Abstract
Microcin E492 (MccE492) is a bactericidal protein secreted by Klebsiella pneumoniae that is active against various species of Enterobacteriaceae. Interaction of MccE492 with target cells leads to the depolarization and permeabilization of their inner membranes. Several MccE492-specific proteins are required for the maturation and secretion of active MccE492. Surprisingly, the expression of only MceA, the polypeptide backbone of MccE492, is shown here to be toxic by itself. We refer to this phenomenon as endogenous MceA bactericidal activity to differentiate it from the action of extracellularly secreted MccE492. The toxicity of endogenous MceA is enhanced by an efficient targeting to the inner membrane. However, a periplasmic intermediate state is not required for MceA toxicity. Indeed, endogenous MceA remains fully active when it is fused to thioredoxin-1, a fast-folding protein that promotes retention of the C terminus of MceA in the cytoplasm. The C-terminal domain of MccE492 is required only for delivery from the extracellular environment to the periplasm, and it is not required for inner membrane damage. A common component is absolutely essential for the bactericidal activity of both endogenous MceA and extracellular MccE492. Indeed, toxicity is strictly dependent on the presence of ManYZ, an inner membrane protein complex involved in mannose uptake. Based on these findings, we propose a new model for cell entry, inner membrane insertion, and toxic activity of MccE492.
Bacteriocins are antibacterial proteins secreted into the medium to kill competing bacteria. Microcin E492 (MccE492) is a low-molecular-weight bacteriocin produced by Klebsiella pneumoniae RYC492 that is active against several species of Enterobacteriaceae, including Escherichia, Salmonella, Enterobacter, and Klebsiella spp. (12, 13, 55). The genetic determinants responsible for MccE492 synthesis, secretion, and immunity are encoded by a 13-kb segment of the Klebsiella pneumoniae chromosome. This gene cluster of 10 open reading frames, mceA to mceJ, has been cloned and shown to be functional in Escherichia coli (32).
The structural gene for MccE492, mceA, is expressed as a 99- or 103-residue preprotein, preMceA (33), which is posttranslationally modified under certain growth conditions through a poorly understood pathway involving MceC, MceI, and MceJ. This modification consists in the covalent attachment to the C-terminal serine of a glucose moiety linked to a trimer of 2,3-dihydroxybenzoylserine. The modified form of MccE492 purified from culture supernatants has been shown to be four to eight times as potent as the nonmodified form (55). However, mutations in either mceC, mceI, or mceJ have been reported to completely abolish the bactericidal activity of the secreted microcin (32), suggesting that the posttranslational modification undergone by preMceA is essential for activity on target cells. More extensive and systematic work will be required to determine whether this modification only improves activity or is absolutely required for antibacterial action.
preMceA is cleaved at its N terminus before or during export through a dedicated ABC exporter made of MceF, MceG, and MceH to yield the active, modified, 84-residue microcin MccE492 (32, 33). MccE492 purified from a culture supernatant has an apparent molecular mass of 22 kDa in physiological buffer, which would correspond to either a dimer or a trimer (16).
MccE492 is recognized by three receptors, FepA, Fiu, and Cir, on the outer membrane of E. coli (43, 54). All are well-described catechol-type siderophore receptors. The specificity of this recognition has been proposed to be enhanced by the posttranslational modification of MccE492, which mimics a catechol-type siderophore (55). MccE492 translocation across the outer membrane requires TonB (45), an inner membrane protein that transduces energy from the proton motive force to various outer membrane proteins, including iron chelate receptors such as FepA, Fiu, and Cir and the vitamin B12 receptor (40).
Several lines of evidence indicate that MccE492 inserts into the inner membrane to form toxic pore-like structures responsible for membrane depolarization and permeabilization. Indeed, in vitro experiments have shown that purified MccE492 forms voltage-independent cation-selective single channels in planar phospholipid bilayers (16, 34). In vivo, MccE492 has been shown to induce a depolarization of the inner membrane, as evidenced by amino acid uptake and tetraphenylphosphonium ion accumulation measurements (14), as well as a permeabilization of the inner membrane, as shown by o-nitrophenyl-β-d-galactopyranoside diffusion assay (16).
MccE492-producing bacteria synthesize an immunity protein, MceB, which renders the cell resistant to the microcin. Although MceB has been shown to be associated with the inner membrane (33), the molecular mechanism of this immunity remains enigmatic.
Since MccE492 is a small protein that is unlikely to form a pore structure as a monomer, an oligomerization process or association with host proteins is probably required for its activity. However, the molecular identity of this toxic structure, as well as the mechanism underlying its formation, is still unknown.
We recently reported that MccE492 has the unusual ability to assemble into amyloid fibrils that exhibit the same morphological, structural, and biochemical properties as the aggregates observed in amyloid-associated diseases, such as Alzheimer's, Parkinson's, or prion diseases. Since the formation of amyloid fibrils is simultaneously associated with a loss of MccE492 solubility and bactericidal activity, it was proposed that bacteria could employ the membrane-permeabilizing properties of small oligomeric prefibrillar amyloid aggregates to kill cells (2).
Here, we report that the expression of MceA, the polypeptide backbone of MccE492, is toxic by itself. We refer to this phenomenon as endogenous MceA bactericidal activity to differentiate it from the action of extracellular MccE492, which is secreted in the medium. We have taken advantage of this property to further characterize the mechanism of action of this microcin. We find that the toxicity of endogenous MceA is enhanced by an efficient targeting to the inner membrane. Endogenous MceA remains fully active when it is C-terminally fused to thioredoxin-1, a fast-folding protein that ensures retention in the cytoplasm. Thus, endogenous MceA probably integrates in the inner membrane with its C terminus in the cytoplasm without being fully exported to the periplasm. In addition, to determine which portions of MceA are required for cell entry or for inner membrane damage, we have engineered a series of constructs expressing full-length or truncated MceA. By comparing their bactericidal activities as endogenous proteins or as secreted proteins acting on target cells, we show that the C-terminal domain of MccE492 is required only for delivery from the extracellular environment to the periplasm and that it is not involved in inner membrane damage. Finally, through a random mutagenesis approach, we find that an inner membrane protein complex involved in mannose uptake, ManYZ, is essential for the bactericidal activity of both endogenous MceA and extracellular MccE492.
MATERIALS AND METHODS
Reagents.
Liquid and solid media were prepared as described previously (39). Antibiotics were used at the following concentrations: ampicillin (Ap), 200 μg/ml; chloramphenicol (Cm), 30 μg/ml; kanamycin (Kn), 40 μg/ml; spectinomycin (Sp), 50 μg/ml; and tetracycline (Tc), 7.5 μg/ml. The anti-MceA rabbit antiserum (AnaSpec) was raised against a C-terminal fragment of MceA (SGSGYNSATSSSGSGS; synthesized at AnaSpec). The anti-OmpA rabbit antiserum was raised against sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-purified OmpA (Covalab).
Bacterial strains and plasmids.
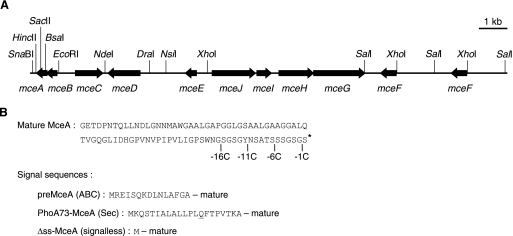
The E. coli strains and plasmids used in this study are described in Tables 1 and 2, respectively. Strain AB2 was obtained by phage P1vir-mediated transduction (39) of AB1, using a lysate grown on MC4100 and selecting for Trp+ recombinants. AB3 was obtained by transduction of the ΔmanXYZ::cat deletion of DPE271ΔLPM into DB503. AB8 was obtained by transduction of the ΔcyaA1400::Kn deletion of SP850 into DB503. Phage M13 mp18 growth was assessed for strain JM101 and for an isogenic strain bearing a trxA14::Kn mutation (49) in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The genes involved in MccE492 production and immunity used for the constructions employed in this study were all derived from pJEM15 (57). In contrast to other plasmids harboring the MccE492 genes, pJEM15 does not interfere with cell growth and was therefore chosen for this study. Note that the orientation of the 6.9-kb XhoI fragment of pJEM15 is inverted relative to that for the K. pneumoniae chromosome and that this plasmid contains a duplication of the 2.5-kb SalI fragment comprising mceF (Fig. 1A). The mceA coding sequence in pJEM15 is identical to that originally obtained from plasmid pJAM434 (33) and deposited in Swiss-Prot under accession number Q9Z4N4. pAB3 (Apr) was constructed by ligating EcoRI/BspEI-digested pBR322 with an oligonucleotide containing an NsiI site. pAB4 was obtained by ligating an NsiI fragment