Abstract
The draft genome sequence of Mannheimia haemolytica A1, the causative agent of bovine respiratory disease complex (BRDC), is presented. Strain ATCC BAA-410, isolated from the lung of a calf with BRDC, was the DNA source. The annotated genome includes 2,839 coding sequences, 1,966 of which were assigned a function and 436 of which are unique to M. haemolytica. Through genome annotation many features of interest were identified, including bacteriophages and genes related to virulence, natural competence, and transcriptional regulation. In addition to previously described virulence factors, M. haemolytica encodes adhesins, including the filamentous hemagglutinin FhaB and two trimeric autotransporter adhesins. Two dual-function immunoglobulin-protease/adhesins are also present, as is a third immunoglobulin protease. Genes related to iron acquisition and drug resistance were identified and are likely important for survival in the host and virulence. Analysis of the genome indicates that M. haemolytica is naturally competent, as genes for natural competence and DNA uptake signal sequences (USS) are present. Comparison of competence loci and USS in other species in the family Pasteurellaceae indicates that M. haemolytica, Actinobacillus pleuropneumoniae, and Haemophilus ducreyi form a lineage distinct from other Pasteurellaceae. This observation was supported by a phylogenetic analysis using sequences of predicted housekeeping genes.
Mannheimia haemolytica serotype A1 is the principle bacterial pathogen associated with bovine respiratory disease complex, also known as bovine shipping fever (25, 31). M. haemolytica resides in the upper respiratory tracts of healthy ruminants, although in immunocompromised animals, such as those with a preexisting viral infection, M. haemolytica can descend into the lungs, leading to pneumonia. The mortality and morbidity associated with this disease cause substantial losses to the cattle industry (25, 31). Hence, much research on M. haemolytica is directed at understanding its virulence and designing vaccines to protect cattle (31).
M. haemolytica is a member of the family Pasteurellaceae, which is classified among the γ-proteobacteria and includes human and animal pathogens of the genera Mannheimia, Pasteurella, Haemophilus, Actinobacillus, and Histophilus (31). M. haemolytica (formerly Pasteurella haemolytica) is classified in the genus Mannheimia based upon 16S rRNA sequence phylogeny and DNA-DNA hybridizations (3). The family Pasteurellaceae includes several members whose genomes have been sequenced: Haemophilus influenzae, Pasteurella multocida, Mannheimia succiniciproducens, Actinobacillus pleuropneumoniae, Histophilus somni, Haemophilus ducreyi, and Actinobacillus actinomycetemcomitans.
The primary virulence factor of M. haemolytica is considered the leukotoxin. It is a calcium-dependent toxin that is a member of the RTX family of toxins. Cytolytic at low concentrations and apoptotic at high concentrations, the leukotoxin provokes an inflammatory response that can lead to intense edema and necrosis in the lungs of infected cattle (36). Despite the importance of the leukotoxin, leukotoxin-defective strains are only partially attenuated, suggesting that other virulence factors are important in pathogenesis (31). Furthermore, because M. haemolytica undergoes a niche change from commensal to pathogenic, the control of the expression of its virulence factors is also of significant interest.
M. haemolytica BAA-410, which was isolated from a calf with bovine respiratory disease complex, was sequenced to draft coverage. Annotation and analysis of the genome provided an opportunity to discover new features potentially related to virulence. It also permitted identification of possible transcriptional regulatory networks and a natural competence system. The genome sequence also allowed comparisons to other sequenced Pasteurellaceae genomes and an evaluation of M. haemolytica's place within the Pasteurellaceae lineage.
MATERIALS AND METHODS
Bacterial growth and DNA isolation.
M. haemolytica strain BAA-410 was selected for sequencing because it is virulent in cattle and is the parent of strain SH1217, which is used for genetic studies (23). SH1217 was created by repeated passage of BAA-410 in the presence of acridine orange to render the strain streptomycin sensitive, and SH1217 has been used to make chromosomal mutations (47). Template DNA was harvested from BAA-410 grown in brain heart infusion broth at 37°C with shaking. Cells from overnight cultures were lysed using lysozyme and Triton X-100, and chromosomal DNA was isolated by ultracentrifugation in a CsCl gradient (62).
Library construction and sequencing.
Library construction, DNA sequencing, and assembly were performed as previously described (48). A cosmid library was also created for end sequencing and scaffolding. BAA-410 DNA was partially digested with Sau3A. Digested DNA was size fractionated by agarose gel electrophoresis, and 30- to 40-kb fragments were ligated into pJG2560, a pBeloBAC11-derived vector with a lac-inducible P1 lytic replicon (J. Gioia, G. M. Weinstock, and S. K. Highlander, unpublished). Cosmids were packaged using the Gigapack III XL packaging kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The ends of the ca. 35-kb bacterial artificial chromosome inserts were sequenced from primers flanking the insertion site in pJG2560.
Sequence assembly.
A total of 39,949 reads were assembled after using phred for base calling (21, 22) and the Atlas assembly system (30). The reads were assembled into 152 contigs of >1 kb, totaling 2,569,125 bases (Table 1). Read pairs were used to assemble the contigs into 103 scaffolds. The scaffolds were arbitrarily ordered into one artificial superscaffold, using Ns to bridge the gaps.
TABLE 1.
M. haemolytica genome general features
| Feature | Value |
|---|---|
| Total length of contigs (bp) | 2,569,125 |
| Coverage (fold) | 8 |
| No. of contigs | 152 |
| No. of scaffolds | 103 |
| No. of gaps | 148 |
| G+C content (%) | 41 |
| No. of tRNAs | 42 |
| No. of CDS | 2,839 |
| No. (%) of CDS with assigned function | 1,966 (69) |
| No. (%) of Pasteurellaceae conserved hypothetical proteins | 113 (4) |
| No. (%) of CDS unique to M. haemolytica | 436 (15) |
| No. (%) of CDS broken at contig ends | 162 (5.7) |
| No. (%) of pseudogenesa | 72 (3) |
| Average CDS size(bp) | 833 |
| Coding density (%) | 92 |
Given that this is a draft sequence, some pseudogenes may be the result of sequencing errors.
Genome annotation.
The superscaffold was used for gene identification and annotation. Glimmer (17) and GeneMark (45) were used to predict protein-coding sequences (CDS), and several databases were used for comparison of sequences. GenBank nr and CDD databases (70) were used for all CDS, and specialized databases such as ExPASy ENZYME (27), the Transporter Classification Database (60), PSORTb (59), TIGR Transporter BLAST Search (http://tigrblast.tigr.org/er-BLAST/index.cgi?project=transporter), MEROPS the Peptidase Database (56), SignalP (7), LipoP (37), and Helix-Turn-Helix Predictor (19) were used when applicable. Sequences of tRNAs were identified using tRNAscan-SE (44). All CDS were annotated manually by two annotators, and discrepancies were resolved to reach a consensus annotation. Annotation was done using previously described conventions (48). The authors believe that a labor-intensive manual evaluation of each CDS is more accurate than automated annotation, as effort is devoted to assigning accurate definitions to each CDS and extant errors within databases are more likely to be avoided.
Phylogenetic tree construction.
