Abstract
We previously reported the X-ray structures of wild-type Escherichia coli AcrB, a proton motive force-dependent multidrug efflux pump, and its N109A mutant. These structures presumably reflect the resting state of AcrB, which can bind drugs. After ligand binding, a proton may bind to an acidic residue(s) in the transmembrane domain, i.e., Asp407 or Asp408, within the putative network of electrostatically interacting residues, which also include Lys940 and Thr978, and this may initiate a series of conformational changes that result in drug expulsion. Herein we report the X-ray structures of four AcrB mutants, the D407A, D408A, K940A, and T978A mutants, in which the structure of this tight electrostatic network is expected to become disrupted. These mutant proteins revealed remarkably similar conformations, which show striking differences from the previously known conformations of the wild-type protein. For example, the loop containing Phe386 and Phe388, which play a major role in the initial binding of substrates in the central cavity, becomes prominently extended into the center of the cavity, such that binding of large substrate molecules may become difficult. We believe that this new conformation may mimic, at least partially, one of the transient conformations of the transporter during the transport cycle.
The Escherichia coli AcrB multidrug efflux pump (10, 11) is a member of the resistance-nodulation-division transporter family (18). It recognizes many structurally unrelated toxic compounds and actively engages to extrude them from cells. Its crystallographic structure was solved by Murakami et al. (13) in 2002. We previously reported the X-ray structures of AcrB in the presence of four different ligands (21, 22). The structures showed that these ligands bind to the wall of the extremely large central cavity in the transmembrane region of the pump. This binding presumably corresponds to the first step in the drug extrusion process, since drug molecules then have to pass through the periplasmic domain of AcrB and eventually reach the outer membrane channel TolC. A subsequent study of the efflux pump by crystallization of a mutant AcrB protein with an N109A mutation with five structurally diverse ligands (20) indicated that AcrB contains at least two distinct binding sites. These five ligands not only bind to various positions of the central cavity but also bind to residues lining the deep external depression formed by the C-terminal periplasmic domain.
AcrB is a proton motive force-dependent multidrug efflux pump that functions via a drug/proton antiport mechanism (23). Coupled with the outward movement of drug molecules, protons have to flow inward (towards the cytoplasm) to energize the efflux process. AcrB contains two acidic residues, Asp407 and Asp408, in the transmembrane (TM) helix TM4 and one basic residue, Lys940, in TM10, and these three residues appear to constitute a salt-bridged (and/or hydrogen-bonded) network (13, 22) (Fig. 1). The presence of such residues often means that they play an important functional role, presumably in the translocation of protons.
FIG. 1.
Putative salt bridge/H-bonding network (D407-D408-K940-T978) in the wild-type AcrB protomer, based on PDB file 1IWG (15). The view is along the line perpendicular to the membrane surface, from the periplasmic side. In spite of the modest resolution of the overall structure, the electron densities of some side chains can be seen clearly (see Fig. 2, top panel). The stick model in this figure as well as that in Fig. 6B was produced by the program PyMol (W. L. Delano, PyMol Graphic System [www.pymol.org]). The locations and sizes of the cross sections of TM4, TM10, and TM11 shown are crude approximations added simply to aid understanding.
For MexB (a homolog of AcrB) of Pseudomonas aeruginosa, it has been shown that Asp407, Asp408, and Lys939 (corresponding to Asp407, Asp408, and Lys940 of AcrB) are indeed essential for transport function (7), and this Asp-Lys-Asp triad was also found to be essential in E. coli AcrB (14). Recently, we found that Thr978 of AcrB TM11, located close to the triad, is also essential for function (17); this residue may also be a component of the putative network of tightly interacting residues just mentioned (Fig. 1).
During the translocation of the ligand, active transporters must go through significant conformational changes, which are coupled to the expenditure of energy. With transporters that use ATP hydrolysis as an energy source, one can attempt to trap the transporter in one of the transient conformations by using vanadate-ADP (5, 16). However, similar approaches are not feasible with transporters such as AcrB, which is energized by proton motive force. We reasoned that proton translocation may perturb the salt bridge/H-bonding interactions within the D407-K940-T978-D408 complex and that this transient state of AcrB might be mimicked by replacing one of these residues with alanine, which cannot be protonated or deprotonated. We report here that the D407A, D408A, K940A, and T978A mutations cause remarkably similar and extensive alterations in the conformation of AcrB.
