Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that produces the siderophore pyoverdine, which enables it to acquire the essential nutrient iron from its host. Formation of the iron-chelating hydroxamate functional group in pyoverdine requires the enzyme PvdA, a flavin-dependent monooxygenase that catalyzes the N5 hydroxylation of l-ornithine. pvdA from P. aeruginosa was successfully overexpressed in Escherichia coli, and the enzyme was purified for the first time. The enzyme possessed its maximum activity at pH 8.0. In the absence of l-ornithine, PvdA has an NADPH oxidase activity of 0.24 ± 0.02 μmol min−1 mg−1. The substrate l-ornithine stimulated this activity by a factor of 5, and the reaction was tightly coupled to the formation of hydroxylamine. The enzyme is specific for NADPH and flavin adenine dinucleotide (FAD+) as cofactors, as it cannot utilize NADH and flavin mononucleotide. By fluorescence titration, the dissociation constants for NADPH and FAD+ were determined to be 105.6 ± 6.0 μM and 9.9 ± 0.3 μM, respectively. Steady-state kinetic analysis showed that the l-ornithine-dependent NADPH oxidation obeyed Michaelis-Menten kinetics with apparent Km and Vmax values of 0.58 mM and 1.34 μmol min−1 mg−1. l-Lysine was a nonsubstrate effector that stimulated NADPH oxidation, but uncoupling occurred and hydrogen peroxide instead of hydroxylated l-lysine was produced. l-2,4-Diaminobutyrate, l-homoserine, and 5-aminopentanoic acid were not substrates or effectors, but they were competitive inhibitors of the l-ornithine-dependent NADPH oxidation reaction, with Kics of 3 to 8 mM. The results indicate that the chemical nature of effectors is important for simulation of the NADPH oxidation rate in PvdA.
Iron is an essential nutrient for most organisms. In humans, iron is sequestered by iron binding proteins such as transferrin, lactoferrin, and hemoglobin and is therefore unavailable to invading microbial pathogens. Microorganisms can counter this nonspecific host defense mechanism by the production of low-molecular-weight, high iron(III) affinity molecules termed siderophores (4, 25). Siderophores are synthesized intracellularly and then secreted into the environment, where they chelate and deliver the complexed iron back into the cell via specific membrane transporters. Most siderophores are peptides, and the functional groups that chelate iron(III) include catecholates, carboxylates, and hydroxamates (15).
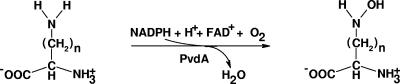
The hydroxamate functional groups are usually derived from diamino acids such as l-lysine and l-ornithine. A flavin-dependent monooxygenase (EC 1.14.13) catalyzes the first step in the formation of hydroxamates by oxidizing the terminal amino group of the respective substrates to produce the corresponding hydroxylamines (Fig. 1). On the basis of studies with the Escherichia coli l-lysine N6-hydroxylase (IucD) and by analogy to other flavin monooxygenases (1), this reaction is proposed to proceed in two steps. In the reductive half reaction, flavin adenine dinucleotide (FAD+) is reduced by NADPH, while in the oxidative half reaction, the reduced flavin reacts with oxygen to form a putative flavin-peroxy intermediate that subsequently oxidizes the terminal amino group of the substrate, l-lysine (9). l-Ornithine N5-hydroxylase, on the other hand, has never been purified or characterized, although it is an important enzyme involved in the biosynthesis of siderophores of Pseudomonas aeruginosa (24), Burkholderia cepacia (18), and Aspergillus fumigatus (5). These are major opportunistic pathogens that infect immunocompromised individuals, including those with the genetic disease cystic fibrosis. Knockout mutations of genes encoding l-ornithine N5-hydroxylases in each of these microorganisms lead to attenuation of virulence because of the inability to synthesize the respective major siderophores (17, 18, 21).
FIG. 1.
General reaction catalyzed by flavin-dependent monooxygenase involved in hydroxamate siderophore biosynthesis. n = 3 for l-ornithine, and n = 4 for l-lysine.