Sequences for tree construction were obtained from this work, NCBI (http://www.ncbi.nlm.nih.gov/), and the A. actinomycetemcomitans genome sequencing project (http://www.genome.ou.edu/act.html). Fifty housekeeping genes common to all nine genomes were selected for analysis. Genes were selected based upon use in previous phylogenetic studies (11, 73). Sequences were aligned using the European Bioinformatics Institute ClustalW (10) server with default parameters to produce alignments in PHYLIP format. The sequences of each gene were aligned individually to confirm alignments before concatenation. Tree construction was done using the SEQBOOT, DNAPARS, CONSENSE, RETREE, and DNAML programs of PHYLIP (24). Phylodendron (28) was used to display the trees. The genes used were pykA, pgk, atpA, atpB, atpD, atpF, atpG, atpH, holB, dnaE, dnaG, uvrB, uvrC, rpoA, rpoB, infB, rluA, truA, truB, truC, dnaJ, groEL, rplA, rplF, rplK, rplJ, rplL, rplO, rplP, rplQ, rplR, rplW, rpmC, rplD, rpsB, rpsD, rpsC, rpsF, rpsG, rpsH, rpsI, rpsJ, rpsK, rpsN, rpsO, rpsP, rpsQ, rpsR, rpsS, and rpsT.
Project accession numbers.
This Whole Genome Shotgun project has been deposited at GenBank under project accession no. AASA00000000. The version described in this paper is the first version, accession no. AASA01000000.
RESULTS AND DISCUSSION
Draft genome sequence.
Sequencing of the M. haemolytica BAA-410 genome was slated only to draft coverage, and thus the quality of the draft product was evaluated (Table 1). The draft genome sequence has 152 contigs, linked into 103 scaffolds, spanning 2.57 Mb. There are 148 gaps in the sequence (46 captured gaps within scaffolds and 102 gaps between scaffolds), amounting to one gap per 17.3 kb of sequence. This degree of contiguity is adequate for gene prediction, and 2,839 predicted CDS and 42 tRNAs were annotated. The average contig length is 16.9 kb, containing 18.7 CDS, and the average scaffold length is 24.9 kb, containing 27.6 CDS. Blastn analysis of the sequence against itself indicates that a maximum of 35 gaps (24%) are associated with repeated sequences at contig ends. Therefore, other factors besides repeated sequences, such as cloning bias, may contribute to the gaps. More than half of the contig ends are within coding sequences, and the gaps within the assembled sequence disrupt 162 CDS, all of which are labeled “incomplete.” Most CDS (69%) have an assigned function or definition, although 436 are hypothetical proteins unique to M. haemolytica. Additionally, the draft genome encodes 113 conserved hypothetical proteins that have orthologs only in the family Pasteurellaceae. Additional features of the genome are listed in Table 1.
The quality of the draft M. haemolytica BAA-410 sequence was addressed by a comparison to previously sequenced genes. All 91 previously sequenced M. haemolytica A1 genes or gene fragments in GenBank are present in the draft genome of strain BAA-410, indicating that the data are comprehensive. The aggregate DNA sequence identity between the previously determined sequences and those from BAA-410 is 99.2%. Nevertheless, the draft nature of the genome means that some features are missing because they lie in nonsequenced gaps. For example, tRNAs for tyrosine, asparagine, and phenylalanine are not found in the draft sequence, and the rRNA operons are all incomplete. Additionally, it is not known if the 162 CDS broken at contig ends are actually intact genes.
Genome content.
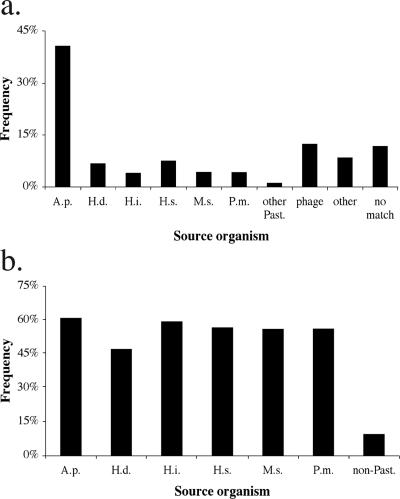
A Blastx analysis of every M. haemolytica CDS allowed a general comparison of M. haemolytica to other organisms. The source organism of the best match of each BLAST report was recorded for every CDS (Fig. 1a). For over 40% of CDS the best match was to a predicted A. pleuropneumoniae protein; this value was far greater than that for any other species. However, when the analysis was extended beyond the best match, it became apparent that the number of M. haemolytica CDS with A. pleuropneumoniae orthologs is not substantially greater than the number of orthologs in related Pasteurellaceae species (Fig. 1b). Slightly more than half of all M. haemolytica CDS have orthologs in the other sequenced Pasteurellaceae genomes, with the exception of H. ducreyi. Only 9.3% of M. haemolytica CDS have only non-Pasteurellaceae orthologs.
FIG. 1.
Distribution of M. haemolytica CDS orthologs. The Blastx reports of all M. haemolytica CDS were analyzed with respect to source organisms of alignments. a. Percentages of M. haemolytica CDS with best match to specific source organisms or classes. b. Percentages of CDS with any alignment to specific source organisms. High-scoring pairs with expect values of >e−5 were not considered. Abbreviations: A.p., A. pleuropneumoniae; H.d., H. ducreyi; H.i., H. influenzae; H.s., H. somni; M.s., M. succiniciproducens; P.m., P. multocida; other Past., other Pasteurellaceae species; non-Past., only non-Pasteurellaceae organisms.
Although the number of M. haemolytica CDS with A. pleuropneumoniae orthologs is not remarkable with respect to other source organisms analyzed here (Fig. 1b), there is a clear distinction between A. pleuropneumoniae and other bacteria when only the best match is considered (Fig. 1a). These observations indicate that there is widespread gene conservation among the Pasteurellaceae but that on an amino acid sequence level, M. haemolytica is closely related to A. pleuropneumoniae. The similarity between M. haemolytica and A. pleuropneumoniae is expected, as they occupy similar niches, although in different hosts. Both cause acute pneumonia associated with massive edema and necrosis, and they use similar virulence factors, including leukotoxin and iron acquisition proteins (8, 35). The similarities between these genomes are discussed in further detail below.
The data in Fig. 1a also revealed that 8.4% of M. haemolytica CDS match best to predicted proteins from 110 non-Pasteurellaceae species. Among these 237 CDS, the largest group consists of 27 CDS with top hits to Escherichia coli proteins. Another 15 have best matches to Neisseria meningitidis proteins (see Table S1 in the supplemental material). Among these are several putative virulence factors, including catalase (MHA_0814), the iron receptor protein Irp (MHA_1629), two immunoglobulin A (IgA)-specific metalloendopeptidase/possible adhesin proteins (MHA_0563 and MHA_2800), a possible adhesin-associated protein (MHA_2262), and two predicted hemoglobin receptor proteins (MHA_1639 and MHA_2261). All of these except MHA_2262 have orthologs in other Pasteurellaceae species. The similarity of virulence factor proteins in M. haemolytica and the β-proteobacterium N. meningitidis suggests that the two share common mechanisms of pathogenesis. This is not surprising considering that both colonize the upper respiratory tract. There are two reasons to consider these similarities the result of convergent evolution rather than horizontal gene transfer. The orthologs described here share only moderate identity on the amino acid level and virtually no similarity on the nucleotide level, except for catalase. Additionally, with the exception of hemoglobin receptor HmbR2 and the adhesin-associated protein, the CDS are neither clustered nor colinear.
Bacteriophages.