MATERIALS AND METHODS
Construction of D407A, D408A, K940A, and T978A mutants.
Mutations were introduced by the method described in the accompanying paper (17). All of the mutant acrB genes contained a sequence coding for four additional histidine residues at the C terminus, and each of the proteins therefore had a hexahistidine sequence at the end (together with two histidine residues supplied by the native AcrB protein).
Purification of AcrB.
The mutant AcrB proteins were overproduced in E. coli BL21-Gold(DE3) cells (Stratagene), using a plasmid derived from pSPORT1 (Invitrogen). Cells were grown in 6 liters of LB medium with 100 μg/ml ampicillin. Cells were disrupted with a French pressure cell. The membrane fraction was collected and washed twice with high-salt buffer containing 20 mM sodium phosphate (pH 7.2), 2 M KCl, 10% glycerol, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride and once with 20 mM HEPES-NaOH buffer (pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride. The membrane proteins were then solubilized in 1% n-dodecyl-β-d-maltoside (wt/vol). Insoluble material was removed by ultracentrifugation at 370,000 × g. The extracted protein was purified with hydroxyapatite and Cu2+-affinity columns (23).
Crystallography.
Crystals of the D407A, D408A, K940A, and T978A mutants were grown by hanging-drop vapor diffusion at 25°C. A protein solution containing 30 mg/ml mutant protein in 20 mM Tris (pH 7.5), 0.1% n-dodecyl-β-d-maltoside, and 20 mM dithiothreitol was mixed with an equal volume of a reservoir solution containing 6.5 to 8.5% polyethylene glycol 3000, 8% glycerol, and 30 mM sodium citrate (pH 5.6) or 30 mM potassium citrate (pH 6.5). Crystals appeared in the drops within 4 days. Cryoprotection was achieved by raising the glycerol concentration stepwise to 35%, with a 5% increment at each step. The conditions used for crystallization were essentially identical to those used earlier for the wild-type AcrB protein (22), except for the pH of the Tris buffer used. When the wild-type protein was crystallized under the conditions described here, a structure identical to that reported earlier was obtained (unpublished results).
All X-ray intensity data sets were collected at the Advanced Light Source at Lawrence Berkeley National Laboratory (beamline 8.2.1) at a cryogenic temperature (100 K). The diffraction data were processed with DENZO and scaled with SCALEPACK (15). The crystals of the AcrB mutants took the R32 space group, with the unit cell dimensions listed in Table 1. Initially, the overall structures of the AcrB mutants were determined by molecular replacement, using the MolRep program (19) in the CCP4 package. The wild-type AcrB structure (Protein Data Bank no. 1OY6) was used as a search model. Before refinement, 5% of all data were set aside for cross-validation (2). The model refinements were performed using CNS (3) and CCP4 (4). Model rebuilding was conducted using the program O (8).
TABLE 1.
Data collection and crystallographic analysis of AcrB mutantsa
| Mutant | Cell constants (Å) | Resolution (Å) | Completeness (%) | Rsym (%) | Rwork/Rfree (%) | No. of reflections
|
1/σ (I) | |
|---|---|---|---|---|---|---|---|---|
| Total | Unique | |||||||
| D407A mutant | a = b = 145.4, c = 514.5, α = β = 90, γ = 120 | 3.56 (3.78-3.56) | 100 (94.7) | 10.2 (35.9) | 27.1/29.4 | 1,086,717 | 41,936 | 3.9 |
| D408A mutant | a = b = 145.0, c = 513.7, α = β = 90, γ = 120 | 3.65 (3.78-3.65) | 94.7 (92.3) | 10.6 (46.5) | 26.1/30.3 | 618,639 | 23,658 | 1.3 |
| K940A mutant | a = b = 145.6, c = 519.7, α = β = 90, γ = 120 | 3.38 (3.56-3.38) | 99.9 (99.0) | 7.8 (45.8) | 25.1/28.0 | 880,107 | 43,054 | 1.4 |
| T978A mutant | a = b = 144.9, c = 518.6, α = β = 90, γ = 120 | 3.38 (3.56-3.38) | 99.1 (94.6) | 9.3 (49.1) | 25.4/27.5 | 1,112,163 | 42,367 | 2.7 |
All mutants belong to space group R32. Numbers in parentheses are for the highest-resolution shells.