Despite a sequence similarity of about 35% between l-lysine and l-ornithine hydroxylases, l-ornithine is not a substrate of l-lysine N6-hydroxylase (9). This suggests a fundamental difference in the substrate binding sites of the two enzymes and further indicates the limited potential of using lysine N6-hydroxylase to design antimicrobials targeting l-ornithine N5-hydroxylase.
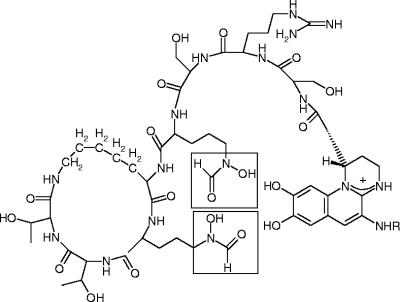
P. aeruginosa strains produce the siderophore pyoverdine, which contain hydroxamates derived from l-ornithine (Fig. 2) (12). The l-ornithine hydroxylase is encoded by the gene pvdA (24). A knockout mutant of pvdA is pyoverdine negative, but production of this siderophore can be restored by feeding the strain exogenous N5-hydroxyornithine (24). Hydroxylation of l-ornithine therefore occurs at an early step of pyoverdine production, prior to further modifications and incorporation into the pyoverdine peptide by nonribosomal peptide synthetases (8, 11). We report here the first successful overexpression and purification of active recombinant l-ornithine N5-hydroxylase, PvdA, from P. aeruginosa PAO1. This enabled us to perform a detailed steady-state kinetic analysis to determine the substrate and coenzyme specificities of PvdA. We further demonstrate that substrate analogues can compete with l-ornithine for the same active site of the enzyme, although these analogues are not hydroxylated.
FIG. 2.
Structure of pyoverdine produced by P. aeruginosa PAO1. The hydroxamate iron ligands derived from l-ornithine are boxed.
MATERIALS AND METHODS
Chemicals.
Amino acids were from Sigma-Aldrich (Oakville, ON). Restriction enzymes, T4 DNA ligase, and Pfx polymerase were from Invitrogen (Burlington, ON) or New England Biolabs (Pickering, ON). All other chemicals were of analytical grade and were obtained from Sigma-Aldrich and Fisher Scientific (Nepean, ON).
DNA manipulation.
DNA was purified, digested, and ligated by using standard protocols (16). The pvdA gene was amplified by PCR with primers with the sequences GACCCATATGACACAGGCAACTGCAACCG and CTATAAGCTTCAGCTGGCCAGGGCGTGC-3′. Introduced NdeI and HindIII restriction sites are underlined. PCR mixtures consisted of 1× Expand High Fidelity Buffer containing 1.5 mM MgCl2 (Roche Diagnostics), 1× Pfx Enhancer Solution (Invitrogen), 0.4 mM each deoxynucleoside triphosphate, 1 pmol of each primer, 22 ng of P. aeruginosa PAO1 chromosomal DNA, and 0.5 U of Platinum Pfx DNA polymerase (Invitrogen). The amplification profile was a hot start at 94°C (2 min); 30 cycles of 94°C (30 s), 50°C (30 s), and 68°C (1 min); and then 68°C for 10 min. PCR fragments were purified and digested with NdeI and HindIII. Following gel purification, the amplified gene was ligated into pT7-7 (20) that had been digested with the same enzymes. The resulting construct was transformed into E. coli JM109. Positive transformants were verified by restriction enzyme digestion and nucleotide sequencing at the Guelph Molecular Supercenter, University of Guelph. The pvdA gene was then excised from pT7-7 by digestion with NdeI and HindIII and then ligated to similar restriction sites in the plasmid vector pET28a (Novagen).
Purification of PvdA.
Recombinant E. coli Rosetta 2(DE3) cells (Novagen) containing pvdA inserted into pET28a were cultured in LB broth containing 34 μg/ml chloramphenicol and 100 μg/ml kanamycin. At log phase, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce expression of the protein. After 3 h of induction, the cells were harvested by centrifugation.