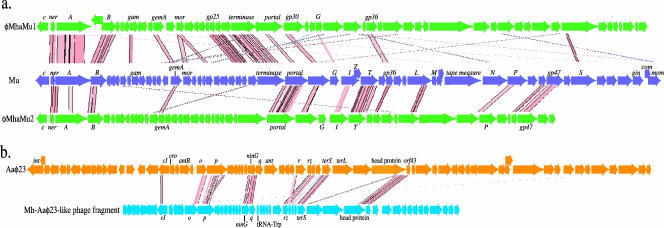
A substantial fraction (12.3%) of M. haemolytica genes are bacteriophage-related (Fig. 1a). There are at least two intact prophages in the genome. Two additional large regions (>20 kb) truncated by contig ends may also be prophages.
Strain BAA-410 contains an intact P2-like phage similar to φMhaA1-PHL101 of strain PHL101 (33). Both phages are integrated within a valine tRNA gene (MHA_t0001), located between the bis(5′-nucleosyl)-tetraphosphatase (MHA_0390) and glyceraldehyde-3-phosphate dehydrogenase (MHA_0442) genes. φMhaA1-PHL101 is a 34.5-kb inducible bacteriophage that encodes 50 CDS. φMhaA1-BAA410 and φMhaA1-PHL101 are very similar and differ only in 22 single-base-pair substitutions, most of which lie within the variable tail fiber gene (MHA_0423), and a 75-bp insertion. The 75-bp insertion lies within a hypothetical CDS (MHA_0419) containing a repeated TTGC tetrad. In φMhaA1-BAA410 this tetrad is repeated 19 more times than in φMhaA1-PHL101. Though this gene appears to be subject to phase variation by slipped mispairing, the relevance is unclear because the protein has no known function.
The M. haemolytica genome contains two Mu-like bacteriophages, φMhaMu1 and φMhaMu2 (Fig. 2a), which are similar to the enteric bacteriophage Mu and to the H. influenzae bacteriophage FluMu. Mu is a 36.7-kb prophage consisting of a 5′ early region containing regulatory genes and a 3′ late region containing mostly structural genes (50). The early region is highly conserved, containing transcriptional regulatory genes c and ner and transposase genes a and b. Genes encoding these regulatory proteins were identified in both φMhaMu1 and φMhaMu2 (Fig. 2a). Both prophages encode several other Mu orthologs, but there is greater mosaicism within the structural proteins, which is common among other Mu-like bacteriophages (50). The 36-kb φMhaMu1 prophage sequence is complete, contains 54 CDS (MHA_1966 to MHA_2019), and is integrated into a pseudogene of an immunoglobulin metalloendopeptidase (MHA_2020). φMhaMu2 is present on a 30-kb fragment encoding 46 CDS (MHA_0901 to MHA_0946), and it is integrated downstream of a possible nucleoside triphosphate-binding transposition protein. φMhaMu2 appears to be incomplete, since its terminal gene encodes a product similar to tail protein S and is located at a contig end. φMhaMu1 and φMhaMu2 share very little nucleotide similarity (81% identity over only 1,100 bp), although many of the predicted proteins share sequence similarity.
FIG. 2.
Bacteriophage maps. The Artemis comparison tool (9) was used to plot Tblastx matches of M. haemolytica prophage sequences to Mu and Aaφ23. Red and pink lines indicate similar regions; darker lines represent higher-quality matches. a. Comparison of φMhaMu1 and φMhaMu2 to Mu. b. Comparison of the Aaφ23-like prophage to Aaφ23.
A fourth bacteriophage fragment, similar to A. actinomycetemcomitans λ-like Aaφ23, was detected in the genome. Aaφ23, which contains 66 CDS on its 43-kb sequence (58), is not associated with virulence but can transduce antibiotic resistance genes (71). A 22.5-kb scaffold of the M. haemolytica genome containing 49 CDS (MHA_2429 to MHA_2477) includes several orthologs of Aaφ23 proteins (Fig. 2b). Compared to the Aaφ23 sequence, the M. haemolytica sequence is missing a ca. 5-kb region (9 CDS) at the 5′ end and ca. 10 kb (11 CDS) at the 3′ end. If M. haemolytica contains a complete Aaφ23-like phage, the remaining fragments must be on other contigs. The first gene of Aaφ23 is int (58); there are several integrases annotated in the M. haemolytica genome that could be candidates. The 3′ end of Aaφ23 encodes tail proteins. The scaffold containing the Aaφ23-like sequence ends with a CDS resembling λ protein M, while a second scaffold encodes orthologs of λ tail proteins L, K, I, and J (MHA_0358 to MHA_0362), which are downstream of M in λ. This scaffold may be part of the M. haemolytica Aaφ23-like prophage. Unlike Aaφ23, the Aaφ23-like sequence in M. haemolytica carries a tRNATrp gene. A second tRNATrp gene is found elsewhere on the M. haemolytica chromosome. The two sequences are each 76 nucleotides long but share only 89% identity over a 37-bp span. Despite this divergence, analysis with tRNAscan-SE indicates that both sequences encode cloverleaf tRNA structures with CCA anticodons. As there is only one tryptophan anticodon, it is unclear what advantage a second tRNATrp might provide.
Virulence factors.
M. haemolytica produces a number of virulence factors that have been the subject of much research. All previously known virulence factor genes have been located in the draft genome, and these have been reviewed previously (31). The M. haemolytica leukotoxin is perhaps the most significant virulence factor. The leukotoxin gene lktA and associated genes lktC, lktB, and lktD are known to have a highly mosaic structure among different M. haemolytica strains (15). The leukotoxin locus of strain BAA-410 carries alleles lktC1.1, lktA1.1, lktB1.1, and lktD1.1 (MHA_0253 to MHA_0256), which are common among serotype A1 strains. M. haemolytica A1 produces a capsule whose role in virulence may include adherence and resistance to serum-mediated killing and phagocytosis (31, 42). The capsule consists of disaccharide repeats of N-acetylmannosaminuronic acid and N-acetylmannosamine (42). The 12 genes (MHA_0519 to MHA_0530) in the capsule locus of strain BAA-410 share 99% or greater identity with the sequence previously reported by Lo et al. (42). Lipopolysaccharide (LPS) is a component of the gram-negative bacterial cell wall and is associated with virulence. M. haemolytica LPS induces an inflammatory cytokine response and expression of the of β2-integrin LFA-1 leukotoxin receptor in the host (31, 40). M. haemolytica serotypes are clonal with respect to LPS type, and the variation occurs primarily in the lipid A core/oligosaccharide region rather than within the O antigen (16). Serotype A1 was previously shown to produce an O-antigen consisting of repeated trisaccharides of d-galactose-N-acetyl-d-galactosamine-d-galactose (31). A total of 38 genes functioning in the synthesis of LPS components, including several genes common to enterobacterial common antigen biosynthesis, and related genes located within LPS gene clusters are listed in Table S2 in the supplemental material. Also included are genes for O-antigen assembly of mannose moieties and epimerases that presumably convert mannose to galactose.
Bacterial adhesins function in colonization by binding to receptor molecules on host cell surfaces. Many adhesins are pili, and the type IV pilus locus pilABCD (MHA_0662 to MHA_0665) was annotated in M. haemolytica. Type IV pili are known to function in DNA uptake, adhesion, and motility in H. influenzae, P. aeruginosa, and Neisseria species (5) and may perform these functions in M. haemolytica. Several predicted nonpilus adhesion proteins and additional proteins that could modify host mucosal surfaces are also present in M. haemolytica.