Labeling of purified CL-F386C AcrBHis mutant proteins with MIANS and fluorescence measurements.
The acrB gene was first modified by converting the codons for two intrinsic cysteines (Cys493 and Cys887) to those for serines by site-directed mutagenesis, producing cysteine-less (CL) AcrB; this mutant protein appeared to be fully functional in providing drug resistance, as reported earlier for proteins with alanine mutations of the same residues (6). Phe386 in CL AcrB was then converted to cysteine, and another mutation in the proton relay region was introduced, when necessary, in the same way. The purified CL-F386C AcrBHis proteins (3 to 4 mg/ml in 20 mM HEPES-KOH buffer containing 50 mM NaCl, 0.02% n-dodecyl-β-d-maltoside, and 10% glycerol [pH 7.5]) were diluted in the assay buffer (20 mM HEPES-KOH [pH 7.5] containing 50 mM NaCl and 0.02% n-dodecyl-β-d-maltoside) at a concentration of 0.5 μM (57 μg/ml). The reaction was initiated by the addition of 2-(4′-maleimidylanilino)naphthalene 6-sulfonic acid sodium salt (MIANS; Molecular Probes) to a final concentration of 5 μM, and fluorescence was monitored continuously at room temperature with an RF-5301PC spectrofluorophotometer (Shimadzu) at an emission wavelength of 430 nm (excitation, 330 nm). A similar labeling experiment was also carried out by using 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM; Molecular Probes).
Protein structure accession numbers.
The coordinates for the protein structures have been deposited in the Protein Data Bank (PDB) under accession numbers 2HQC (D407A mutant), 2HQD (D408A mutant), 2HQF (K940A mutant), and 2HQG (T978A mutant).
RESULTS
Crystallographic structures of mutants of the putative proton relay system.
We solved the three-dimensional structures of the AcrB D407A, D408A, K940A, and T978A mutants by X-ray crystallography through molecular replacement. The structures were determined with a maximum resolution of 3.38 to 3.65 Å. The overall Rwork values were between 25.1 and 27.1%, and Rfree values were between 27.5 and 30.3%. Refinement statistics are shown in Table 1. The overall structures of these mutant proteins were very similar to each other. Each mutant protein formed a homotrimer, with its threefold symmetry axis passing through the center of the trimer. Compared with wild-type AcrB, the mutant transporters showed some unique conformational changes, as described below.
Short-range alterations.
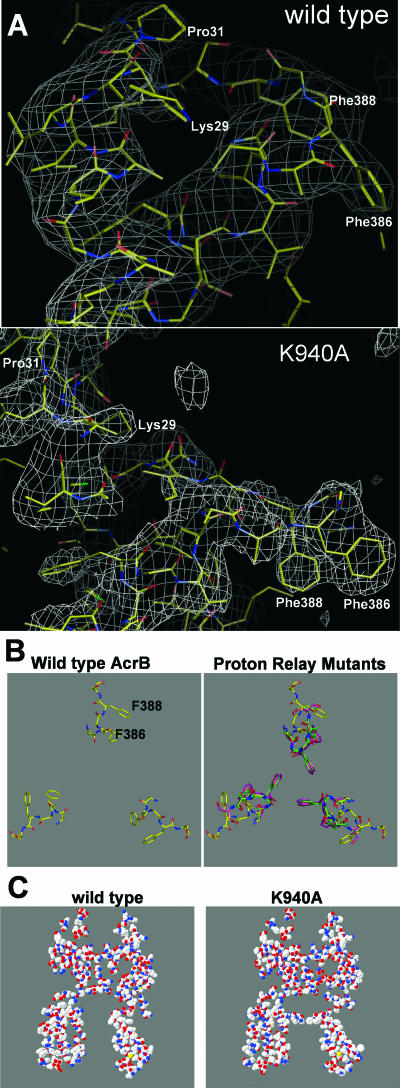
Electron density omit maps showed that some of the tight interactions among four residues, namely, Asp407, Asp408, Lys940, and Thr978 (Fig. 1), became profoundly disturbed in the mutants. As shown in Fig. 2 (top panel), in the wild-type protein the side chains of some of these residues show clearly interpretable electron densities, even at the modest overall resolution of our structures, presumably because they are immobilized by the strong electrostatic interactions between them. However, in the mutant proteins (Fig. 2, bottom panel, shows the same region in the D408A mutant protein as an example), much less detail of the side chain density is seen, most likely because the tight interaction network was disturbed.