of pJEM15 containing mceE, mceJ, mceI, mceH, mceG, and mceF (one NsiI site is in an unmapped region upstream of the two mceF genes) into pAB3. pAB5 (Tcr) was produced by ligating an SnaBI-DraI fragment of pJEM15 containing mceA, mceB, mceC, and mceD into pACYC184 digested with ScaI and DraI (the DraI site was recovered). A BglII site was inserted in pAB5 by PCR downstream of mceA to yield pAB6. pAB7, containing mceB, mceC, and mceD, was obtained by digesting pAB6 with BglII and BsaI, filling in with Klenow fragment, and self-ligating. pAB8, containing mceC and mceD, was obtained by digesting pAB6 with BglII and EcoRI, filling in with Klenow fragment, and self-ligating. To construct pBAD101 (Spr), a fragment containing the promoter of the arabinose BAD operon (PBAD) and the araC gene was obtained by digesting pBAD24 (25) with BspHI and filling in with Klenow fragment. This fragment was cloned in EcoRI/HindIII-digested and Klenow fragment-treated pGB2, a low-copy-number vector with a pSC101 replication origin (8). pAB10, expressing preMceA with its 15-residue ABC signal sequence (ABC-MceA) under the control of the PBAD promoter, was constructed by PCR amplification of pJEM15, using as primers 5′-CGCGTCATGAGAGAAATTAGTC and MCEADON (5′-GGAATAGATCTAGGCATAAGGATCTCGCACC). After treatment with Klenow fragment, the amplified DNA was digested with BspHI and cloned in NcoI/HincII-digested pBAD101. Note that it is unknown whether the original ABC signal sequence of preMceA consists of 15 or 19 residues in K. pneumoniae, due to the presence of two possible start codons. However, the presence of a better Shine-Dalgarno consensus would favor the 15-residue signal sequence in E. coli (33). We originally cloned both coding sequences in pBAD101. Since the plasmid corresponding to the 15-residue signal sequence (Fig. 1B), pAB10, was associated with an MccE492 activity in culture supernatants much higher than that of the plasmid expressing preMceA with the 19-residue signal sequence (data not shown), the former was chosen for this study. To construct pAB9, expressing a fusion of the alkaline phosphatase (PhoA) signal sequence to the mature portion of preMceA (PhoA-MceA) under the control of the PBAD promoter, a region comprising the end of the PBAD promoter and the beginning of the phoA coding sequence was amplified by performing a PCR on pBAD24phoA (1), using as primers 5′-CCTGACGCTTTTTATCGC (PBAD promoter) and 5′-GGATCGGTCTCTCCTGCTTTTGTCACAGG. The amplified DNA was digested with NheI and BsaI and ligated into pAB10 partially digested with NheI/BsaI. pAB11, expressing a fusion of the signal sequence of PhoA73 (Fig. 1B) to the mature portion of preMceA (PhoA73-MceA) under the control of the PBAD promoter, was constructed similarly to pAB9, except that the PCR was performed on pBAD24phoA73 (lab collection), a plasmid with a mutation (L14Q) in the PhoA signal sequence (38). pAB12, expressing the mature portion of preMceA immediately following the N-terminal methionine without any signal sequence (Δss-MceA) under the control of the PBAD promoter, was constructed by performing a PCR on pAB6, using as primers 5′-CGTAGCTAGCAGGAGGAATTCACCATGGGAGAGACCGATC and primer MCEADON, treating with Klenow fragment and NheI, and ligating with NheI/HincII-digested pBAD101. To construct pAB13, expressing the mature portion of preMceA with an N-terminal hexahistidine tag (6H-MceA) under the control of the PBAD promoter, a fragment encoding mature MceA was obtained by digesting pAB10 with BsaI and treating with Klenow fragment and HindIII. This fragment was cloned in NcoI-, Klenow fragment-, and HindIII-digested pBADHisEYGFD (pBAD24 vector with a hexahistidine tag; a kind gift of F. Duong). This resulted in the insertion of one alanine between the N-terminal hexahistidine tag and the mature portion of preMceA. pAB10-1C, expressing ABC-MceA without the C-terminal serine (Fig. 1B), was obtained by performing a PCR on pAB10 by use of primers 5′-CTATTAACCACTACCGGAACTGGATG and UPSACII (5′-CGGATTAGGATCAGCAGC), treating with Klenow fragment and SacII, and ligating with HincII/SacII-digested pAB10. pAB10-6C, -11C, and -16C, expressing ABC-MceA without the 6, 11, and 16 C-terminal residues (Fig. 1B), were constructed similarly, using primers 5′-CTATTAGGATGTTGCGCTGTTATAACC, 5′-CTATTAATAACCACTACCGCTACCATTCC, 5′-CTATTAACCATTCCAGCTTGGCC, and 5′-CTATTACCCGATGAGTACAGGGATGG, respectively, with primer UPSACII. pAB13-1C, -6C, -11C, and -16C, expressing 6H-MceA without the 1, 6, 11, and 16 C-terminal residues, were obtained by replacing the SacII-EagI fragment of pAB13 containing the 3′ portion of mceA by the cognate fragments from pAB10-1C, -6C, -11C, and -16C, respectively. To construct pAB16, expressing TrxA fused to the C terminus of Δss-MceA, the trxA coding sequence was obtained by performing a PCR on DB503 genomic DNA, using as primers 5′-CCGATAAAATTATTCACCTGACTGACGAC and 5′-GACAGAGATCTCGAAGTCAACACTAAGTTAGCATGAC. The amplified DNA was digested with BglII and ligated with HincII/BglII-digested pAB12. pAB15, expressing TrxA fused to the C terminus of PhoA73-MceA, was constructed by replacing the SpeI-SacII fragment of pAB16 containing the 5′ portion of mceA by the cognate fragment from pAB11. pAB17, expressing ManZ under the control of the PBAD promoter, was constructed by PCR amplification of pJFP-H6M, using as primers 5′-CAGCGAGCTCAGGAGGAATTCACCATGAGACACCATCACCATCAC and 5′-GTCTAGATATTATACCCGGGCCAGTCCCAGCAGGCCGCAAGCGTAAC. After treatment with Klenow fragment, the amplified DNA was digested with SacI and cloned in SacI/HincII-digested pBAD33.
TABLE 1.
E. coli strains
| Strain | Genotype/description | Source/reference |
|---|---|---|
| DH5α | F−deoR recA1 endA1 hsdR17 supE44 λ−thi-1 gyrA96 relA1 | 26 |
| DH10B-T1 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara-leu)7697 galU galK λ−rpsL nupG fhuA | Invitrogen |
| BL21 | F−ompT hsdS(rB− mB−) gal dcm | Amersham |
| MC4100 | F−araD139 relA1 thi rpsL150 flbB5301 Δ(argF-lac)U169 deoC1 ptsF25 rbsR | 6 |
| DB503 | MC4100 malE16-1 Δara714 | Lab collection |
| AB4 | DB503 manY::Tn10 | This study |
| DB512 | MC4100 malE18-1 Δara714 rpoA341 | Lab collection |
| AB1 | λ− Δ(tonB-trpB)873 his IN(rrnD-rrnE)1 rph-1 | W3110hisΔtrpAB in reference 59 |
| AB2 | AB1 trp+tonB+ | Lab collection |
| MC1000 | F−araD139 Δ(ara-leu)7697 Δ(lac)X74 rpsL150 galU galK thi | 7 |
| AD85 | MC1000 malFΔ3 phoAΔ(PvuII) phoR F′lacIQpro with pACYClacIQ | 15 |
| AD413 | AD85 prlA4 | 15 |
| DPE271ΔLPM | ΔmanXYZ::cat | 24 |
| AB3 | DB503 ΔmanXYZ::cat | This study |
| MG1655 | λ−ilvG rfb-50 rph-1 | 3 |
| AB5 | MG1655 manY::Tn10 | This study |
| JCB606 | E. coli RV Δnir dipZ | 9 |
| AB6 | JCB606 manY::Tn10 | This study |
| H1443 | MC4100 aroB | 27 |
| H1876 | H1443 fepA::Tn10 fiu::MudX cir | 27 |
| AB7 | DH10B-T1 with pAB4 and pAB7 | This study |
| SP850 | λ− e14−relA1 spoT1 ΔcyaA1400::Kn thi-1 | 53 |
| AB8 | DB503 ΔcyaA1400::Kn | This study |
TABLE 2.
Plasmids
| Plasmid | Description | Source/reference |
|---|---|---|
| pJEM15 | Microcin E492 genes (Fig. 1A) | 57 |
| pAB3 | pBR322 with NsiI site, Apr | This study |
| pAB4 | pAB3 with mceE mceJ mceI mceH mceG mceF | This study |
| pAB5 | pACYC184 with mceA mceB mceC mceD; Tcr | This study |
| pAB6 | pAB5 with BglII site | This study |
| pAB7 | pAB6 ΔmceA | This study |
| pAB8 | pAB6 ΔmceAB | This study |
| pBAD24 | Vector with pBR322 replication origin, PBAD promoter and araC; Apr | 25 |
| pGB2 | Vector with pSC101 replication origin; Spr | 8 |
| pBAD101 | PBAD promoter and araC in pGB2 | This study |
| pBAD24phoA73 | pBAD24 expressing PhoA73 | Lab collection |
| pAB9 | pBAD101 expressing PhoA-MceA | This study |
| pAB10 | pBAD101 expressing ABC-MceA | This study |
| pAB11 | pBAD101 expressing PhoA73-MceA | This study |
| pAB12 | pBAD101 expressing Δss-MceA | This study |
| pAB13 | pBAD24 expressing MceA with an N-terminal hexahistidine tag (6H-MceA) | This study |
| pAB10-1C, -6C, -11C, and -16C | pAB10 expressing ABC-MceA without 1, 6, 11, and 16 C-terminal residues | This study |
| pAB13-1C, -6C, -11C, and -16C | pAB13 expressing 6H-MceA without 1, 6, 11, and 16 C-terminal residues | This study |
| pAB15 | pAB11 expressing TrxA fused to the C terminus of PhoA73-MceA | This study |
| pAB16 | pAB12 expressing TrxA fused to the C terminus of Δss-MceA | This study |
| pJFL | manX driven by Ptac; Apr | 52 |
| pTSP11 | manY driven by Ptac; Apr | 20 |
| pJFP-H6M | manYZ driven by Ptac; Apr | 21 |
| pJFLPM | manXYZ driven by Ptac; Apr | 21 |
| pBAD33 | Vector with pACYC184 replication origin, PBAD and araC; Cmr | 25 |
| pAB17 | pBAD33 with manZ driven by PBAD | This study |
FIG. 1.
(A) Microcin E492 gene cluster from plasmid pJEM15. Open reading frames are represented by arrows. The restriction sites relevant for plasmid constructions are indicated. (B) Amino acid sequence of mature 84-residue MceA. The positions of the stop codons inserted to generate the C-terminally truncated MceA mutants are indicated (-16C, -11C, -6C, and -1C). The posttranslational modification of the C-terminal serine is symbolized by an asterisk. The original ABC signal sequence of preMceA and the signal sequence of PhoA73-MceA are shown underneath. The mutated amino acid in PhoA73-MceA (Q14) is underlined. The signalless construct (Δss-MceA) consists of a methionine residue followed by the mature portion of preMceA.