Many pathogens express a filamentous hemagglutinin that functions in adhesion to host mucosa (14, 64). In Bordetella pertussis, the filamentous hemagglutinin gene, fhaB, encodes a 3,591-amino-acid preprotein that is found both on the cell surface and in the extracellular milieu. In B. pertussis, fhaB lies in a cluster with fhaC, which encodes an outer membrane transporter specific for FhaB (64). M. haemolytica encodes a 3,215-amino-acid FhaB ortholog (MHA_0866) (Fig. 3a) with moderate amino acid similarity to B. pertussis FhaB (40% over 1,587 amino acids) and FhaB orthologs in N. meningitidis, A. pleuropneumoniae, M. succiniciproducens, and Pseudomonas syringae. M. haemolytica fhaB is adjacent to an fhaC ortholog (MHA_0867), so it likely uses the two-partner secretion pathway. Unlike B. pertussis FhaB, the M. haemolytica ortholog does not contain an integrin-binding RGD motif. If the M. haemolytica FhaB functions as an adhesin, it must have a different binding physiology. A second physical feature distinguishing M. haemolytica FhaB from B. pertussis FhaB is the presence of a bacterial intein-like (BIL) domain within the carboxy terminus of M. haemolytica FhaB. BIL domains are similar to protein-splicing domains found in eukaryotic proteins such as Hedgehog and Hedgehog-like proteins (1). An active BIL domain may allow the protein to undergo autoproteolysis. The cleavage and release of the BIL domain may be advantageous by allowing variability in receptor recognition or receptor release. Since cleavage of the BIL domain changes the structure of FhaB, it may also interfere with the host immune response.
FIG. 3.
Domain maps of predicted colonization proteins. a. Filamentous hemagglutinin MHA_0866. b. autotransporter/adhesin MHA_2701. c. autotransporter/adhesin MHA_1367. d. IgA peptidase/adhesin MHA_0563. e. IgA peptidase/adhesin MHA_2800. f. neuraminidase MHA_1532 (incomplete CDS). The sketches are to scale. Domains are labeled below each sketch. Triangle, signal sequence cleavage site. Asterisk, IgA protease active serine residue.
M. haemolytica also encodes a protein (MHA_2262) similar to an adhesin complex protein found only in N. meningitidis and the periodontal pathogen Eikenella corrodens. The E. corrodens protein is believed to be associated with an adhesin protein and located near the carbohydrate recognition domain, but its function is unknown (4).
Nontypeable H. influenzae produces HMW, an adhesin that requires accessory proteins HMWB and HMWC for maturation. HMWB is an outer membrane protein that translocates HMW across the outer membrane (65), while HMWC functions in glycosylation of HMW (29). Neither HMW nor HMWB orthologs were found in M. haemolytica, but an HMWC ortholog was annotated (MHA_0708). It is unknown what adhesin M. haemolytica HMWC glycosylates.
The serotype A1-specific antigen (Ssa1) is also thought to function as an adhesin (43). The amino acid sequence of Ssa1 from BAA-410 (MHA_2492) is identical to that of the previously sequenced Ssa1, except for the carboxy-terminal 14 amino acids (43). The sequence contains an amino-terminal signal sequence and carboxy-terminal autotransporter β domain. An internal serine protease domain is also present, but no domain typical of adhesins was identified.
Autotransporter proteins can be involved in adhesion to host mucosal surfaces. M. haemolytica encodes two proteins, MHA_2701 and MHA_1367, resembling the autotransporter adhesins of H. influenzae and N. meningitidis (13). Both predicted proteins contain amino-terminal signal sequence cleavage sites for inner membrane translocation and carboxy-terminal translocator domains for outer membrane pore formation (63). MHA_2701 and MHA_1367 contain ATP-binding sequences and carboxy-terminal YadA-like domains typical of the trimeric subfamily of autotransporters (Fig. 3b and c). This subfamily, which includes the Yersinia enterocolitica YadA and H. influenzae Hia adhesins, is distinguished from conventional autotransporters by the formation of trimers in the periplasmic space (13). Trimeric carboxy-terminal domains form a pore through which the passenger domains traverse the outer membrane. This mechanism results in the presentation of homotrimeric adhesin complexes. It is hypothesized that this structure enhances avidity with its multimeric binding sites and resists extracellular proteases by protecting nonbinding regions of the protein within the core of the trimer (13). MHA_2701 is similar to the Hsf-like adhesions of A. pleuropneumoniae and P. multocida. The MHA_1367 sequence is similar to those of H. influenzae Hia and the Hia orthologs of A. pleuropneumoniae and P. multocida.
M. haemolytica encodes two autotransporters (MHA_0563 and MHA_2800) orthologous to H. influenzae and N. meningitidis Iga1, an IgA peptidase/adhesin (Fig. 3d and e). The primary function of Iga1 is to cleave host mucosal antibody, supporting colonization by immunoevasion (65). MHA_0563 and MHA_2800 have Iga1 domains and conserved serine residues corresponding to the active sites of H. influenzae Iga1 proteases (54). Although these proteins are similar to IgA proteases, they may actually cleave IgG, as this is the secretory antibody of the bovine respiratory tract and IgG protease activity has been reported to be present in M. haemolytica (31). In addition to an Iga1-like domain, MHA_0563 and MHA_2800 have an internally located, 240-amino-acid domain similar to the B. pertussis adhesin pertactin (64). Proteolytic cleavage and release of the Iga1-like domain may expose the pertactin-like domain on the M. haemolytica surface, facilitating a function in adhesion. However, MHA_0563 and MHA_2800 lack the RGD motif common to many bacterial adhesins, including pertactin, so this domain may use an alternative motif for molecular interaction with a host receptor. Like FhaB, pertactin is a B. pertussis vaccine component (64), making these predicted proteins possible M. haemolytica vaccine targets. M. haemolytica encodes a third protein with an Iga1 domain and conserved serine active site; however, the CDS (MHA_1965) does not contain an adhesin domain or an autotransporter domain. MHA_1965 does have an amino-terminal signal sequence, and PSORT predicts it localizes to the outer membrane.
Bacteria can cleave host cell glycoproteins with glycoproteases and neuraminidases (or sialidases), modifying cell surfaces to enhance adhesion (61). The neuraminidase and zinc metalloglycoprotease genes previously described for M. haemolytica are also present in the BAA-410 sequence. The gcp gene (MHA_1559) encodes an M22 family O-sialoglycoprotein endopeptidase that has been associated with enhancement of adhesion to host cells (51). The neuraminidase gene (MHA_1532) is incomplete in the M. haemolytica BAA-410 genome, as it is truncated at a contig end (Fig. 3f). The carboxy-terminal 584 amino acids share 55% identity with the 832-amino-acid P. multocida ortholog NanH. The sequenced fragment of the M. haemolytica neuraminidase contains a carboxy-terminal autotransporter domain and an incomplete amino-terminal sialidase domain. The incomplete sialidase domain contains one aspartic acid box motif. This motif, common among sialidases (12), is found three times in NanH. It is suspected that the unsequenced 5′ end of the M. haemolytica neuraminidase gene encodes two additional aspartic acid boxes orthologous to those of NanH.