FIG. 2.
Side view of the putative proton relay network involving TM4 and TM10. Electron density omit maps were calculated at 1 σ. (Top) In wild-type AcrB, some of the side chains are visible, presumably because of the strong electrostatic interactions among these residues. (Bottom) In contrast, in mutants (the D408A mutant is shown as an example), much less detail can be seen, presumably because of the weakening and abolition of the electrostatic interactions.
This disorder in the network region, introduced by the mutations, apparently spreads into the neighboring regions. Thus, in all four mutants, extensive changes occur in the backbone atoms in the part of TM4 that is proximal to residue 407 (Fig. 3). Thus, the regular H-bonding interactions of the backbone NH and C=O groups characteristic of the α-helix (Fig. 3, left panel) become distorted or disrupted (the right panel of Fig. 3 shows the D407A mutant as an example; similar changes are also found in the other mutants), as also seen in the disappearance of regular helical structures in the upper part of TM4 in Fig. 4 (bottom panel), where the structure of the TM domain of the K940A mutant is given as an example. The disordering of the proximal part of TM4, as illustrated in Fig. 3, makes this segment longer, also creating a drastic change in the positions of residues of the loop between TM helices 3 and 4 (noted by an arrow on the left in Fig. 5). This loop is located close to the ceiling of the central cavity, and the details, which could be established unequivocally because of the electron density of the phenyl groups in Phe386 and Phe388, are shown in Fig. 6B (panel A shows the electron density maps of the wild-type protein and the K940A mutant protein). Compared with the wild-type structure (Fig. 6B, left panel), the loops of all mutants, formed by residues 384 to 393 between TM3 and TM4, intrude prominently into the center of the cavity (Fig. 6B, right panel). The change includes residues Phe386 and Phe388 coming about 6 to 7 Å closer to the center of the cavity, and thus also to their counterparts from the other subunits of the trimer. For example, the distances between the CZ carbon atoms of the phenyl groups of Phe386 and Phe386′ in the D407A, D408A, K940A, and T978A mutants are 6.7 Å, 6.0 Å, 6.2 Å, and 7.4 Å, respectively, in contrast to 16.2 Å in the wild-type protein. These three phenylalanines essentially decrease the size of the upper part of the central cavity by forming a septum-like structure (Fig. 6C), resulting in an opening of only about 6 Å in diameter at this region, in contrast to the nearly 20-Å-diameter opening in the wild-type protein.
FIG. 3.
Structure of the N-terminal portion of TM4. The view is from the side (in a direction parallel to the membrane surface). The hydrogen bonds (green dotted lines) were calculated by the program DeepView, and the picture was drawn by using DeepView and POV-ray. The wild-type structure (left) and the D407A mutant structure (right) were aligned for the best overall fit of the entire protein backbone by DeepView and then horizontally displaced for a better view. In the wild-type AcrB protein (left), the backbone NH of Asp407 is H-bonded to the backbone C=O of Gly403, four residues back, as it should be in the regular α-helical structure. However, in the D407A mutant AcrB protein (right), the corresponding NH of Ala407 appears to H-bond to the C=O of not only Gly403 but also Leu404, distorting the helix. Further upward, the H-bonding interactions involving NH groups of Leu405, Leu404, and Gly403 in the wild-type protein are totally lost in the mutant protein. This distortion of the α-helix extends the chain, such that the top of TM4 (near the top of the figure) becomes higher in position in the mutant protein by nearly 2 Å, as shown.
FIG. 4.
Secondary structures in TM domains. The figure was drawn by using the Cn3D program (National Center for Biotechnology Information) after alignment of the three-dimensional structures of the wild type (PDB file 1IWG) and the K940A mutant (this study) through the Vector Alignment Search Tool (12), available at the National Center for Biotechnology Information website. Note that the assignment of the secondary structure was done in a uniform manner by the Vector Alignment Search Tool program, independent of the annotations in the PDB files. The TM helices are numbered in the wild-type structure.
FIG. 5.
