J. Biol. Chem. 279:9951-9955. [DOI] [PubMed] [Google Scholar]
- 20.Stewart, C., J. Bailey, and C. Manoil. 1998. Mutant membrane protein toxicity. J. Biol. Chem. 273:28078-28084. [DOI] [PubMed] [Google Scholar]
- 21.Su, C.-C., M. Li, R. Gu, Y. Takatsuka, G. McDermott, H. Nikaido, and E. W. Yu. 2006. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 188:7290-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tate, C. G., I. Ubarretxena-Belandia, and J. M. Baldwin. 2003. Conformational changes in the multidrug transporter EmrE associated with substrate binding. J. Mol. Biol. 332:229-242. [DOI] [PubMed] [Google Scholar]
- 24.Tikhonova, E. B., and H. I. Zgurskaya. 2004. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 279:32116-32124. [DOI] [PubMed] [Google Scholar]
- 25.Touzé, T., J. Eswaran, E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol. Microbiol. 53:697-706. [DOI] [PubMed] [Google Scholar]
- 26.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 27.Venter, H., R. A. Shilling, S. Velamakanni, L. Balakrishnan, and H. W. Veen. 2003. An ABC transporter with a secondary-active multidrug translocator domain. Nature 426:866-870. [DOI] [PubMed] [Google Scholar]
- 28.Yerushalmi, H., and S. Schuldiner. 2000. A model for coupling of H+ and substrate fluxes based on “time-sharing” of a common binding site. Biochemistry 39:14711-14719. [DOI] [PubMed] [Google Scholar]
- 29.Yu, E. W., J. R. Aires, G. McDermott, and H. Nikaido. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 187:6804-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zgurskaya, H. I., and H. Nikaido. 1999. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285:409-420. [DOI] [PubMed] [Google Scholar]
- 31.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]