A 50 mM concentration of sodium phosphate buffer (pH 8.0) containing 300 mM sodium chloride was used in the purification of PvdA. Cells from 500 ml of culture with a wet weight of about 1 g were resuspended in 50 ml buffer containing 20 mM imidazole. The resuspended cells were lysed by passage through a French press twice at 12,000 lb/in2. Cell debris were pelleted by centrifugation at 26,500 × g for 20 min, and the supernatant was incubated with 3 ml Ni2+-nitrilotriacetic acid resin suspension (QIAGEN, Inc.) for 1 h at 4°C. The resin was then packed into a glass column and washed with 50 ml buffer containing 20 mM imidazole, followed by 3 ml buffer containing 50 mM imidazole. PvdA was eluted with a stepwise gradient of 3 ml buffer containing 100 mM imidazole and 9 ml buffer containing 200 mM imidazole. A 10-μl portion of each 3-ml fraction collected was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing PvdA were pooled, concentrated by ultrafiltration with a YM10 filter (Millipore), and washed repeatedly with buffer containing 1 mM dithiothreitol to remove imidazole from the solution. The concentrated enzyme (about 1 mg/ml) was stored frozen in aliquots at −80°C.
Determination of protein concentration, purity, and molecular mass.
Protein concentrations were determined by the Bradford assay with bovine serum albumin as the standard (2). SDS-PAGE was performed, and staining was done with Coomassie blue according to established procedures (7). The BenchMark Protein Ladder (Invitrogen) containing proteins ranging from 10 to 220 kDa was used as a molecular size marker. To determine the native molecular weight of PvdA, gel filtration was performed on a Superdex 200 column (GE Biosciences) with 20 mM HEPES buffer (pH 7.5) containing 0.15 M NaCl as the equilibration and elution buffer. The standard curve used consisted of the proteins cytochrome c (Mr = 12,400), carbonic anhydrase (Mr = 29,000), bovine serum albumin (Mr = 66,000), alcohol dehydrogenase (Mr = 150,000), and β-amylase (Mr = 200,000) (all from Sigma-Aldrich).
Kinetic assays and determination of substrate specificity of PvdA.
All kinetic assays were performed at least in duplicate at 25°C with a Varian Cary 3 spectrophotometer equipped with a thermojacketed cuvette holder. Standard assays were carried out with 0.1 M sodium phosphate buffer, pH 8.0, containing NADPH (300 μM), l-ornithine (2.0 mM), and FAD+ (50 μM) at 25°C. Enzyme activity was estimated from the decrease in NADPH absorbance at 340 nm (E = 6,300 M−1 cm−1) and expressed in terms of micromoles of NADPH produced per minute per milligram of protein.
Assays to determine the kinetic parameters of PvdA toward l-ornithine were carried out under similar conditions, except that the l-ornithine concentrations were varied. Data were fitted to a Michaelis-Menten equation by nonlinear regression with the program Leonora (3). The apparent Km value for FAD+ was determined by fixing the concentration of l-ornithine at 10 mM and that of NADPH at 300 μM. The apparent Km value for NADPH was determined by fixing the concentration of l-ornithine at 10 mM and that of FAD+ at 50 μM. Inhibition assays were carried out with 300 μM NADPH, 50 μM FAD+, and various concentrations of l-ornithine and inhibitors. Data for inhibition assays were fitted to a competitive, uncompetitive, or mixed inhibition equation with the Leonora program (3).
The pH dependence of PvdA activity was tested spectrophotometrically with assay mixtures containing NADPH (300 μM), l-ornithine (2.0 mM), and FAD+ (50 μM) at 25°C over a pH range of 6.0 to 9.0 in 0.5 pH increments.
Stoichiometry of hydroxylamine and hydrogen peroxide formation.
Stoichiometry of NADPH utilization and product formation was determined with 2-ml reaction mixtures containing 300 μM NADPH, 50 μM FAD, 75 μg PvdA, and 2 mM l-ornithine or l-lysine in 0.1 M sodium phosphate buffer, pH 8.0. After 5 min, a 1-ml aliquot was removed and quenched with either 0.07 N perchloric acid for quantitation of hydroxylamine or 1.5% trichloroacetic acid for hydrogen peroxide determination (described below).