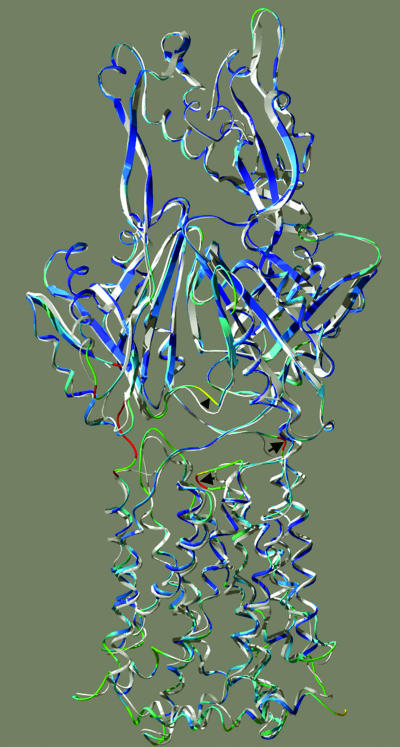
Global conformation of AcrB protomers, showing the deviation of the K940A structure from the wild-type structure. The structures were aligned for the best global fit, using backbone atoms, by the Iterative Magic Fit feature of the DeepView program, and the mutant structure was colored in rainbow colors (with red showing the most deviation) by using the “color by RMS deviation” feature of the program, whereas the wild-type structure is shown in white. The structure is shown so that the helices on the right side correspond to the N-terminal ones (TM1 through TM6) and those on the left side correspond to the C-terminal ones (TM7 through TM12). The structure is tilted slightly from the membrane perpendicular in order to show the loop between TM3 and TM4 more clearly. The shift of this loop between TM3 and TM4 is indicated by an arrow on the left, and that of the loop of residues 29 to 34 is shown by an arrow on the right. The shift of the loop at the bottom of the periplasmic ligand binding site (23) is indicated by an arrowhead. Red sections at the extreme left portion of the figure are an artifact caused by the absence of some residues in the wild-type structure. The drawing was done by using DeepView and POV-ray.
FIG. 6.
Structure of the loop between TM3 and TM4. (A) Simulated annealing composite omit map of the K940A mutant contoured at 1.0 σ. The figure shows the locations of Phe386 and Phe388 in the central cavity and of residues along the left side of the vestibule. (B) Models of the loop (residues 385 to 389) viewed along the threefold axis (perpendicular to the membrane surface) from the periplasmic side. The left panel shows the structure in wild-type AcrB (from PDB file 1IWG) (15), and the right panel shows the structures in mutant AcrB proteins (D407A, green; K940A, mauve; and T978A, orange) overlaid on top of the wild-type structure (yellow). (C) Vertical slab (5 Å thick) of the AcrB trimer, showing the essential closure of the top portion of the central cavity by the loop between TM3 and TM4 in the K940A mutant protein (right). The same slab of the wild-type trimer is shown on the left for comparison. The slice plane goes through the approximate center of the central cavity as well as through the Pro833 residue in protomer A and Thr145 in protomer B. The picture was generated by the Deep View program followed by POV-ray.
Long-range alterations.
The local changes were propagated to produce a number of remarkably extensive long-range alterations, as shown in Fig. 4. Thus, looking at the K940A mutant protein, the kinked TM helix 5 becomes disordered such that the helical structure before the kink totally disappears, and even the portion after the kink becomes strongly disordered. The following TM6 helix also becomes shortened. Furthermore, the helix connecting the N-terminal and C-terminal halves of the transmembrane domain, i.e., Iα (13), again becomes drastically shortened. The TM helices of the C-terminal half (TM7 through TM12) do not undergo extensive disordering, except that helix 11 (containing Thr978) becomes somewhat shorter and the C-terminal end of helix 12 becomes disordered. All of these changes occur with remarkable regularity in all four mutants (K940A, D407A, D408A, and T978A).
As part of this global conformational alteration, we found that the upper half of TM5 shifts upward about 2 Å, just like the corresponding part of TM4. Significant backbone movement was also found in the flexible loop, i.e., residues 29 to 32, above TM1 (the electron density map of this region is also shown in Fig. 6A, and the shift is highlighted with an arrow on the right in Fig. 5). This loop forms one side of the vestibule through which ligands are hypothesized to enter the central cavity (13, 21, 22). The displacement can be interpreted as a rotation of this portion of the loop, with the rotational axis passing through the C-α atoms of residues 28 and 32. The side chain of Leu30, which is at the center of this displaced region, appears to have been flipped about 100° from the central cavity. The rotation also shifts the locations of the C-α atoms of Leu30 and Pro31 about 5 Å away from their original positions. The movement of this region occurs in a direction opposite (that is, away from the cavity) from that of the nearby loop between TM3 and TM4 mentioned earlier (into the cavity).