Purification of MccE492.
MccE492 was purified from E. coli BL21(pJEM15) culture supernatant as described previously (2).
Bacterial viability assay by fluorescence.
Cells were grown in LB medium to an A600 of 0.3, induced with 0.2% arabinose for 1 h, resuspended in the same volume of H2O (live cells) or 70% isopropanol (dead cells used as a control), incubated for 1 h at room temperature, washed, and resuspended in H2O to a final A600 of 0.6. One milliliter of this suspension was mixed with 3 μl of a dye mixture containing 1.67 mM SYTO 9 dye and 10 mM propidium iodide (LIVE/DEAD BacLight Viability Kit L-7012; Molecular Probes). After 15 min of incubation at room temperature, fluorescence emission was measured at 500 and 635 nm, with the excitation wavelength set at 470 nm. Viability is expressed as a ratio of the emission at 500 nm to that at 635 nm. DB503 pBAD101 cells resuspended in H2O were taken as the reference for live cells (100% viability), while the same cells resuspended in 70% isopropanol were taken as the reference for dead cells (0% viability).
MccE492 activity assay by the critical dilution method.
Purified MccE492 (10 mg/ml) was serially diluted in 50% methanol. Four-microliter drops of diluted samples were laid onto LB agar plates previously covered with 3 ml of LB top agar containing 2 × 107 cells. After 15 h of incubation at 37°C, the activity was determined by the critical dilution method and expressed in arbitrary units per ml (37).
Proline uptake assay.
Proline uptake was measured essentially as described previously (1). Cultures were grown in LB medium to an A600 of 0.2 to 0.3 and induced for the indicated times with 0.2% arabinose. Cells were washed three times with MM (M63 salts, 1 mM MgCl2) and resuspended at ∼2 A600. [3H]proline (50 Ci/mmol) (MT531; Hartmann, Germany) was diluted in MM with a 100-fold excess of unlabeled proline. Cells were exposed to 4 μM proline (2 μCi/ml). After 20, 40, and 60 seconds, 200 μl of cells was filtered through nitrocellulose (DAWP, 0.65 μm; Millipore). The filters were immediately washed with 5 ml of MM, and the radioactivity was measured by liquid scintillation. An unfiltered aliquot was used to determine the total radioactivity input. The values of proline uptake were normalized to the A600, and the slope of uptake was calculated by linear regression. Approximately 10 to 25% of the input proline was recovered after 1 minute from uninduced control cells.
Generation and mapping of transposon insertion mutants.
Cells expressing endogenous MceA upon induction with arabinose were grown at 37°C in LB containing the appropriate antibiotics to an A600 of 1.5. Two hundred fifty microliters of culture was centrifuged and resuspended in 1 ml of LB supplemented with 10 mM MgSO4 and λNK1316 phages (31) in a final ratio of 0.3 phage per bacterium. The suspension was incubated for 15 min at 37°C without shaking, and, after addition of 1 ml of LB, further incubated for 90 min under agitation. The total number of cells bearing a transposon was measured by spreading appropriate dilutions onto LB plates supplemented with kanamycin. Cells potentially resistant to MccE492 (arabinose resistant [Arar]) were obtained by diluting the suspension 1:10 in LB medium containing the appropriate antibiotics and incubating overnight at 39.5°C to allow phenotypic expression of mutant genes. Various dilutions of this culture were spread onto LB plates supplemented with the appropriate antibiotics and 0.2% arabinose and incubated overnight at 39.5°C. Arar colonies were then tested for MccE492 sensitivity by streaking bacteria on LB plates across a drop of purified MccE492 (1 mg/ml) and incubating overnight at 37°C. The ability to metabolize mannose was assessed by streaking bacteria on MacConkey plates supplemented with 1% mannose and on minimal plates (M63 salts, 1 mM MgSO4, 4 μg/ml thiamine) containing 0.2% mannose as the only carbon source. The insertions were transferred into DB503 by P1vir phage-mediated transduction.
Transposon insertions were mapped as described previously (29). Briefly, circularized fragments of Sau3AI-digested mutant chromosomal DNA were subjected to inverse PCR (30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s), using a pair of divergently oriented primers designed for the ends of the transposon (ah1 and ah2). The amplified chromosomal region flanking the transposon was sequenced with the same primers.
Pulse-labeling and immunoprecipitation.
Cells from saturated cultures in LB medium were diluted 1:50 and grown overnight in M63 medium containing 0.2% (vol/vol) glycerol as a carbon source and all amino acids except methionine and cysteine at a final concentration of 50 mg/liter, diluted 1:40, and grown at 37°C to an A600 of 0.2. Cells were induced with 0.2% arabinose for 1 h, and 1-ml samples were labeled with 50 μCi/ml of [35S]methionine (IS-103; Hartmann, Braunschweig, Germany) for 1 min. Labeling was stopped by transferring the samples to chilled tubes containing 0.1 ml of 0.2% (wt/vol) methionine. Cells were centrifuged for 2 min at 13,000 × g at 4°C, resuspended in 50 μl of SDS buffer (1% [wt/vol] SDS, 10 mM Tris [pH 8], 1 mM EDTA), and heated for 2 min at 90°C. After 5 min at room temperature, 800 μl of KI buffer (50 mM Tris [pH 8], 150 mM NaCl, 2% [vol/vol] Triton X-100) was added. After 10 min on ice, the lysates were centrifuged for 10 min at 13,000 × g. Anti-MceA or anti-OmpA antiserum was added to 300 μl of lysates. After 12 h on ice, immune complexes were bound to an excess of fixed Staphylococcus aureus cells (IgGsorb) for 1 h on ice. The cells were washed twice with 1 ml of HB (50 mM Tris [pH 8], 1 M NaCl, 1% Triton X-100, 1 mM EDTA) and once with 1 ml of 10 mM Tris (pH 8). Pellets were resuspended in 50 μl of SB+ (50 mM Tris [pH 6.8], 5% [vol/vol] β-mercaptoethanol, 1% SDS, 0.0025% [wt/vol] bromophenol blue, and 8.5% [vol/vol] glycerol) and boiled for 2 min, and eluates were loaded onto 19.6% SDS-polyacrylamide gels, using a “high-Tris” buffer system containing 6 M urea (5). Gels were fixed, dried, and autoradiographed.
Pulse-labeling and detection of hexahistidine-tagged proteins.
Cells were grown, labeled, and lysed as described above, except for the following modifications: cells were induced for 1 min and labeled for 5 min, the SDS buffer was free of EDTA, and the KI buffer contained 300 mM NaCl. Four hundred microliters of lysates was incubated at 4°C for 30 min with 20 μl of nickel-nitrilotriacetic acid agarose matrix (QIAGEN) and washed twice with 10 mM Tris (pH 8), 1 M NaCl, 1% Triton X-100 and once with 1 ml of 10 mM Tris (pH 8). Pellets were resuspended in 40 μl of SBX (80 mM Tris [pH 6.8], 1% [vol/vol] β-mercaptoethanol, 2% SDS, 0.01% [wt/vol] bromophenol blue, 10% [vol/vol] glycerol, and 10 mM EDTA) and incubated for 20 min at 37°C, and eluates were loaded onto 19.6% SDS-polyacrylamide gels. Gels were fixed, dried, and autoradiographed.
Immunoblotting.
Samples were diluted in SB+ (see “Pulse-labeling and immunoprecipitation” above), boiled for 5 min, and loaded onto 15% SDS-polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (Protran BA 85; Schleicher & Schuell, Keene, NH). MceA was detected with an enhanced chemiluminescence kit (Amersham) by use of anti-MceA antiserum (1:3,000) and horseradish peroxidase-linked anti-rabbit donkey antibody (1:5,000; Amersham).
RESULTS
The expression of MceA alone is bactericidal.
The mceA gene encodes preMceA, which is modified and cleaved before or during secretion into the extracellular medium. The secreted, modified protein is called MccE492, and its polypeptide backbone is referred to as MceA. Several specific proteins are absolutely required for the maturation and secretion of functional MccE492 into the extracellular medium. These proteins are believed to be involved either in export (MceFGH) or in a posttranslational modification important for MccE492 recognition by outer membrane receptors on target cells (MceCIJ). Therefore, they may not be required for self-killing of a cell that expresses MceA without the immunity protein MceB. Since MccE492 normally gains access to the inner membranes of target cells from the periplasmic side of the membrane, we tried first to assess the toxicity of MceA targeted to the periplasm to test this hypothesis. To this goal, we constructed a plasmid, pAB9, which expresses the signal sequence of alkaline phosphatase (PhoA) fused to the 84-residue mature portion of preMceA (PhoA-MceA). PhoA is a periplasmic protein that is exported through the general secretory pathway. Translocation via this pathway is achieved by the Sec translocase, a large complex that includes the SecYEG inner membrane proteins (11, 56). The expression of this fusion protein was placed under the control of the arabinose-inducible PBAD promoter (25). This promoter was chosen because of its very low basal expression level, which is of key importance when dealing with toxic proteins. However, pAB9 was found to strongly interfere with cell growth even without arabinose induction. We therefore designed a new plasmid, pAB11, which is identical to pAB9 except that it codes for a fusion of a mutated PhoA signal sequence (PhoA73) to mature MceA (PhoA73-MceA). PhoA73 has a point mutation (L14Q) that reduces export to 30% of that of wild-type PhoA (38). We also designed a plasmid expressing the cleaved, mature portion of preMceA without any signal sequence (Δss-MceA). This plasmid, pAB12, should produce MceA that accumulates in the cytoplasm, which would in principle not be toxic.