Hydroxylamine was assayed by measuring oxidation of hydroxylamine to nitrite with iodine, which in turn catalyzes the azo dye coupling between sulfanilic acid and α-naphthylamine. The procedure was similar to that reported for the assay of lysine N6-hydroxylase activity reported previously (19). The concentration of hydroxylamine produced by the enzyme was calculated from the extinction coefficient of 12,500 M−1 cm−1 determined from a standard curve of known concentrations of hydroxylamine hydrochloride.
Hydrogen peroxide was assayed according to an established procedure (10). Hydrogen peroxide in the assay mixture was calculated from absorbance at 480 nm and an extinction coefficient of 3,500 M−1 cm−1, determined with known concentrations of standard hydrogen peroxide.
Determination of coenzyme dissociation constants.
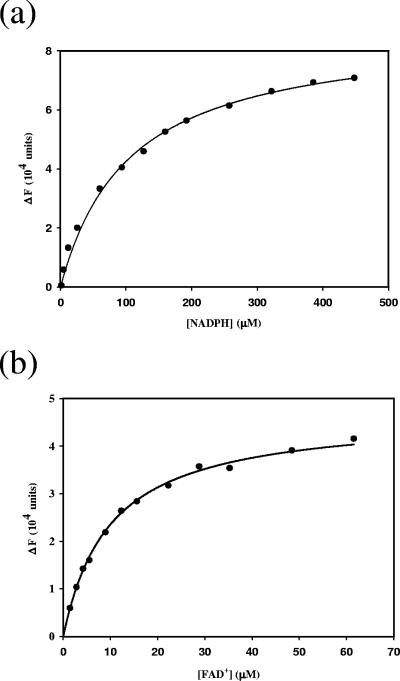
Equilibrium dissociation constants of NADPH and FAD+ were determined by fluorescence titration. Protein fluorescence was excited at 280 nm, and the emission fluorescence was detected at 330 nm. The protein (3 μg PvdA) emission fluorescence was recorded in the presence of various concentrations of NADPH or FAD+ in 100 mM sodium phosphate buffer, pH 8.0. The dissociation constant (Kd) can be determined by fitting the data, by nonlinear regression with the program Leonora (3), to the equations ΔF = (ΔFmax[L])/(Kd + [L]) and ΔF = F0 − F, where F is the protein fluorescence intensity at various concentrations of NADPH or FAD+ (L) and F0 is the fluorescence intensity of the protein in the absence of coenzymes.
RESULTS
Purification and physical properties of PvdA.
Recombinant PvdA was overexpressed as an N-terminal hexahistidine tag fusion protein in E. coli Rosetta 2(DE3) cells. Soluble proteins can be purified from cells induced for 3 h by nickel affinity chromatography with a yield of 5 mg of enzyme per 500 ml of cultured cells. Induction time was important, as cells grown for more than 12 h after addition of the inducer IPTG expressed PvdA as insoluble inclusion bodies.
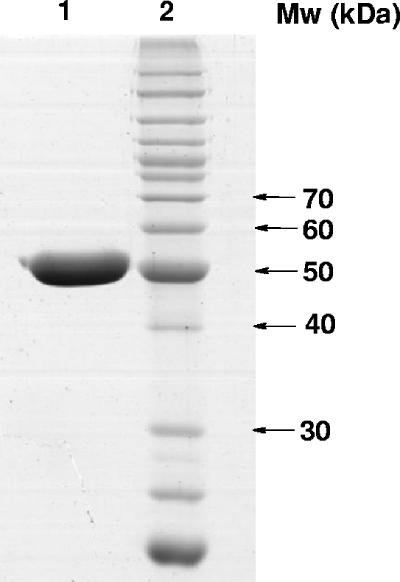
The subunit molecular mass of purified PvdA, as determined by SDS-PAGE, was 51 kDa (Fig. 3). This is in agreement with the predicted molecular mass of 51.6 kDa, based on the amino acid sequence. By gel filtration, the estimated native molecular mass of PvdA was 236.6 kDa, suggesting that the enzyme adopts a tetrameric or pentameric quaternary structure.
FIG. 3.