Many regions of the periplasmic domain are also altered. It is intriguing that the loop that forms the bottom and part of the “left” wall of the periplasmic drug binding site (20) is shifted significantly (indicated by an arrowhead in Fig. 5). The side chain of one of the residues that appear to be involved in ligand binding, Glu673 (see Fig. 5 in reference 20), is part of this shifted region, and in our model its side chain is moved away from the position of bound ligand about 6 Å in comparison with the position in the wild-type protein.
Accessibility of the loop between TM3 and TM4.
The conformational alterations observed in the mutant AcrB proteins were extensive. Although the crystallization conditions were similar to those employed earlier (21, 22), we wanted noncrystallographic evidence to strengthen our conclusion that the region involving Phe386 and Phe388 is indeed altered strongly in its conformation in the mutant proteins. We therefore mutated the Phe386 residue to cysteine and examined the accessibility of this residue for modification with fluorescent maleimide reagents. As shown in Fig. 7, Cys386 mutant AcrB proteins containing additional mutations in the putative proton relay complex (D407A, D408A, K940A, and T978A) were modified by MIANS significantly more rapidly than the protein not containing any second mutation. Because the fluorophore of MIANS (anilinonaphthalene sulfate) is affected by its environment in terms of its fluorescence, we also used a coumarin maleimide (CPM), which is much less sensitive to its environment, and obtained similar results (not shown). These results suggest that the conformation of the region between TM3 and TM4 is indeed altered in the D407A, D408A, K940A, and T978A mutants.
FIG. 7.
Reaction of purified CL-F386C AcrBHis mutants with the fluorescence probe MIANS. MIANS labeling was carried out with 0.5 μM purified protein in 2 ml of 20 mM HEPES-KOH (pH 7.5)-50 mM NaCl-0.02% dodecyl maltoside. Reactions were initiated by the addition of MIANS to a final concentration of 5 μM, and the fluorescence increase was recorded continuously at 430 nm (excitation, 330 nm). Mutants were as follows: 1, CL-F386C; 2, CL386-N109A; 3, CL386-D407A; 4, CL386-D408A; 5, CL386-K940A; and 6, CL386-T978A.
DISCUSSION
In previously determined structures of AcrB, the side chains of Asp407, Asp408, Lys940, and Thr978 are close to each other and appear to form a salt bridge/hydrogen-bonding network (Fig. 1). It may be postulated that AcrB in this “resting-state” conformation is ready to bind substrates, as the central cavity of this conformer has indeed been shown to bind various drugs (22).
As a working hypothesis, we assumed that the ɛ-amino group of Lys940 is protonated and the carboxyl groups of Asp407 and Asp408 are deprotonated, in view of their close proximity (17). During the process of proton translocation, one of the carboxyl groups, perhaps that of Asp407, can be postulated to become protonated. This is similar to what has been assumed for many active transporters energized by proton motive force (for example, see the case of the lactose permease LacY [1]). This protonation of an acidic residue is expected to disturb the tight salt bridge/hydrogen-bonding network of the four residues, distort the positions of these residues relative to each other, and initiate a series of conformational changes that would result in the transport of drugs, as stated in the accompanying paper (17).
We undertook this study on the assumption that disturbing the interactions among Asp407, Asp408, Lys940, and Thr978 might in some way mimic the consequences of protonation of one of the carboxyl residues. Indeed, we found that conversion of any of these residues to alanine produced a widespread conformational alteration (Fig. 3 to 6). It is also remarkable that mutation of any of the four residues produced nearly identical new conformations, suggesting that the trigger for change is the disruption of the same salt bridge/H-bonding network. Although it may be argued that the replacement of a charged residue with an uncharged Ala residue may disturb the protein structure simply because of the unfavorable thermodynamics of insertion of the remaining charged residues into the membrane, without any relation to H+ translocation, the same altered conformation in the T978A protein, where charged residues have not been altered, refutes this interpretation.