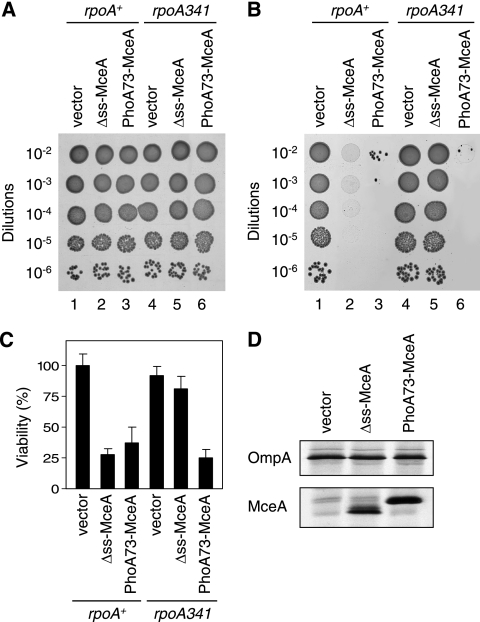
Figure 2A shows that neither pAB11 (PhoA73-MceA) nor pAB12 (Δss-MceA) interferes with colony formation or with the size of the colonies in the absence of arabinose. Upon arabinose induction, pAB11 confers strong toxicity (Fig. 2B, lane 3), which indicates that MceA is toxic when it is targeted to the general secretory pathway. Surprisingly, pAB12 (Fig. 2B, lane 2) is also toxic in the presence of arabinose. In order to determine if the levels of toxicity of pAB11 and pAB12 are similar, these two plasmids were introduced in a strain bearing an rpoA341 mutation to lower expression from the PBAD promoter. rpoA codes for the alpha subunit of the RNA polymerase, and the rpoA341 allele specifically impairs the transcription of several positively regulated operons. In particular, it is associated with a more-than-10-fold decrease in transcription from the PBAD promoter (23). Interestingly, while pAB11 maintains a strong toxicity in an rpoA341 strain (Fig. 2B, lane 6), pAB12 is no longer toxic in such a strain (Fig. 2B, lane 5). Similar conclusions can be drawn from fluorescence viability experiments (Fig. 2C). Figure 2D (lower panel) shows that upon arabinose induction, pAB12 and pAB11 express similar amounts of Δss-MceA and PhoA73-MceA proteins, respectively; the expression of OmpA is shown as a control (Fig. 2D, upper panel). Taken together, these results show that the expression of both Δss-MceA and PhoA73-MceA is strongly bactericidal. However, the specific activity of Δss-MceA is lower than that of PhoA73-MceA, indicating that targeting to the general secretory pathway is associated with a higher bactericidal activity. Strains harboring pAB11 or pAB12 are no longer sensitive to arabinose induction when they express the immunity protein MceB from plasmid pAB7 (data not shown). This indicates that the toxicity mediated by endogenous Δss-MceA or PhoA73-MceA is mechanistically related to that of extracellular MccE492.
FIG. 2.
Bactericidal activity of endogenous MceA in the absence of all the other proteins encoded by the MccE492 gene cluster. (A and B) Serial dilutions of overnight cultures of DB503 (rpoA+) with pBAD101 (vector), pAB12 (Δss-MceA), or pAB11 (PhoA73-MceA) and of DB512 (rpoA341) with pBAD101 (vector), pAB12 (Δss-MceA), or pAB11 (PhoA73-MceA) were spotted (5 μl) on LB plates without arabinose (A) or with 0.2% arabinose (B) and incubated overnight at 37°C. Results shown here are representative of three independent experiments. (C) Viability assay of cultures of DB503 (rpoA+) with pBAD101 (vector), pAB12 (Δss-MceA), or pAB11 (PhoA73-MceA) and of DB512 (rpoA341) with pBAD101 (vector), pAB12 (Δss-MceA), or pAB11 (PhoA73-MceA) after 1 h of arabinose (0.2%) induction. Each value represents the average of three independent cultures. Error bars indicate standard deviations. (D) Synthesis levels of Δss-MceA and PhoA73-MceA. Cultures of AB3 (MccE492-resistant strain; see below) with pBAD101 (vector), pAB12 (Δss-MceA), or pAB11 (PhoA73-MceA) were pulse-labeled as described in Materials and Methods. Proteins were immunoprecipitated with anti-MceA antiserum (lower panel) or with anti-OmpA antiserum (upper panel) and detected by SDS-PAGE and autoradiography.
One interpretation for the unexpected toxicity associated with the expression of Δss-MceA would be that Δss-MceA is somehow exported to the periplasm, for instance because it may harbor an internal signal sequence that allows targeting to the Sec translocase. Alternatively, Δss-MceA could be toxic without being exported by inserting into the inner membrane from the cytoplasm.
The bactericidal activity of endogenous MceA is enhanced by an efficient targeting to the inner membrane.
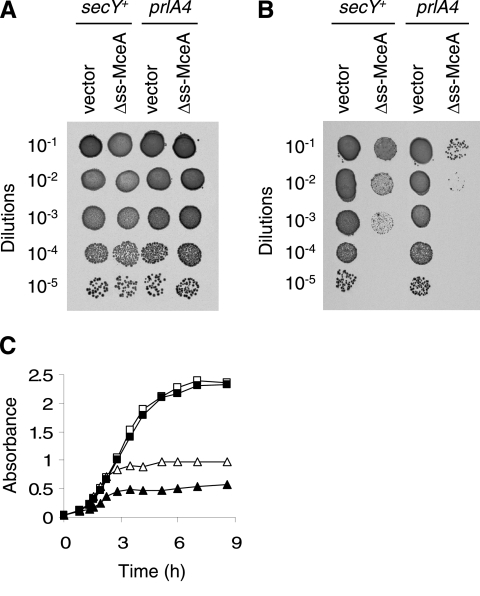
In order to better understand why Δss-MceA expression is toxic, we tried to determine whether Δss-MceA toxicity was dependent on membrane targeting by comparing the bactericidal activities of this protein in wild-type (AD85) and prlA4 (AD413) strains. prlA4 is a secY allele that improves the export of proteins with a variety of defective signal sequences and even allows secretion of proteins entirely devoid of signal sequences (15). Figure 3B shows that Δss-MceA expression is more toxic in a prlA4 strain than in a secY+ strain. A similar conclusion can be drawn from growth experiments performed with a low level of arabinose induction (0.002%). Indeed, Δss-MceA expression arrests cell growth more rapidly in a prlA4 strain than in a secY+ strain (Fig. 3C). Since a more efficient recognition by the Sec translocase (prlA4) leads to increased toxicity, it appears that Δss-MceA toxicity is enhanced by targeting to the inner membrane or to the periplasm. This conclusion is in good agreement with the observations (i) that PhoA73-MceA has a stronger bactericidal activity than Δss-MceA (Fig. 2) and (ii) that the mutation in the signal sequence of PhoA73, which is known to reduce targeting to the Sec translocase compared to wild-type PhoA, is associated with a toxicity of PhoA73-MceA lower than that of PhoA-MceA (data not shown). However, these experiments do not determine whether Δss-MceA toxicity in a secY+ strain is due to partial recognition by the Sec translocase or to membrane targeting by another route.
FIG. 3.
Toxicity of Δss-MceA expression in secY+ and prlA4 strains. (A and B) Serial dilutions of overnight cultures of AD85 (secY+) with pBAD101 (vector) or pAB12 (Δss-MceA) and of AD413 (prlA4) with pBAD101 (vector) or pAB12 (Δss-MceA) were spotted (5 μl) on LB plates without arabinose (A) or with 0.2% arabinose (B) and incubated overnight at 37°C. Results shown here are representative of three independent experiments. (C) Growth curves of AD85 (secY+) with pBAD101 (vector) (□), AD413 (prlA4) with pBAD101 (vector) (▪), AD85 (secY+) with pAB12 (Δss-MceA) (▵), and AD413 (prlA4) with pAB12 (Δss-MceA) (▴) in LB supplemented with 0.002% arabinose. Cell growth was followed by recording the A600 of the cultures at the indicated times.
The bactericidal activity of endogenous MceA does not require complete export to the periplasm.
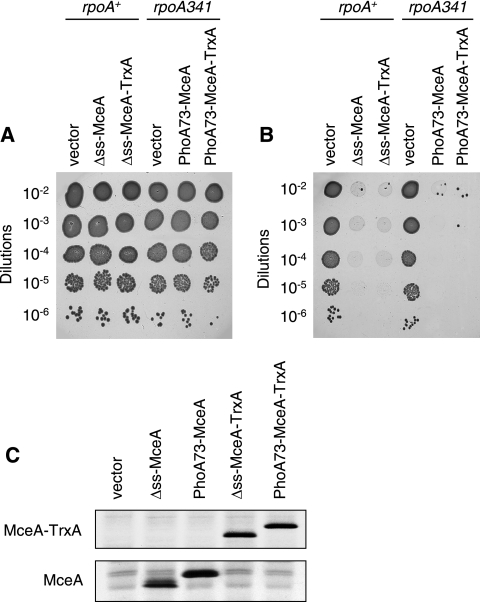
Two distinct mechanisms could account for the toxic activity of endogenous Δss-MceA and PhoA73-MceA shown in Fig. 2. MceA may first need to be fully translocated to the periplasm and then integrate into the inner membrane from the outer side of the membrane. In the case of Δss-MceA, export could be due to partial recognition by the Sec translocase in the absence of a well-defined N-terminal signal sequence. Alternatively, endogenous MceA could insert into the inner membrane from the cytoplasm and be active without first being exported to the periplasm. To discriminate between these two possibilities, we fused thioredoxin-1 (TrxA) to the C terminus of MceA. TrxA is a cytoplasmic protein that rapidly folds and is therefore unable to be exported, even when it is fused to the signal sequence of PhoA (51). Figure 4B shows that fusing TrxA does not alter the toxicity mediated by Δss-MceA or PhoA73-MceA. To rule out that these two MceA-TrxA fusion proteins are toxic due to cleavage and release of free MceA, we assessed the presence of free and TrxA-fused MceA in the corresponding cells. Figure 4C shows that no free MceA can be detected in the cultures expressing the Δss-MceA-TrxA and PhoA73-MceA-TrxA fusions. In addition, to ensure that the TrxA domains of the MceA-TrxA fusions are properly folded and remain in the cytoplasm, the fusions were assessed for their abilities to complement a trxA strain. TrxA has been shown to be required for the assembly of filamentous phages such as f1 and M13 (35). As expected, we found that while a trxA strain was totally unable to support phage M13 replication, it recovered a phage M13 plating efficiency similar to that of the isogenic trxA+ strain when it was transformed with either pAB16 (Δss-MceA-TrxA) or pAB15 (PhoA73-MceA-TrxA) (data not shown). Thus, since the TrxA domains of the MceA-TrxA fusions are functional, they must be normally folded and localized in the cytoplasm. Taken together, these results indicate that endogenous MceA does not need to be entirely translocated to the periplasm to be active but remains fully functional when its C terminus is retained in the cytoplasm by TrxA. As expected, the bactericidal activity caused by these MceA-TrxA fusions is also abolished in strains that express MceB (data not shown).