Coomassie blue-stained SDS-PAGE of PvdA purified by Ni2+-nitrilotriacetic acid chromatography. The gel was loaded with 5 ng of purified PvdA (lane 1). The molecular masses (Mw) of the proteins in the standard (lane 2) are indicated beside the gel.
Activity and pH optimum of PvdA.
At pH 8.0 and in the presence of 50 μM FAD+ and 300 μM NADPH, PvdA catalyzed the oxidation of NADPH at a rate of 0.24 ± 0.02 μmol min−1 mg−1. This NADPH oxidase activity was enhanced by about fivefold in the presence of 4 mM l-ornithine, with a specific activity of 1.24 ± 0.04 μmol min−1 mg−1. Stimulation of flavin reduction by substrates is commonly observed in other flavoprotein monooxygenases, such as p-hydroxybenzoate and phenol hydroxylases (6). This is attributed to a conformation change in the enzyme upon substrate binding that positions the cofactors at an optimal distance for hydride transfer (1). Cleavage of the histidine tag by thrombin had negligible effects on the specific activity of PvdA (1.16 ± 0.01 μmol min−1 mg−1). The histidine tag was therefore not removed in the subsequent analysis.
With the iodine oxidation assay, the coupling efficiency of NADPH oxidation and hydroxylamine formation from 2 mM l-ornithine was determined to be 96% ± 2%. There was no detectable hydrogen peroxide produced under the same condition.
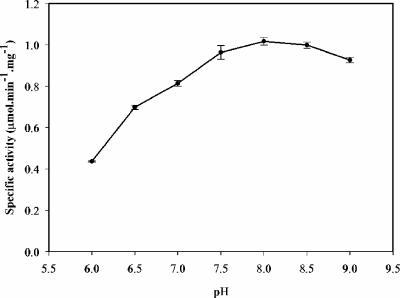
Activity of PvdA under standard reaction conditions (50 μM FAD+ and 300 μM NADPH, 2 mM l-ornithine) was determined in a pH range of 6 to 9 (Fig. 4). The pH-rate profile shows an activity optimum at about pH 8.0. The subsequent steady-state kinetic analysis was therefore conducted at this pH.
FIG. 4.
pH dependence of PvdA activity. Assay mixtures contained NADPH (300 μM), l-ornithine (2.0 mM), and FAD+ (50 μM) in 0.1 M sodium phosphate buffer at pHs of 6.0 to 9.0.
Coenzyme specificity.
PvdA was found to exhibit stringent coenzyme specificity since NADH and flavin mononucleotide cannot be utilized in place of NADPH and FAD+.
The intrinsic fluorescence of PvdA was quenched in the presence of NADPH or FAD+. This enabled the determination of dissociation constants for the coenzymes by fluorescence titration (Fig. 5). At 25°C, the NADPH and FAD+ equilibrium dissociation constants were 105.6 ± 6.0 μM and 9.9 ± 0.3 μM, respectively. By contrast, flavoenzymes that contained tightly bound FAD+, such as p-hydroxybenzoate hydroxylase and pyruvate oxidase, have dissociation constants in the nanomolar range for FAD+ (13, 26). The relatively low affinity of PvdA for FAD+ accounts for the lack of characteristic flavin absorbance between 400 and 500 nm in purified recombinant PvdA. Similarly, lysine N6-hydroxylase from E. coli has been reported to possess high dissociation constants for FAD+, with values of 30 μM and 100 μM at respective temperatures of 4°C and 25°C (9).
FIG. 5.
Fluorescence titration of PvdA with FAD+ (a) and NADPH (b). The changes in fluorescence of PvdA (ΔF) were plotted against the total concentrations of NADPH and FAD+, respectively. The solid line represents the best-fitted titration curve obtained with the program Leonora (3).
By steady-state kinetic analysis with fixed concentrations of l-ornithine (10 mM) and one of the two cofactors (300 μM NADPH or 50 μM FAD+), the apparent Km values for NADPH and FAD+ were determined to be 160 ± 20 μM and 21.9 ± 1.1 μM, respectively.
Substrate specificity.