Iron is a vital nutrient for most bacteria, including M. haemolytica (32). The role of iron in M. haemolytica pathogenesis is unclear, as there are conflicting reports on the role of iron in the transcription and expression of leukotoxin (47, 67). M. haemolytica genes encoding iron acquisition proteins and several related iron homeostasis proteins are listed in Table S3 in the supplemental material. Expression of these proteins should allow M. haemolytica to acquire iron from host hemoglobin, hemopexin, and transferrin. M. haemolytica encodes two hemoglobin receptors, HmbR1 (MHA_1639) and HmbR2 (MHA_2261), that are more similar to N. meningitidis HmbR than the hemoglobin receptors of other Pasteurellaceae. M. haemolytica also has a gene encoding the hemophore HxuA (MHA_1004), indicating that it can acquire iron from hemopexin. The previously described transferrin-binding proteins TbpA (MHA_0196) and TbpB (MHA_0197) were also annotated in strain BAA-410 (18, 52). Other iron-regulated outer membrane proteins may also participate in iron acquisition. Although M. haemolytica has no known siderophores, it may use the iron scavengers produced by other bacteria, as it encodes two siderophore receptors. One receptor, which has several Pasteurellaceae orthologs, is similar to FhuE (MHA_2388), the E. coli ferric hydroximate siderophore receptor, while a second ferric hydroximate siderophore receptor (MHA_1541) is unique among Pasteurellaceae species. To complement these TonB-dependent outer membrane receptors, M. haemolytica also encodes ATP-binding cassette (ABC) superfamily transporters for transporting heme, hemin, siderophores, and iron across the inner membrane. The number of ABC transporters suggests diversity in substrates, as there are four ferric transporters and four siderophore transporters. M. haemolytica also encodes a ferric uptake regulator (Fur; MHA_2790), which is a transcriptional repressor that regulates iron acquisition genes (2). Recent experiments suggest that M. haemolytica Fur is atypical, as its transcription is not repressed by iron (J. Gioia and S. K. Highlander, unpublished data).
Resistance to antibiotics such as β-lactams, tetracycline, streptomycin, and sulfanomides is well documented in M. haemolytica (31). Although there is increasing control of the practice in some nations, administration of antibiotics in cattle feed still occurs and is likely to contribute to antibiotic resistance. Several antimicrobial resistance genes were found in the BAA-410 genome (see Table S4 in the supplemental material). These include the TetH tetracycline resistance protein (MHA_1158) and its regulator TetR (MHA_1159), a possible Zn-β-lactamase (MHA_2752), and a major facilitator superfamily permease possibly associated with efflux of bicyclomycin (MHA_1575), an antibiotic that targets transcription terminator factor ρ (38). Orthologs of the resistance-nodulation-cell division superfamily multidrug efflux pump AcrAB, as well as the regulatory protein AcrR (MHA_0370 to MHA_0372), are also present (41). A possible MarC family multiple antibiotic resistance transporter (MHA_0052) was also identified. The gene encoding this transporter is upstream of genes encoding an ABC superfamily binding protein and an ATPase. They may function together in antibiotic resistance. Additionally, there are 13 drug metabolite transporter family transporters predicted in the genome, though their substrates are unknown.
Genes encoding resistance to antimicrobial metals are also present. TehA (MHA_0652) and TehB (MHA_2274) function in the efflux of tellurite and several additional antiseptic compounds (68). M. haemolytica also encodes a tellurium resistance protein, TerC (MHA_1456). Strain BAA-410 may also be resistant to mercury, since it encodes a mercury transporter, MerT (MHA_2090); two MerP mercury transport periplasmic proteins (MHA_2089 and MHA_2120); and the MerR transcriptional regulator (MHA_2119) (53).
Transcriptional regulation.
As an opportunistic pathogen, M. haemolytica changes its relationship with its host from commensal to pathogenic. Transcriptional regulation may be central to niche versatility and the production of virulence factors. Based on the annotation, 125 CDS (4.4%) were classified as transcriptional regulators. These include CDS orthologous to transcriptional regulators, CDS encoding helix-turn-helix motifs as predicted using the method of Dodd and Egan (19), and sigma factors. The ratio of percentage of transcriptional regulators to total number of CDS (1.55 × 10−3) is similar to that in Pseudomonas aeruginosa (1.69 × 10−3), which has the highest proportion of transcriptional regulators described for any bacterial genome (66). In contrast, the ratio for E. coli is 1.35 × 10−3 (66). Transcriptional regulators identified are listed in Table S5 in the supplemental material. While it is not possible to predict the effectors of transcriptional regulators by annotation, there are several proteins of interest for future study. Ten regulators belong to the LysR family, which is associated with various functions such as virulence, nitrogen fixation, and oxidative stress response (72). There are five members of the AraC family, which is associated with virulence, metabolism, and stress response (26). There are two AcrR family transcriptional regulators, which putatively regulate drug efflux genes (46). There are also three members of the MerR family of metal detoxification transporter regulators (53). The ferric uptake regulator (Fur; MHA_2790), associated with virulence in other pathogens (2), is also present in M. haemolytica. Finally, the NarPQ two-component regulatory system (MHA_2193 and MHA_1462), though normally associated with nitrite and nitrate metabolism (68), may also be associated with virulence, as NarP was identified in an experiment to select transcriptional regulators of leukotoxin (J. Gioia, A. M. Marciel, L. E. Alvarez, J. M. Criglar, and S. K. Highlander, unpublished data).
M. haemolytica encodes at least six different RNA polymerase sigma factors. These include the major housekeeping sigma factor RpoD (σ70; MHA_2291), the heat shock sigma factor RpoH (σ32; MHA_2650), and four RpoE-like alternative sigma factors (MHA_0296, MHA_0739, MHA_1691, and MHA_2095). The alternative sigma factors belong to a subclass of σ70 proteins called extracytoplasmic sigma factors that function in response to a variety of conditions, such as periplasmic protein misfolding, heat stress, and nutrient stress (49). The function of the M. haemolytica alternative sigma factors remains to be determined. In E. coli RpoE is controlled by the RseABC proteins. These proteins sequester RpoE under nonstimulatory conditions to prevent it from transcribing the genes in its regulon (49, 55). Orthologs of RseABC (MHA_2570 to MHA_2572) are present in M. haemolytica, suggesting that a similar mechanism of regulation occurs in this organism.
Natural competence.
Many bacteria are naturally competent, meaning that they are able to acquire extracellular DNA for nucleic acid supply or gene transfer (20). Although natural competence is well characterized in H. influenzae, there are no published reports of natural competence in M. haemolytica. The M. haemolytica genome contains several features indicating the capacity for natural competence. In H. influenzae pilin protein PilA and secretin outer membrane pore protein ComE transport DNA across the outer membrane (20, 57). The M. haemolytica genome encodes predicted ComE (MHA_0164), PilA (MHA_0662), and pilus assembly proteins PilB (MHA_0663) and PilC (MHA_0664). The genome also encodes orthologs of proteins that function in the transport of DNA from the periplasm to the cytoplasm, including lipoprotein ComL (MHA_1560), transmembrane protein ComEA (MHA_2354), and periplasmic protein ComF (MHA_1838) (20, 57). Orthologs of cytoplasmic competence proteins DprA (MHA_1048), which functions in transport of DNA across the inner membrane, and ComM (MHA_0963), a protease believed to function in preventing cytoplasmic degradation of DNA, are also present (20, 57), as are competence regulatory proteins CRP (MHA_1000) and Sxy (MHA_1438) (57).