FIG. 4.
Bactericidal activity of endogenous MceA-TrxA chimeric proteins. (A and B) Serial dilutions of overnight cultures of DB503 (rpoA+) with pBAD101 (vector), pAB12 (Δss-MceA), or pAB16 (Δss-MceA-TrxA) and of DB512 (rpoA341) with pBAD101 (vector), pAB11 (PhoA73-MceA), or pAB15 (PhoA73-MceA-TrxA) were spotted (5 μl) on LB plates without arabinose (A) or with 0.2% arabinose (B) and incubated overnight at 37°C. DB512 (rpoA341) was used instead of DB503 (rpoA+) to better monitor a possible change in bactericidal activity between PhoA73-MceA and PhoA73-MceA-TrxA. Results shown here are representative of three independent experiments. (C) Synthesis levels of Δss-MceA and PhoA73-MceA fused or not to TrxA. Cultures of AB3 (MccE492-resistant strain; see below) with pBAD101 (vector), pAB12 (Δss-MceA), pAB11 (PhoA73-MceA), pAB16 (Δss-MceA-TrxA), or pAB15 (PhoA73-MceA-TrxA) were pulse-labeled as described in Materials and Methods. Proteins were immunoprecipitated with anti-MceA antiserum and separated by SDS-PAGE. The lower panel corresponds to the gel portion where Δss-MceA and PhoA73-MceA alone migrate. The upper panel corresponds to the gel portion where Δss-MceA and PhoA73-MceA fused to TrxA migrate.
The C-terminal domain of MceA is necessary for delivery from the extracellular environment to the periplasm but is not required for bactericidal activity.
The posttranslational modification of the C-terminal residue (serine 84) of MceA that is found in secreted MccE492 has been proposed to be involved in the recognition by the outer membrane receptors on target cells (55). Based on this assumption, we tried to determine whether the C-terminal portion of MceA was involved only in outer membrane recognition and was not necessary for inner membrane insertion and toxic activity. MceA variants lacking 1, 6, 11, or 16 C-terminal residues were generated from the full-length MceA protein by adding two stop codons (one ochre and one amber) at the desired positions (Fig. 1B). We first assessed the effect of such truncations on the MceA protein synthesized with its original ABC signal sequence (ABC-MceA; see Materials and Methods) to direct export to the extracellular medium. Figure 5A shows that while the full-length ABC-MceA is functional, removing only the C-terminal serine residue (ABC-MceA-1C) is sufficient to abolish the activity of secreted MceA on target cells. As expected, the other truncated ABC-MceA proteins lacking 6, 11, or 16 C-terminal residues are also inactive (data not shown). To ensure that this loss of activity was not due to a defect in MceA production or transport to the extracellular medium, we performed an immunoblot on the medium of the cultures producing either full-length ABC-MceA or the mutant lacking the C-terminal serine. Figure 5B shows that both proteins accumulate to similar levels.
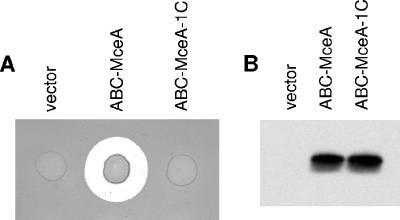
FIG. 5.
Bactericidal activity of extracellular full-length MccE492 and of C-terminally truncated MceA. (A) Growth inhibition assay of a lawn of MccE492-sensitive cells [BL21(pBR322, pBAD101)] overlaid with 5 μl of overnight culture of AB7 with pBAD101 (vector), pAB10 (ABC-MceA), or pAB10-1C (ABC-MceA-1C) on an LB plate supplemented with 0.2% arabinose. Strain AB7 contains all the genes of the MccE492 system, except mceA, in plasmids pAB4 and pAB7. The plate was incubated overnight at 37°C. Note that both the lawn and the tested cultures are growing on this plate. (B) MceA immunoblot of the media of cultures of AB7 with pBAD101 (vector), pAB10 (ABC-MceA), or pAB10-1C (ABC-MceA-1C). Media were obtained from 24-h cultures in M9 minimal medium supplemented with 0.2% arabinose.
The MceA mutants lacking 1, 6, 11, or 16 amino acids were investigated for their bactericidal activity against the cells that produce them, in the absence of the immunity protein MceB. Since several of the truncations remove the epitope recognized by our antiserum, a hexahistidine tag was introduced at the N terminus of each of the proteins. This does not interfere with the bactericidal activity of MceA. Figure 6B shows that cells harboring the plasmids encoding either full-length MceA or the mutants lacking 1, 6, or 11 C-terminal residues are all sensitive to arabinose. By contrast, no toxicity is observed upon induction of the mutant lacking 16 residues. The synthesis levels of full-length MceA and of the truncated MceA mutants were assessed to make sure that the differences in activity among these proteins were not due to variations in the amount of proteins being produced (Fig. 6C). These experiments show that at least 11 amino acids can be removed from the C terminus without preventing toxicity if the microcin is synthesized directly in the target cell, when no extracellular intermediate is required. The bactericidal activity caused by these endogenous truncated MceA proteins is abolished in strains that express MceB (data not shown). Thus, their final mechanism of action cannot be distinguished from that of full-length MceA.
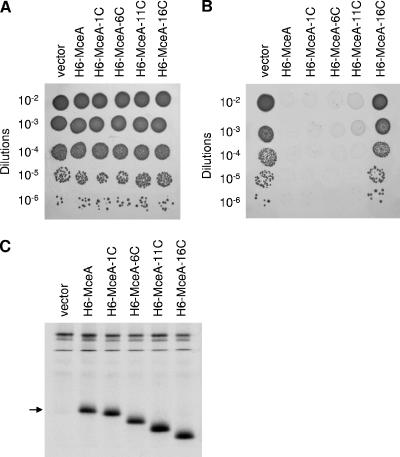
FIG. 6.
Bactericidal activity of endogenous full-length and C-terminally truncated MceA. (A and B) Serial dilutions of overnight cultures of DH5α with pBAD101 (vector), pAB13 (6H-MceA), pAB13-1C (6H-MceA-1C), pAB13-6C (6H-MceA-6C), pAB13-11C (6H-MceA-11C), or pAB13-16C (6H-MceA-16C) were spotted (5 μl) on LB plates without arabinose (A) or with 0.2% arabinose (B) and incubated overnight at 37°C. Results shown here are representative of three independent experiments. (C) Synthesis levels of full-length and C-terminally truncated MceA. Cultures of DH5α with pBAD101 (vector), pAB13 (6H-MceA), pAB13-1C (6H-MceA-1C), pAB13-6C (6H-MceA-6C), pAB13-11C (6H-MceA-11C), or pAB13-16C (6H-MceA-16C) were pulse-labeled as described in Materials and Methods. 6H-tagged proteins were purified and separated by SDS-PAGE. The position of full-length 6H-MceA is indicated by an arrow.
The results obtained by comparing the bactericidal activities of full-length and truncated extracellular MccE492 and endogenous MceA strongly suggest that the C-terminal domain of MccE492 is involved in outer membrane recognition and/or transport to the periplasm. By contrast, at least 11 residues can be removed from the C terminus of endogenous MceA without preventing bactericidal activity.
The ManYZ inner membrane complex is necessary for the bactericidal action of both extracellular MccE492 and endogenous MceA.
MccE492 killing may require interaction with another inner membrane component to form a toxic structure. For instance, this other component could either help MccE492 adopt its final conformation in the membrane or, alternatively, be part of the toxic structure with MccE492. Sensitivity to MccE492 has been shown to depend on the presence of several proteins. These include three outer membrane proteins, FepA, Fiu, and Cir, as well as TonB. Since these four proteins are involved in the transport of MccE492 from the extracellular medium to the periplasm, their absence does not abolish the sensitivity for endogenous MceA expressed from pAB11 or pAB12 (data not shown). We took advantage of this property to look for mutants that would be resistant to MccE492 because of a missing component necessary for bactericidal activity. Indeed, mutants resistant to endogenous MceA may also be resistant to extracellular MccE492 but should not correspond to outer membrane receptors or TonB mutants. To this goal, DB503 cells expressing endogenous MceA were randomly mutagenized by Tn10 transposition. Arar colonies were obtained at a frequency of 10−3 among Tn10-containing colonies. From 100 Arar colonies, 52 were found to be resistant to purified MccE492. Sequencing two of these mutants revealed that the transposons were inserted in the manXYZ operon (8th codon of manX and 96th codon of manY). The manY::Tn10 insertion was chosen for further studies. Table 3 shows that while several wild-type E. coli K-12 strains (DB503, MG1655, and JCB606) are sensitive to purified MccE492, they become fully resistant to the highest available concentrations of purified MccE492 when they contain the manY::Tn10 insertion (strains AB4, AB5, and AB6). The same result was obtained with BL21, an E. coli B strain (data not shown). As expected, a strain with a deletion of the manXYZ operon (strain AB3) is also resistant to MccE492 (Table 3).