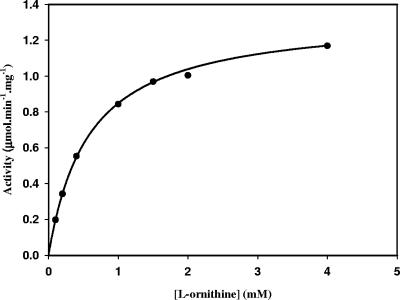
The specificity of PvdA was tested with a variety of amino acids. At saturating concentrations of NADPH and FAD+, the activity of PvdA at various concentrations of l-ornithine fits a classical Michaelis-Menten kinetic model, yielding an apparent Vmax value of 1.34 ± 0.01 μmol min−1 mg−1 and an apparent Km value of 0.58 ± 0.01 mM for l-ornithine (Fig. 6). d-Ornithine did not enhance the NADPH oxidation rate and was not hydroxylated by PvdA, thus indicating that the enzyme is specific for the l isomer of the substrate.
FIG. 6.
Plot of initial velocity versus concentration of l-ornithine. The assay mixture contained 300 μM NADPH, 50 μM FAD+, and various concentrations of l-ornithine in 0.1 M sodium phosphate buffer, pH 8.0. The data were fitted by nonlinear regression to the Michaelis-Menten equation with the program Leonora (3).
Surprisingly, l-lysine (2 mM) was able to enhance the NADPH oxidation rate of PvdA by 3.9-fold. However, the NADPH consumed in the presence of 2 mM l-lysine was channeled toward formation of hydrogen peroxide (90% ± 1%) and no hydroxylamine was detected with the iodine oxidation assay. l-Lysine is therefore a nonsubstrate effector of PvdA. Since l-lysine is one carbon longer than l-ornithine, the terminal amine group is probably not at an optimal position to be hydroxylated by the hydroperoxyflavin intermediate. This unstable flavin intermediate would then decay to form hydrogen peroxide.
l-2,4-Diaminobutyrate, which is one carbon atom shorter than l-ornithine, and 5-aminopentanoic acid, which differs from l-ornithine by the absence of a 2-amino group, were tested at concentrations of 0.1 to 10 mM and found to have no effect on the NADPH oxidation rate of PvdA, and no hydroxylated products were detected. PvdA was similarly inactive toward l-glutamine and l-asparagine, which contain terminal amide instead of amine nitrogens.
Inhibition assays.
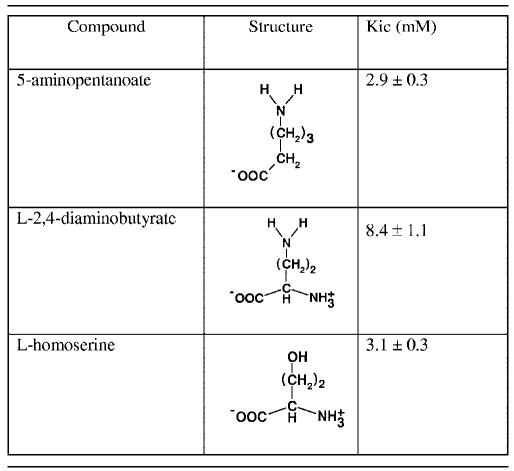
Steady-state inhibition assays were performed to determine if compounds that were not substrates or effectors of PvdA were able to bind to the enzyme, and the results are summarized in Table 1.
TABLE 1.
Inhibition constants derived from steady-state kinetics with l-ornithine as a substratea
Assay mixture contains contained 300 μM NADPH, 50 μM FAD+, and various concentrations of l-ornithine. The data were fitted to a competitive-inhibition model with the program Leonora (3).