The DNA uptake signal sequence (USS) is a feature of natural competence in bacteria of the Neisseriaceae and Pasteurellaceae families (6, 69). The cell surface DNA-binding proteins of these bacteria bind to their specific USS, facilitating DNA uptake. In H. influenzae the USS is an AAGTGCGGT nonamer that is overrepresented in the genome (Table 2). This same nonamer is common among several other Pasteurellaceae bacteria (6, 69). Although this H. influenzae nonamer is also overrepresented in M. haemolytica (98 times), the most common nonamer has the sequence ACAAGCGGT and is present at 931 copies in the draft genome (Table 2). This putative USS does confer a 100-fold increase in uptake when cloned as a dimer onto a plasmid lacking the sequence (M. Dillon and S. K. Highlander, unpublished data).
TABLE 2.
Nonamer repeats in Pasteurellaceae genomes
| Organism | Most common 9-mer | Frequency | Frequency/Mb |
|---|---|---|---|
| H. influenzae | AAGTGCGGT | 1,471 | 803 |
| P. multocida | AAGTGCGGT | 927 | 412 |
| H. somni | AAGTGCGGT | 1,345 | 640 |
| A. actinomycetemcomitans | AAGTGCGGT | 1,473 | 736 |
| M. succiniciproducens | AAGTGCGGT | 1,485 | 642 |
| M. haemolytica | ACAAGCGGT | 931 | 362 |
| A. pleuropneumoniae | ACAAGCGGT | 755 | 329 |
| H. ducreyi | ACAAGCGGT | 199 | 117 |
The M. haemolytica USS is also the most common nonamer in A. pleuropneumoniae and H. ducreyi. Bakkali et al. postulate that USSs are the result of sequence uptake bias, or molecular drive, and that shared USSs among different species indicate a close evolutionary relationship (6). Thus, it can be hypothesized that the divergence of USS among the Pasteurellaceae represents different lineages in which M. haemolytica, A. pleuropneumoniae, and H. ducreyi are divergent with respect to the species in which the H. influenzae nonamer dominates. The common DNA USS in these organisms also implies the possibility of horizontal gene transfer with one another as the species diverged.
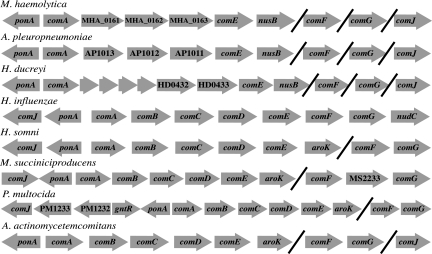
The divergence of M. haemolytica, A. pleuropneumoniae, and H. ducreyi from the H. influenzae lineage within the Pasteurellaceae is also supported by gene conservation in loci related to competence. In H. influenzae the com locus consists of the genes comABCDEFG (Fig. 4). The comABCDE gene order is conserved among all Pasteurellaceae species except M. haemolytica, A. pleuropneumoniae, and H. ducreyi. These three genomes contain com loci with comA and comE orthologs, but they do not contain orthologs of comB, comC, and comD. Instead, M. haemolytica encodes Pasteurella conserved hypothetical proteins MHA_0161, MHA_0162, MHA_0163, which are orthologous to AP1013, AP1012, and AP1011, respectively, in A. pleuropneumoniae. In H. ducreyi, HD0432 and HD0433 are orthologs of MHA_0162 and MHA_0163, respectively, but there is no intact ortholog of MHA_0161. Instead, there are four small hypothetical proteins encoded between comA and HD0432. One of these proteins has weak similarity (33% identity over 66 amino acids) to a portion of MHA_0161, so it is possible that H. ducreyi had an MHA_0161 ortholog at one time. Thus, a comparison of the com loci of these species supports the hypothesis that M. haemolytica, A. pleuropneumoniae, and H. ducreyi are members of a distinct subgroup of the Pasteurellaceae. Other features of the com locus are consistent with this point. As shown in Fig. 4, in H. influenzae and other Pasteurellaceae, comF and comG are adjacent to one another, and comJ is usually located upstream of comA. However, these genes are unlinked in M. haemolytica, A. pleuropneumoniae, and H. ducreyi. Additionally, the com locus is adjacent to nusB in M. haemolytica, A. pleuropneumoniae, and H. ducreyi, whereas it is adjacent to aroK or nudC in other Pasteurellaceae.
FIG. 4.
Pasteurellaceae competence loci. The com loci and flanking genes of eight Pasteurellaceae are sketched (not to scale). Arrows indicate genes and orientation. Hash marks separate noncontiguous sequences. Four small arrows preceding HD0432 represent small hypothetical proteins.
A divergence in gene conservation is also observed in the competence regulatory element (CRE) locus (57). In H. influenzae the four-gene CRE operon is induced under conditions favorable for natural competence, and mutation of HI0938, the first gene in the locus, prevents DNA uptake (57). Similar CRE operons are found in M. haemolytica (MHA_1011 to MHA_1014), A. pleuropneumoniae, and H. ducreyi, but these organisms have no ortholog to HI0940, the third gene of the locus. Instead, they have a different gene that is conserved among these three species.
M. haemolytica phylogeny.
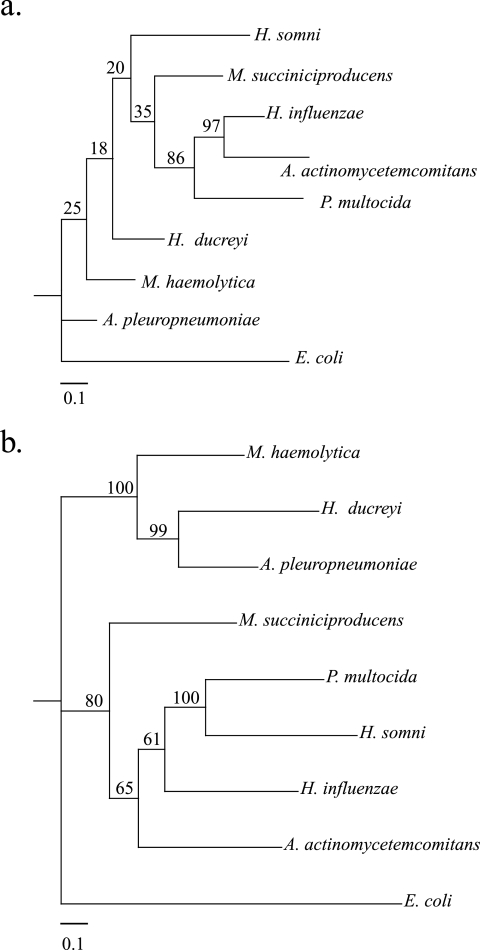
M. haemolytica, A. pleuropneumoniae, and H. ducreyi appear to be closely related based on examination of competence genes and features, but phylogenetic analysis requires a more general approach. Phylogenetic tree construction based upon 16S rRNA gene sequences is a common tool that has been used for classification of the Pasteurellaceae (11), including the creation of the genus Mannheimia (3). Analysis of 16S rRNA genes of the sequenced Pasteurellaceae species is of limited use because the bootstrap values are very low (Fig. 5a). However, the annotated genomes of these organisms allowed the construction of a phylogenetic tree based upon the sequences of 50 highly conserved housekeeping genes (Fig. 5b). Here, M. haemolytica, A. pleuropneumoniae, and H. ducreyi form a group that is divergent from the other Pasteurellaceae. Notably, the other Mannheimia species, M. succiniciproducens is found in a cluster with other members of the Pasteurellaceae family, distinct from M. haemolytica. M. succiniciproducens was placed in the genus Mannheimia because of phylogenetic analysis of 16S rRNA sequences (34, 39), though the analysis of Lee et al. (39) did not include sequences of H. ducreyi and A. pleuropneumoniae. Both the 16S rRNA gene tree and the housekeeping gene tree presented here indicate that M. haemolytica is more closely related to A. pleuropneumoniae and H. ducreyi than it is to the other Pasteurellaceae included in this analysis. This observation is similar to previously reported analyses using 16S rRNA gene sequences and the sequences of three individual housekeeping genes (11), although M. succiniciproducens was not included in those analyses. The data presented here suggest that the taxonomy of the group is not necessarily in agreement with the phylogeny of the group.