TABLE 3.
MccE492 sensitivities of mannose permease-deficient strains and other strainsa
| Strain | Relevant genotype | Activity ± SD (AU/ml)b |
|---|---|---|
| DB503 | Wild type | 960 ± 111 |
| MG1655 | Wild type | 939 ± 148 |
| JCB606 | Wild type | 896 ± 222 |
| AB4 | manY::Tn10 | No activity |
| AB5 | manY::Tn10 | No activity |
| AB6 | manY::Tn10 | No activity |
| AB3 | ΔmanXYZ | No activity |
| AB3(pJFL) | ΔmanXYZ/manX+ | No activity |
| AB3(pTSP11) | ΔmanXYZ/manY+ | No activity |
| AB3(pAB17) | ΔmanXYZ/manZ+ | No activity |
| AB3(pJFP-H6M) | ΔmanXYZ/manYZ+ | 1045 ± 225 |
| AB3(pJFLPM) | ΔmanXYZ/manXYZ+ | 896 ± 222 |
| MG1655(pAB8) | Wild type | 939 ± 148 |
| MG1655(pAB7) | mceB+ | No activity |
| H1443 | Wild type | 960 ± 111 |
| H1876 | fepA fiu cir | No activity |
| AB2 | Wild type | 1045 ± 225 |
| AB1 | tonB | 48 ± 16 |
The bactericidal activity of purified MccE492 (10 mg/ml) was assessed in triplicate for the indicated strains by the critical dilution method (see Materials and Methods). Expression from the indicated plasmids was induced with 600 μM IPTG or 0.2% arabinose for Ptac or PBAD promoters, respectively.
AU, arbitrary units.
The manXYZ operon (formerly referred to as ptsLPM; manY is formerly referred to as pel) encodes one cytoplasmic protein, ManX, and two inner membrane proteins, ManY and ManZ. These three proteins form the mannose permease, a complex involved in the uptake of mannose and related hexoses (20, 58). In order to determine which of these proteins is/are required for MccE492 activity, we performed complementation experiments. As can be seen in Table 3, when strain AB3 (ΔmanXYZ) harbors pJFL, which contains only manX, no sensitivity to MccE492 can be detected. In contrast, when it harbors either pJFP-H6M or pJFLPM, which contain manYZ and manXYZ, respectively, cells become as sensitive as DB503 (wild type). Identical results were obtained using AB4 (manY::Tn10) instead of AB3 (data not shown). To determine if ManY or ManZ alone would be sufficient for the action of MccE492, we expressed both proteins separately. Table 3 shows that strain AB3 (ΔmanXYZ) remains fully resistant to MccE492 when it expresses either ManY (from pTSP11) or ManZ (from pAB17). In contrast, when the strain harbors both plasmids, cells become as sensitive as DB503 (data not shown). Taken together, these observations indicate that while ManX is not involved in MccE492 sensitivity, both ManY and ManZ are required for the bactericidal action of the microcin. This double requirement for ManY and ManZ is consistent with the fact that these two proteins are stable in the inner membrane only as a complex and cannot accumulate as individual proteins (B. Erni, personal communication).
Expression of the manXYZ operon is regulated both positively and negatively (44). Positive regulation is mediated by binding to the promoter region of cyclic AMP-bound catabolite gene activator protein (CAP). A deletion of cyaA, the gene coding for adenylate cyclase, lowers the expression of LacZ fused to manXYZ operon-encoded proteins by a factor of 6 (42). Negative regulation occurs through binding of the Mlc and NagC repressors to operators located upstream of manX. While a mutation in nagC has only very little effect, a mutation in mlc was shown to result in a threefold derepression of the expression of a ManX-LacZ fusion protein (44). To determine whether the level of ManXYZ expression has any influence on the degree of sensitivity to MccE492, the bactericidal activity of purified MccE492 (10 mg/ml) was assessed for various strains by the critical dilution method (see Materials and Methods). No significant difference was observed between strain DB503 and strain AB8, an isogenic strain with a deletion of cyaA (data not shown). Thus, abolishing cyclic AMP synthesis and as a consequence CAP-mediated positive regulation is not sufficient to lower ManXYZ expression to a level which is limiting for the action of MccE492. On the other hand, overexpressing ManXYZ from the pJFLPM plasmid, even with a strong induction (600 μM IPTG), did not result in any measurable change in MccE492 sensitivity (data not shown). Taken together, these results indicate that the ManYZ inner membrane complex is already present in excess for the bactericidal action of MccE492 when positive regulation is blocked and negative regulation is intact.
In order to verify that a ManYZ-deficient strain is always resistant to the bactericidal activity of MceA regardless of the route by which it gains access to the inner membrane, the toxicities of various plasmids were assessed in strain AB4 (manY::Tn10). In contrast to DB503, AB4 is fully Arar when it is transformed with pAB9 (PhoA-MceA), pAB11 (PhoA73-MceA), pAB12 (Δss-MceA), or pAB10 (ABC-MceA) together with the plasmids encoding all the genes involved in ABC-MceA posttranslational modification and export (pAB4 and pAB8) (data not shown). Thus, whatever the route by which MceA reaches the inner membrane, ManYZ are essential for its bactericidal activity.
Table 3 also shows that resistance to the highest available concentrations of purified MccE492 seen with the manXYZ mutations is equivalent to that conferred either by the expression of MceB [MG1655(pAB7)] or by the inactivation of the three outer membrane receptors for MccE492, namely, FepA, Fiu, and Cir (H1876). In contrast, a tonB deletion (AB1) does not render cells fully resistant to MccE492, which indicates that some transport of MccE492 from the extracellular medium to the periplasm occurs in the absence of TonB.
The 52 MccE492-resistant mutants were tested for their abilities to metabolize mannose. While the nonmutagenized strain is Man+, all the mutants were found to be Man−. This suggests that our insertion mutagenesis was saturating and that mannose permease mutants were the only ones that could be identified with this approach.
To ensure that we were not missing mutations in other genes, we performed another Tn10 transposon insertion mutagenesis with an isogenic strain transformed with pJFLPM (manXYZ). Since this strain contains the manXYZ operon in multiple copies, the probability of obtaining mutants without any functional ManYZ is extremely low. As expected, from 80 Arar colonies obtained, all were found to be Man+ and sensitive to purified MccE492. These clones are probably Arar due to chromosomal mutations affecting expression from the PBAD promoter (30) or to mutations in the plasmid expressing MceA and have not been further characterized.
As transposon insertion mutagenesis usually yields loss-of-function mutations, we tried to obtain spontaneous mutants in order to determine if other targets could be identified, particularly in essential genes. From DB503 pAB11 cultures, 50 independent spontaneous mutants that were Arar and resistant to purified MccE492 were identified. Again, all mutants were Man−, supporting the notion that only mannose permease mutations can completely prevent a cell from being killed by both endogenous MceA and extracellular MccE492.
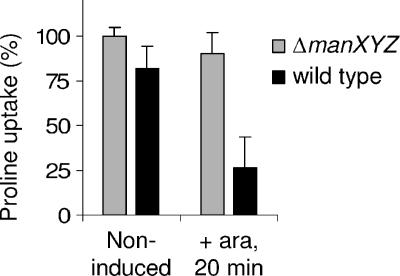
Since MccE492 is known to cause a depolarization of the inner membrane of target cells (14), we wanted to determine whether our mannose permease mutants possess an intact membrane potential upon exposure to the microcin. To this goal, we assessed the capacity of PhoA73-MceA-expressing cells to accumulate proline, whose uptake by the H+/proline symporter is strictly dependent on the proton motive force (PMF). As can be seen in Fig. 7, while MccE492-sensitive cells (wild type) exhibited a dramatic loss of PMF when PhoA73-MceA is expressed from pAB11, no significant change in the level of PMF was observed for the mannose permease mutant strain (ΔmanXYZ). Therefore, it appears that the mannose permease is critical for an essential step in MccE492 toxicity that has not been detected in previous studies.
FIG. 7.
Proline uptake in a mannose permease-deficient strain and in a wild-type strain when PhoA73-MceA is produced. Uptake was measured in AB3 (ΔmanXYZ) and in DB503 (wild type) transformed with pAB11 (PhoA73-MceA) either in the absence of arabinose (ara) or after 20 min of arabinose (0.2%) induction. Proline uptake is expressed as the percentage of the slope of uptake measured in uninduced AB3 cultures. Each value represents the average of three independent cultures. Error bars indicate standard deviations.
DISCUSSION
Although it is well established that MccE492 inserts in lipid bilayers and exerts deleterious effects on the inner membrane of target cells, it has never been demonstrated whether this is due to a direct action of inner membrane-associated MccE492 or alternatively to a more indirect mechanism involving periplasmic or cytoplasmic MccE492, for example. The results presented here support the view that MccE492 toxicity relies primarily on its localization in the inner membrane. First, the observation that endogenous MceA remains fully active when it is fused to TrxA indicates that the bactericidal action of MceA does not require a complete export to the periplasm or to the extracellular medium. Second, our experiments show that Δss-MceA is more toxic in a prlA4 strain than in a secY+ strain. Since prlA4 allows inner membrane targeting and export of proteins devoid of signal sequence (15), this indicates that membrane-targeted MceA is more toxic than cytoplasmic MceA. This conclusion is supported by the fact that PhoA73-MceA is more active than Δss-MceA in a secY+ strain. The relatively weak toxicity mediated by Δss-MceA in a secY+ strain could be due to partial recognition of a cryptic signal sequence by the Sec translocase. Alternatively, Δss-MceA could insert into the inner membrane either spontaneously or with the help of other proteins. For instance, YidC has been shown to mediate Sec-independent membrane insertion of proteins, such as the major coat protein of bacteriophage M13 (50).