The new conformation shows that the TM domains of the three protomers still form a large central cavity with a diameter of about 35 Å. However, the diameter of this central cavity decreases to about 6 Å in a region close to the ceiling due to the protrusion of the loop containing Phe386 and Phe388 (Fig. 6). This is the loop that forms a multidrug binding site in wild-type AcrB (22), but this region may become too tight to bind large drugs in these AcrB mutants. If we assume that the new conformation mimics one of the transient stages after the binding of drugs by resting-state AcrB, it may be that the conformational alteration induced by protonation pushes the drug molecules away from the initial site to a new position closer to the final stage of their expulsion. Drug binding studies, both biochemical and crystallographic, are obviously needed to follow up this hypothesis.
Compared with the trimer of wild-type AcrB, there is an increase in intersubunit distance in the mutant homotrimers. For example, the distance between the top regions of TM1 and TM8′ from adjacent subunits is increased about 2 Å in the K940A structure. This change opens the vestibule located above these TM helices some 3 Å. It also enlarges the bottom of the cavity opened to the cytoplasm by 2 Å. Similar changes were found in the other mutant AcrB proteins. Anisotropic network model calculations (9) showed that the motion in the central cavity of AcrB is dominated by breathing vibrational modes. We suspect that the expansion of the AcrB mutants mimics this breathing motion.
Our hope was that the conformation of the mutant proteins would give us not only a glimpse of the transitional states of the AcrB transporter during a normal cycle, but also hints on the pathway of drug molecules during export, especially within the periplasmic domain. However, alterations were not extensive in the conformation of the periplasmic domain (Fig. 5), and the reasons for this outcome are not known.
ADDENDUM IN PROOF
After this paper was accepted for publication, two papers (M. A. Seeger, A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos, Science 313:1295-1298, 2006; S. Murakami, R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi, Nature, on-line 16 August 2006) describing an asymmetric trimer structure of AcrB have appeared. One of the AcrB protomers in this asymmetric structure, called the O or extrusion unit, is described as having a disrupted interaction in the D407-D408-K940-T978 network. Some features of our mutant structure model resemble this subunit: for example, there are, in both structures, an extension of the N-terminal (upper) end of the TM helix 8 (Fig. 4), the reorientation of the Lys940 side chain, and the movement of the PC2 periplasmic domain toward the PC1 domain.
Acknowledgments
This study was supported in part by U.S. Public Health Service grants GM074027 (to E.W.Y.) and AI09644 (to H.N.). Y. Takatsuka was supported by a postdoctoral fellowship from the Japan Society for Promotion of Science during the early stages of this study.
REFERENCES
- 1.Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2002. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610-615. [DOI] [PubMed] [Google Scholar]
- 2.Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. [DOI] [PubMed] [Google Scholar]
- 3.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 4.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:764-767. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. [DOI] [PubMed] [Google Scholar]
- 6.Fujihira, E., N. Tamura, and A. Yamaguchi. 2002. Membrane topology of a multidrug efflux transporter AcrB in Escherichia coli. J. Biochem. (Tokyo) 131:145-151. [DOI] [PubMed] [Google Scholar]
- 7.Guan, L., and T. Nakae. 2001. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 183:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-120. [DOI] [PubMed] [Google Scholar]
- 9.Lu, W. C., C. Z. Wang, E. W. Yu, and K. M. Ho. 2006. Dynamics of the trimeric AcrB transporter protein inferred from a B-factor analysis of the crystal structure. Proteins 62:152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma, D., D. A. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 12.Madej, T., J. F. Gibrat, and S. H. Bryant. 1995. Threading a database of protein cores. Proteins 23:356-369. [DOI] [PubMed] [Google Scholar]
- 13.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 14.Murakami, S., and A. Yamaguchi. Multidrug-exporting secondary transporters. Curr. Opin. Struct. Biol. 13:443-452. [DOI] [PubMed]
- 15.Otwinowski, Z., and M. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 16.Reyes, C. L., and G. Chang. 2005. Structure of the ABC transporter MsbA in complex with ADP. vanadate and lipopolysaccharide. Science 308:1028-1031. [DOI] [PubMed] [Google Scholar]
- 17.Takatsuka, Y., and H. Nikaido. 2006. Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J. Bacteriol. 188:7284-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 19.Vagin, A., and A. Teplyakov. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D 56:1622-1624. [DOI] [PubMed] [Google Scholar]
- 20.Yu, E. W., J. R. Aires, G. McDermott, and H. Nikaido. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 187:6804-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu, E. W., J. R. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185:5657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 23.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:7190-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]