FIG. 5.
Phylogenetic trees. PHYLIP (24) was used to make trees, using sequences from eight sequenced Pasteurellaceae species. E. coli was used as an outgroup. Numbers at nodes represent bootstrap values (out of 100). a. Tree derived from 16S rRNA gene. b. Tree derived from 50 highly conserved housekeeping genes totaling ca. 40 kb.
The high degree of similarity between M. haemolytica and A. pleuropneumoniae is not unexpected given the analogous niches of the two bacteria. Both are commensal upper respiratory tract flora that are also opportunistic lower respiratory tract pathogens. Although they infect different host species, they share tissue tropism and similar virulence factors (8, 35). Hence, it is reasonable to hypothesize that these bacteria may be derived from a common Pasteurellaceae ancestor. The relationship to H. ducreyi is less obvious, as it is a pathogen of the human genital epithelium. Furthermore, the number of M. haemolytica CDS with H. ducreyi orthologs is the lowest of all Pasteurellaceae species examined. The relationships of H. ducreyi to M. haemolytica and A. pleuropneumoniae may reflect a case in which H. ducreyi has undergone greater divergence because of its adaptation to a different host species and tissue. Because housekeeping genes are highly conserved, this divergence may be slow to appear in an analysis such as the phylogenetic tree presented in Fig. 5b, despite being apparent in a genome-wide examination (Fig. 1b). It is also noteworthy that the DNA USS is present only 199 times in H. ducreyi, a far lower frequency than in other Pasteurellaceae. Considering the theory of molecular drive, this comparatively low frequency may reflect a lack of interaction with other bacteria sharing the same DNA USS because of diversification in a different niche. It appears that M. haemolytica and M. succiniciproducens are more closely related to other Pasteurellaceae than to each other, despite being placed in the same genus. This lesser degree of similarity may also be understandable given the difference in niche—M. succiniciproducens lives within the rumen of cattle (34). Given the phylogenetic relationships among the species examined here, a reevaluation of the taxonomy of one or more species may be in order.
Supplementary Material
Acknowledgments
Funding for this work was provided by USDA National Research Grant Initiative 00-35204-9229.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Amitai, G., O. Belenkiy, B. Dassa, A. Shainskaya, and S. Pietrokovski. 2003. Distribution and function of new bacterial intein-like protein domains. Mol. Microbiol. 47:61-73. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Angen, O., R. Mutters, D. A. Caugant, J. E. Olsen, and M. Bisgaard. 1999. Taxonomic relationships of the [Pasteurella] haemolytica complex as evaluated by DNA-DNA hybridizations and 16S rRNA sequencing with proposal of Mannheimia haemolytica gen. nov., comb. nov., Mannheimia granulomatis comb. nov., Mannheimia glucosida sp. nov., Mannheimia ruminalis sp. nov. and Mannheimia varigena sp. nov. Int. J. Syst. Bacteriol. 49:67-86. [DOI] [PubMed] [Google Scholar]
- 4.Azakami, H., H. Yumoto, H. Nakae, T. Matsuo, and S. Ebisu. 1996. Molecular analysis of the gene encoding a protein component of the Eikenella corrodens adhesin complex that is close to the carbohydrate recognition domain. Gene 180:207-212. [DOI] [PubMed] [Google Scholar]
- 5.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 73:1635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakkali, M., T. Y. Chen, H. C. Lee, and R. J. Redfield. 2004. Evolutionary stability of DNA uptake signal sequences in the Pasteurellaceae. Proc. Natl. Acad. Sci. USA 101:4513-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Bosse, J. T., H. Janson, B. J. Sheehan, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4:225-235. [DOI] [PubMed] [Google Scholar]
- 9.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 10.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, H., P. Kuhnert, J. E. Olsen, and M. Bisgaard. 2004. Comparative phylogenies of the housekeeping genes atpD, infB and rpoB and the 16S rRNA gene within the Pasteurellaceae. Int. J. Syst. Evol. Microbiol. 54:1601-1609. [DOI] [PubMed] [Google Scholar]
- 12.Copley, R. R., R. B. Russell, and C. P. Ponting. 2001. Sialidase-like Asp-boxes: sequence-similar structures within different protein folds. Protein Sci. 10:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. [DOI] [PubMed] [Google Scholar]
- 14.Coutte, L., S. Alonso, N. Reveneau, E. Willery, B. Quatannens, C. Locht, and F. Jacob-Dubuisson. 2003. Role of adhesin release for mucosal colonization by a bacterial pathogen. J. Exp. Med. 197:735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, R. L., S. Campbell, and T. S. Whittam. 2002. Mosaic structure and molecular evolution of the leukotoxin operon (lktCABD) in Mannheimia (Pasteurella) haemolytica, Mannheimia glucosida, and Pasteurella trehalosi. J. Bacteriol. 184:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, R. L., and W. Donachie. 1996. Intra-specific diversity and host specificity within Pasteurella haemolytica based on variation of capsular polysaccharide, lipopolysaccharide and outer-membrane proteins. Microbiology 142:1895-1907. [DOI] [PubMed] [Google Scholar]
- 17.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deneer, H. G., and A. A. Potter. 1989. Iron-repressible outer-membrane proteins of Pasteurella haemolytica. J. Gen. Microbiol. 135:435-443. [DOI] [PubMed] [Google Scholar]
- 19.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 21.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 22.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 23.Fedorova, N. D., and S. K. Highlander. 1997. Plasmids for heterologous expression in Pasteurella haemolytica. Gene 186:207-211. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein, J. 2005. PHYLIP (phylogeny inference package) version 3.6. Department of Genome Sciences, University of Washington, Seattle.
- 25.Frank, G. H. 1989. Pasteurellosis of cattle, p. 197-221. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press, New York, N.Y..
- 26.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert, D. G. 1999. Phylodendron version 0.8, beta. Department of Biology, Indiana University, Bloomington.