In contrast to the accumulation of MceA, the accumulation of the structural proteins of other pore-forming bacteriocins in the cytoplasm in the absence of the corresponding immunity proteins is not bactericidal. For instance, the expression of colicin A without its corresponding immunity protein and lysis protein leads to colicin A accumulation in the cytoplasm but does not result in loss of viability; similar results were obtained with colicins E1 and Ib (17). In addition, colicin V was shown to be bactericidal only when provided to the inner membrane from its periplasmic face (60). Since endogenous MceA does not need to be entirely translocated to the periplasm to be active and remains fully functional when its C terminus is retained in the cytoplasm by TrxA, it seems to represent an exception among pore-forming bacteriocins.
The data shown here identify a C-terminal domain of MceA that is not required for bactericidal activity, although this region is essential for transport from the extracellular medium to the inner membrane. Interestingly, this domain exhibits a strong similarity to the C-terminal domains of microcin M and microcin H47 (43). Indeed, the 10 C-terminal amino acids of these microcins show an 80% identity between MceA and microcin M and a 70% identity between MceA and microcin H47; virtually no homology can be detected outside of the C-terminal domains of these proteins. Strikingly, the activities of microcins M and H47 on target cells are dependent on the presence of the same three outer membrane receptors (FepA, Fiu, and Cir) that are required for MccE492 action (43). By contrast, the bactericidal activities of these proteins appear to be mediated by different mechanisms. While ManYZ is essential for the action of MccE492, the Fo proton channel of the ATP synthase is necessary for microcin H47 activity (48); no information is currently available on an inner membrane component involved in microcin M toxicity. Thus, the C-terminal domains of microcins E492, H47, and M are likely to be responsible for the specific binding to FepA, Fiu, and Cir and possibly also for TonB-mediated import in the periplasm. This notion is in good accordance with the observation that numerous bacteriocins, in particular colicins, have a modular structure that presumably arose through evolution by different combinations of toxic activity and receptor specificity domains (4).
Our results demonstrate that the bactericidal activity of MccE492 is dependent on the presence of ManYZ, the inner membrane components of the mannose permease. This permease couples sugar transport with sugar phosphorylation. Two cytosolic proteins, namely, enzyme I and HPr, sequentially transfer a phosphoryl group from phosphoenolpyruvate to ManX, which donates the phosphate to the sugar substrate. In contrast to ManX, ManYZ are not detectably phosphorylated during these reactions (19). Since only ManYZ are required for MccE492 activity, the phosphate transfer reactions involved in sugar transport by the mannose permease are not related to the mechanism of action of MccE492.
Several inner membrane proteins have been shown to be essential for the bactericidal activities of other bacteriocins. For instance, SdaC, a serine transporter, is required for the activity of colicin V (22), while the Fo proton channel of the ATP synthase is necessary for the antibacterial action of microcin H47 (48). However, it is unknown whether these inner membrane proteins act as docking proteins to promote the membrane attachment and insertion of the bacteriocins, if they help the bacteriocins form their active structures, or if they are part of the structures responsible for toxicity.
Mannose permease mutants are resistant both to extracellular MccE492, delivered to the inner membrane from the periplasmic side, and to endogenous MceA, delivered from the cytoplasmic side. If ManYZ had a membrane docking function for the microcin, then ManYZ would need to have redundant domains facing the periplasm and the cytoplasm. Considering also that PhoA73-MceA, which is targeted to the Sec translocase and therefore to the inner membrane, is not toxic to mannose permease mutants, it appears unlikely that ManYZ act as an inner membrane docking complex. However, it remains to be determined whether ManYZ play only a transient role in helping MccE492 form a toxic structure or if they play a structural role in the final toxic structure with MccE492.
The mannose permeases of various gram-positive bacteria have previously been reported to be required for the activities of several bacteriocins. In Listeria monocytogenes, the mannose permease, encoded by the mptACD operon, has been shown to be necessary for the bactericidal activity of leucocin A and mesentericin Y105, two closely related bacteriocins (10, 47). In addition, the mannose permease of Enterococcus faecalis is also involved in mesentericin Y105 sensitivity (28). The mechanism of action of mesentericin Y105 is similar to that of MccE492, since it also dissipates the proton motive force of susceptible cells (36). Interestingly, MptC, which is homologous to the E. coli ManY, is sufficient to confer leucocin A sensitivity when it is expressed in Lactococcus lactis (46). It is therefore tempting to speculate that ManY, and not ManZ, is the protein directly involved in MccE492 sensitivity. Thus, our findings with MccE492 suggest that the mannose permease could be a target for diverse bacteriocins, not only among gram-positive species, but also in gram-negative ones.
Interestingly, the ManYZ complex has also been shown to be required for the penetration of bacteriophage λ DNA across the cytoplasmic membrane by an as-yet-unknown mechanism (18, 20). A ManYZ mutagenesis approach should help determine whether the entry of bacteriophage λ DNA and the toxic activity of MccE492 rely on interactions with the same portions of the mannose permease.
Another class of MccE492-resistant mutants, the semA mutants, has been described previously (45). Two types of semA mutants were identified, namely, Tn5 insertion mutants and spontaneous mutants. Genetic data indicate that the spontaneous semA mutants may correspond to mutations close to or in the manXYZ operon. By contrast, mapping data rule out the possibility that semA12, a Tn5 insertion mutant, represents a mutation close to the manXYZ operon. Since semA12 is associated with only partial resistance to MccE492, it could correspond to a mutation in a regulatory gene controlling the expression of FepA, Fiu, Cir, TonB, or ManXYZ.
Among all known bacteriocins, microcin 24 exhibits the highest degree of similarity to MccE492, and its amino acid sequence is 50% identical to that of MccE492. Microcin 24 is produced by a uropathogenic E. coli strain and is active against Escherichia and Salmonella spp. (41). Although its mechanism of action is totally unknown, this high similarity may indicate a shared mechanism. In particular, it would be interesting to determine if the action of microcin 24 also requires the presence of ManYZ in the inner membrane.
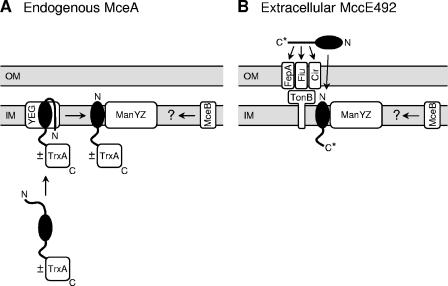
In conclusion, we propose in Fig. 8 models for the bactericidal mechanisms of endogenous MceA (Fig. 8A) and extracellular MccE492 (Fig. 8B). Endogenous MceA fused to the PhoA73 signal sequence (PhoA73-MceA) is targeted to the SecYEG complex, where it might be N-terminally cleaved (by homology with PhoA) and laterally released into the inner membrane to form a toxic structure that leads to membrane depolarization and permeabilization. Toxicity is strictly dependent on the presence of the ManYZ complex in the inner membrane. The presence of a folded TrxA domain fused to the C terminus of MceA does not impair its bactericidal activity. In cells containing all the genes from the original MccE492 gene cluster, MccE492 is first synthesized as a precursor which is posttranslationally modified at its C terminus, cleaved, and secreted through its dedicated ABC exporter to the extracellular medium. The C-terminal domain of MccE492, including the posttranslational modification, is responsible for specific binding to the three outer membrane receptors on target cells, FepA, Fiu, and Cir. MccE492 is then translocated through the outer membrane by a mechanism whose efficiency depends on TonB, although MccE492 can be translocated to some degree in the absence of TonB. Upon inner membrane insertion with its C terminus on the cytoplasmic side of the membrane (by homology with endogenous MceA), MccE492 forms a toxic structure. The ManYZ inner membrane complex is also absolutely essential for MccE492 toxicity.
FIG. 8.
Models for the bactericidal mechanisms of endogenous MceA (A) and extracellular MccE492 (B). MceA and MccE492 are represented as black ellipses with protruding N-terminal signal sequences and C-terminal domains. OM, outer membrane; IM, inner membrane; YEG, SecYEG complex; C*, posttranslationally modified MccE492 C terminus. ±TrxA indicates that the bactericidal activity of endogenous MceA is not affected by the presence of a C-terminal TrxA domain. Instead of Δss-MceA, PhoA73-MceA was chosen here as an example of endogenous MceA. Details are described in the text.
Two major issues remain to be addressed. First, the structure responsible for the bactericidal activity of MccE492 is still unknown. In particular, the respective contributions of MccE492 and ManYZ to a putative channel-like structure are yet to be determined. Second, the mechanism by which MceB blocks the action of MccE492 remains unsolved. Since MceB is able to prevent the toxicity mediated by endogenous MceA targeted to the Sec translocase, as well as extracellular MccE492 that is delivered to the inner membrane from its periplasmic side, it is unlikely that MceB would exert its action by preventing the membrane insertion of MccE492. Further experimental work will be required to determine if MceB functions by blocking an interaction between MccE492 and ManYZ or by destabilizing the final toxic structure.
Acknowledgments
This work was supported by the Swiss National Science Foundation, the Roche Research Foundation, and the State of Geneva.
We thank C. Georgopoulos, P. Linder, L. Bernheim, and P. Genevaux for helpful discussions and a critical reading of the manuscript; K. Abid and J. Castilla for technical advice; and B. Erni for strains and plasmids.