- 29.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 30.Havlak, P., R. Chen, K. J. Durbin, A. Egan, Y. Ren, X. Z. Song, G. M. Weinstock, and R. A. Gibbs. 2004. The Atlas genome assembly system. Genome Res. 14:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Highlander, S. K. 2001. Molecular genetic analysis of virulence in Mannheimia (Pasteurella) haemolytica. Front. Biosci. 6:D1128-D1150. [DOI] [PubMed] [Google Scholar]
- 32.Highlander, S. K., O. Garza, B. J. Brown, S. Koby, and A. B. Oppenheim. 1997. Isolation and characterization of the integration host factor genes of Pasteurella haemolytica. FEMS Lett. 146:181-188. [DOI] [PubMed] [Google Scholar]
- 33.Highlander, S. K., S. Weissenberger, L. E. Alvarez, G. M. Weinstock, and P. B. Berget. 2006. Complete nucleotide sequence of a P2 family lysogenic bacteriophage, φMhaA1-PHL101, from Mannheimia haemolytica serotype A1. Virology 350:79-89. [DOI] [PubMed] [Google Scholar]
- 34.Hong, S. H., J. S. Kim, S. Y. Lee, Y. H. In, S. S. Choi, J. K. Rih, C. H. Kim, H. Jeong, C. G. Hur, and J. J. Kim. 2004. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat. Biotechnol. 22:1275-1281. [DOI] [PubMed] [Google Scholar]
- 35.Jacques, M. 2004. Surface polysaccharides and iron-uptake systems of Actinobacillus pleuropneumoniae. Can. J. Vet. Res. 68:81-85. [PMC free article] [PubMed] [Google Scholar]
- 36.Jeyaseelan, S., S. Sreevatsan, and S. K. Maheswaran. 2002. Role of Mannheimia haemolytica leukotoxin in the pathogenesis of bovine pneumonic pasteurellosis. Anim. Health Res. Rev. 3:69-82. [DOI] [PubMed] [Google Scholar]
- 37.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohn, H., and W. Widger. 2005. The molecular basis for the mode of action of bicyclomycin. Curr. Drug Targets Infect. Disord. 5:273-295. [DOI] [PubMed] [Google Scholar]
- 39.Lee, P. C., S. Y. Lee, S. H. Hong, and H. N. Chang. 2002. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl. Microbiol. Biotechnol. 58:663-668. [DOI] [PubMed] [Google Scholar]
- 40.Leite, F., S. Gyles, D. Atapattu, S. K. Maheswaran, and C. J. Czuprynski. 2003. Prior exposure to Mannheimia haemolytica leukotoxin or LPS enhances beta(2)-integrin expression by bovine neutrophils and augments LKT cytotoxicity. Microb. Pathog. 34:267-275. [DOI] [PubMed] [Google Scholar]
- 41.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 42.Lo, R. Y., L. J. McKerral, T. L. Hills, and M. Kostrzynska. 2001. Analysis of the capsule biosynthetic locus of Mannheimia (Pasteurella) haemolytica A1 and proposal of a nomenclature system. Infect. Immun. 69:4458-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo, R. Y., C. A. Strathdee, P. E. Shewen, and B. J. Cooney. 1991. Molecular studies of Ssa1, a serotype-specific antigen of Pasteurella haemolytica A1. Infect. Immun. 59:3398-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 47.Marciel, A. M., and S. K. Highlander. 2001. Use of operon fusions in Mannheimia haemolytica to identify environmental and cis-acting regulators of leukotoxin transcription. Infect. Immun. 69:6231-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLeod, M. P., X. Qin, S. E. Karpathy, J. Gioia, S. K. Highlander, G. E. Fox, T. Z. McNeill, H. Jiang, D. Muzny, L. S. Jacob, A. C. Hawes, E. Sodergren, R. Gill, J. Hume, M. Morgan, G. Fan, A. G. Amin, R. A. Gibbs, C. Hong, X. J. Yu, D. H. Walker, and G. M. Weinstock. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 186:5842-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 50.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 51.Nyarko, K. A., B. L. Coomber, A. Mellors, and P. A. Gentry. 1998. Bovine platelet adhesion is enhanced by leukotoxin and sialoglycoprotease isolated from Pasteurella haemolytica A1 cultures. Vet. Microbiol. 61:81-91. [DOI] [PubMed] [Google Scholar]
- 52.Ogunnariwo, J. A., T. K. Woo, R. Y. Lo, G. C. Gonzalez, and A. B. Schryvers. 1997. Characterization of the Pasteurella haemolytica transferrin receptor genes and the recombinant receptor proteins. Microb. Pathog. 23:273-284. [DOI] [PubMed] [Google Scholar]
- 53.Pennella, M. A., and D. P. Giedroc. 2005. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals 18:413-428. [DOI] [PubMed] [Google Scholar]
- 54.Poulsen, K., J. Reinholdt, and M. Kilian. 1992. A comparative genetic study of serologically distinct Haemophilus influenzae type 1 immunoglobulin A1 proteases. J. Bacteriol. 174:2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 56.Rawlings, N. D., D. P. Tolle, and A. J. Barrett. 2004. MEROPS: the peptidase database. Nucleic Acids Res. 32:D160-D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redfield, R. J., A. D. Cameron, Q. Qian, J. Hinds, T. R. Ali, J. S. Kroll, and P. R. Langford. 2005. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J. Mol. Biol. 347:735-747. [DOI] [PubMed] [Google Scholar]
- 58.Resch, G., E. M. Kulik, F. S. Dietrich, and J. Meyer. 2004. Complete genomic nucleotide sequence of the temperate bacteriophage Aaφ23 of Actinobacillus actinomycetemcomitans. J. Bacteriol. 186:5523-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rey, S., M. Acab, J. L. Gardy, M. R. Laird, K. deFays, C. Lambert, and F. S. Brinkman. 2005. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 33:D164-D168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34:D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakarya, S., and S. Oncu. 2003. Bacterial adhesins and the role of sialic acid in bacterial adhesion. Med. Sci. Monit. 9:RA76-RA82. [PubMed] [Google Scholar]
- 62.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 63.Scarselli, M., R. Rappuoli, and V. Scarlato. 2001. A common conserved amino acid motif module shared by bacterial and intercellular adhesins: bacterial adherence mimicking cell cell recognition? Microbiology 147:250-252. [DOI] [PubMed] [Google Scholar]
- 64.Smith, A. M., C. A. Guzman, and M. J. Walker. 2001. The virulence factors of Bordetella pertussis: a matter of control. FEMS Microbiology Rev. 25:309-333. [DOI] [PubMed] [Google Scholar]
- 65.St. Geme, J. W., III. 2000. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19(Suppl. 1):S41-S50. [DOI] [PubMed] [Google Scholar]
- 66.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 67.Strathdee, C. A., and R. Y. Lo. 1989. Regulation of expression of the Pasteurella haemolytica leukotoxin determinant. J. Bacteriol. 171:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turner, R. J., D. E. Taylor, and J. H. Weiner. 1997. Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob. Agents Chemother. 41:440-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, Y., S. D. Goodman, R. J. Redfield, and C. Chen. 2002. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 184:3442-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, W. Helmberg, D. L. Kenton, O. Khovayko, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, J. U. Pontius, K. D. Pruitt, G. D. Schuler, L. M. Schriml, E. Sequeira, S. T. Sherry, K. Sirotkin, G. Starchenko, T. O. Suzek, R. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2005. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 33:D39-D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willi, K., H. Sandmeier, E. M. Kulik, and J. Meyer. 1997. Transduction of antibiotic resistance markers among Actinobacillus actinomycetemcomitans strains by temperate bacteriophages Aaφ23. Cell. Mol. Life Sci. 53:904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, R. G., Y. Kim, T. Skarina, S. Beasley, R. Laskowski, C. Arrowsmith, A. Edwards, A. Joachimiak, and A. Savchenko. 2002. Crystal structure of Thermotoga maritima 0065, a member of the IclR transcriptional factor family. J. Biol. Chem. 277:19183-19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao, Y., R. E. Davis, and I. M. Lee. 2005. Phylogenetic positions of ‘Candidatus Phytoplasma asteris ’ and Spiroplasma kunkelii as inferred from multiple sets of concatenated core housekeeping proteins. Int. J. Syst. Evol. Microbiol. 55:2131-2141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.