REFERENCES
- 1.Belin, D., L. M. Guzman, S. Bost, M. Konakova, F. Silva, and J. Beckwith. 2004. Functional activity of eukaryotic signal sequences in Escherichia coli: the ovalbumin family of serine protease inhibitors. J. Mol. Biol. 335:437-453. [DOI] [PubMed] [Google Scholar]
- 2.Bieler, S., L. Estrada, R. Lagos, M. Baeza, J. Castilla, and C. Soto. 2005. Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 280:26880-26885. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 5.Brundage, L., J. P. Hendrick, E. Schiebel, A. J. Driessen, and W. Wickner. 1990. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62:649-657. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 9.Crooke, H., and J. Cole. 1995. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol. Microbiol. 15:1139-1150. [DOI] [PubMed] [Google Scholar]
- 10.Dalet, K., Y. Cenatiempo, P. Cossart, and Y. Héchard. 2001. A sigma-54-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263-3269. [DOI] [PubMed] [Google Scholar]
- 11.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo, V. 1984. Isolation and characterization of microcin E492 from Klebsiella pneumoniae. Arch. Microbiol. 139:72-75. [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., J. L. Martinez, and C. Asensio. 1984. Microcin-mediated interactions between Klebsiella pneumoniae and Escherichia coli strains. J. Gen. Microbiol. 130:391-400. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo, V., and A. P. Pugsley. 1985. Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob. Agents Chemother. 27:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derman, A. I., J. W. Puziss, P. J. Bassford, Jr., and J. Beckwith. 1993. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 12:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Destoumieux-Garzón, D., X. Thomas, M. Santamaria, C. Goulard, M. Barthélémy, B. Boscher, Y. Bessin, G. Molle, A. M. Pons, L. Letellier, J. Peduzzi, and S. Rebuffat. 2003. Microcin E492 antibacterial activity: evidence for a TonB-dependent inner membrane permeabilization on Escherichia coli. Mol. Microbiol. 49:1031-1041. [DOI] [PubMed] [Google Scholar]
- 17.Duché, D. 2002. The pore-forming domain of colicin A fused to a signal peptide: a tool for studying pore-formation and inhibition. Biochimie 84:455-464. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, J., and W. Arber. 1978. E. coli K-12 pel mutants, which block phage lambda DNA injection, coincide with ptsM, which determines a component of a sugar transport system. Mol. Gen. Genet. 161:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Erni, B., B. Zanolari, P. Graff, and H. P. Kocher. 1989. Mannose permease of Escherichia coli. Domain structure and function of the phosphorylating subunit. J. Biol. Chem. 264:18733-18741. [PubMed] [Google Scholar]
- 20.Erni, B., B. Zanolari, and H. P. Kocher. 1987. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J. Biol. Chem. 262:5238-5247. [PubMed] [Google Scholar]
- 21.Esquinas-Rychen, M., and B. Erni. 2001. Facilitation of bacteriophage lambda DNA injection by inner membrane proteins of the bacterial phosphoenol-pyruvate:carbohydrate phosphotransferase system (PTS). J. Mol. Microbiol. Biotechnol. 3:361-370. [PubMed] [Google Scholar]
- 22.Gérard, F., N. Pradel, and L. F. Wu. 2005. Bactericidal activity of colicin V is mediated by an inner membrane protein, SdaC, of Escherichia coli. J. Bacteriol. 187:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giffard, P. M., and I. R. Booth. 1988. The rpoA341 allele of Escherichia coli specifically impairs the transcription of a group of positively-regulated operons. Mol. Gen. Genet. 214:148-152. [DOI] [PubMed] [Google Scholar]
- 24.Gutknecht, R., R. Lanz, and B. Erni. 1998. Mutational analysis of invariant arginines in the IIAB(Man) subunit of the Escherichia coli phosphotransferase system. J. Biol. Chem. 273:12234-12238. [DOI] [PubMed] [Google Scholar]
- 25.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 27.Hantke, K. 1990. Dihydroxybenzolyserine—a siderophore for Escherichia coli. FEMS Microbiol. Lett. 67:5-8. [DOI] [PubMed] [Google Scholar]
- 28.Héchard, Y., C. Pelletier, Y. Cenatiempo, and J. Frère. 2001. Analysis of sigma-54-dependent genes in Enterococcus faecalis: a mannose PTS permease (EIIMan) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology 147:1575-1580. [DOI] [PubMed] [Google Scholar]
- 29.Higashitani, A., N. Higashitani, S. Yasuda, and K. Horiuchi. 1994. A general and fast method for mapping mutations on the Escherichia coli chromosome. Nucleic Acids Res. 22:2426-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khatib, K., and D. Belin. 2002. A novel class of secA alleles that exert a signal-sequence-dependent effect on protein export in Escherichia coli. Genetics 162:1031-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 32.Lagos, R., M. Baeza, G. Corsini, C. Hetz, E. Strahsburger, J. A. Castillo, C. Vergara, and O. Monasterio. 2001. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42:229-243. [DOI] [PubMed] [Google Scholar]
- 33.Lagos, R., J. E. Villanueva, and O. Monasterio. 1999. Identification and properties of the genes encoding microcin E492 and its immunity protein. J. Bacteriol. 181:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagos, R., M. Wilkens, C. Vergara, X. Cecchi, and O. Monasterio. 1993. Microcin E492 forms ion channels in phospholipid bilayer membrane. FEBS Lett. 321:145-148. [DOI] [PubMed] [Google Scholar]
- 35.Lim, C. J., B. Haller, and J. A. Fuchs. 1985. Thioredoxin is the bacterial protein encoded by fip that is required for filamentous bacteriophage f1 assembly. J. Bacteriol. 161:799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maftah, A., D. Renault, C. Vignoles, Y. Héchard, P. Bressollier, M. H. Ratinaud, Y. Cenatiempo, and R. Julien. 1993. Membrane permeabilization of Listeria monocytogenes and mitochondria by the bacteriocin mesentericin Y105. J. Bacteriol. 175:3232-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayr-Harting, A., A. J. Hedges, and R. C. W. Berkeley. 1972. Methods for studying bacteriocins. Methods Microbiol. 7A:315-422. [Google Scholar]
- 38.Michaelis, S., J. F. Hunt, and J. Beckwith. 1986. Effects of signal sequence mutations on the kinetics of alkaline phosphatase export to the periplasm in Escherichia coli. J. Bacteriol. 167:160-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien, G. J., and H. K. Mahanty. 1994. Colicin 24, a new plasmid-borne colicin from a uropathogenic strain of Escherichia coli. Plasmid 31:288-296. [DOI] [PubMed] [Google Scholar]
- 42.Palva, E. T., P. Saris, and T. J. Silhavy. 1985. Gene fusions to the ptsM/pel locus of Escherichia coli. Mol. Gen. Genet. 199:427-433. [DOI] [PubMed] [Google Scholar]
- 43.Patzer, S. I., M. R. Baquero, D. Bravo, F. Moreno, and K. Hantke. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149:2557-2570. [DOI] [PubMed] [Google Scholar]
- 44.Plumbridge, J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369-380. [DOI] [PubMed] [Google Scholar]
- 45.Pugsley, A. P., F. Moreno, and V. de Lorenzo. 1986. Microcin-E492-insensitive mutants of Escherichia coli K12. J. Gen. Microbiol. 132:3253-3259. [DOI] [PubMed] [Google Scholar]
- 46.Ramnath, M., S. Arous, A. Gravesen, J. W. Hastings, and Y. Héchard. 2004. Expression of mptC of Listeria monocytogenes induces sensitivity to class IIa bacteriocins in Lactococcus lactis. Microbiology 150:2663-2668. [DOI] [PubMed] [Google Scholar]
- 47.Ramnath, M., M. Beukes, K. Tamura, and J. W. Hastings. 2000. Absence of a putative mannose-specific phosphotransferase system enzyme IIAB component in a leucocin A-resistant strain of Listeria monocytogenes, as shown by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 66:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez, E., and M. Laviña. 2003. The proton channel is the minimal structure of ATP synthase necessary and sufficient for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 47:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russel, M., and P. Model. 1985. Thioredoxin is required for filamentous phage assembly. Proc. Natl. Acad. Sci. USA 82:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 51.Schierle, C. F., M. Berkmen, D. Huber, C. Kumamoto, D. Boyd, and J. Beckwith. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185:5706-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seip, S., J. Balbach, S. Behrens, H. Kessler, K. Flukiger, R. de Meyer, and B. Erni. 1994. Mannose transporter of Escherichia coli. Backbone assignments and secondary structure of the IIA domain of the IIABMan subunit. Biochemistry 33:7174-7183. [DOI] [PubMed] [Google Scholar]
- 53.Shah, S., and A. Peterkofsky. 1991. Characterization and generation of Escherichia coli adenylate cyclase deletion mutants. J. Bacteriol. 173:3238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strahsburger, E., M. Baeza, O. Monasterio, and R. Lagos. 2005. Cooperative uptake of microcin E492 by receptors FepA, Fiu, and Cir and inhibition by the siderophore enterochelin and its dimeric and trimeric hydrolysis products. Antimicrob. Agents Chemother. 49:3083-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, X., D. Destoumieux-Garzón, J. Peduzzi, C. Afonso, A. Blond, N. Birlirakis, C. Goulard, L. Dubost, R. Thai, J. C. Tabet, and S. Rebuffat. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279:28233-28242. [DOI] [PubMed] [Google Scholar]
- 56.van den Berg, B., W. M. Clemons, Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature 427:36-44. [DOI] [PubMed] [Google Scholar]
- 57.Wilkens, M., J. E. Villanueva, J. Cofre, J. Chnaiderman, and R. Lagos. 1997. Cloning and expression in Escherichia coli of genetic determinants for production of and immunity to microcin E492 from Klebsiella pneumoniae. J. Bacteriol. 179:4789-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams, N., D. K. Fox, C. Shea, and S. Roseman. 1986. Pel, the protein that permits lambda DNA penetration of Escherichia coli, is encoded by a gene in ptsM and is required for mannose utilization by the phosphotransferase system. Proc. Natl. Acad. Sci. USA 83:8934-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanofsky, C., and J. Ito. 1966. Nonsense codons and polarity in the tryptophan operon. J. Mol. Biol. 21:313-334. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, L. H., M. J. Fath, H. K. Mahanty, P. C. Tai, and R. Kolter. 1995. Genetic analysis of the colicin V secretion pathway. Genetics 141:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]