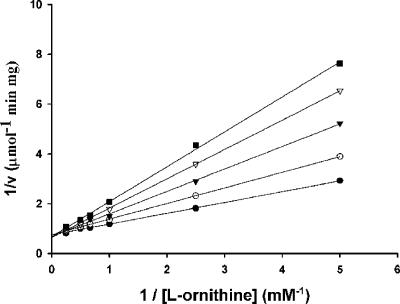
d-Ornithine, l-asparagine, and l-glutamine did not inhibit the l-ornithine-dependent NADPH oxidation rate. 5-Aminopentanoic acid, on the other hand, was a competitive inhibitor of l-ornithine, with a Kic fivefold higher than the apparent Km of l-ornithine (Fig. 7). l-2,4-Diaminobutyrate and l-homoserine, which are structurally similar except that l-homoserine has a terminal hydroxyl instead of a terminal amino group, were also competitive inhibitors, with their respective Kics 14-fold and 5.3-fold higher than the apparent Km of l-ornithine. However, l-serine, which is one carbon shorter than l-homoserine, did not inhibit the l-ornithine-dependent NADPH oxidation rate. The results showed that compounds that are able to bind to the same site of the enzyme as l-ornithine do not necessarily facilitate efficient hydride transfer from NADPH to FAD+. The chemical structures of compounds that bind to the enzyme are therefore important for their function as effectors.
FIG. 7.
Lineweaver-Burk plot of inhibition of PvdA NADPH oxidation activity by 5-aminopentanoic acid. Assays were performed with 0 (•), 2 (○), 3 (▾), 4 (▿), and 5 (▪) mM 5-aminopentanoic acid.
DISCUSSION
Hydroxamates are common functional groups involved in ferric chelation in bacterial and fungal siderophores. Biosynthesis of hydroxamates from diamino acids involves the hydroxylation of the terminal amino group by a flavin monooxygenase to form a hydroxylamine. Only one of these monooxygenases, an l-lysine N6-hydroxylase (IucD) involved in E. coli aerobactin siderophore biosynthesis, has been previously purified. Although previous attempts have been made to overexpress l-ornithine N5-hydroxylase, the recombinant enzyme formed inclusion bodies and was inactive (14).
In this study, an active l-ornithine N5-hydroxylase (PvdA) was expressed in E. coli and purified for the first time. PvdA had its optimum activity at pH 8.0. It utilized NADPH and FAD+ as cofactors. These properties are similar to those reported for IucD (9, 23). Fluorescence titration revealed that PvdA has a dissociation constant of 9.9 μM for the FAD+ cofactor at 25°C, which is about 10-fold lower than the value reported for IucD.
The coenzyme and substrate specificities of PvdA are clearly distinct from those of IucD. IucD was able to utilize NADH as a cofactor, although the activity of the enzyme was twofold lower than the corresponding activity with NADPH as a cofactor (22). PvdA, on the other hand, was not able to use NADH in place of NADPH as a cofactor. The NADPH oxidase activity of PvdA was enhanced in the presence of l-ornithine, and this was tightly coupled to substrate hydroxylation. However, IucD was not activated and did not hydroxylate l-ornithine (9). By contrast, although l-lysine enhanced the NADPH oxidation rate of PvdA, uncoupling occurred and hydrogen peroxide instead of hydroxylamine was produced. This is analogous to the reported observation that homolysine, which is one carbon longer than l-lysine, was a nonsubstrate effector of IucD (9). The dependence of the NADPH oxidation rate of PvdA on the l-ornithine concentration obeyed the classic Michaelis-Menten equation, which is in contrast to the substrate inhibition model described for IucD with l-lysine as the substrate (9). Nevertheless, the apparent Km value of PvdA for l-ornithine (0.58 mM) is of the same order of magnitude as the Km value reported for IucD with l-lysine as the substrate (0.11 mM) (9).
Other compounds tested in this study were found to be neither substrates nor effectors of PvdA. These include two close structural homologues of l-ornithine, l-2,4-diaminobutyrate and 5-aminopentanoic acid. The inability of these compounds to act as effectors or substrates could be due to impaired binding to the active site of the enzyme or the result of unproductive binding. To distinguish between these two possibilities, steady-state kinetic analysis was used to determine if the compounds are inhibitors of the l-ornithine-dependent NADPH oxidase activity of PvdA. l-Asparagine and l-glutamine did not inhibit the activity of PvdA and are therefore unable to bind to the active site of the enzyme. l-2,4-Diaminobutyrate, 5-aminopentanoic acid, and l-homoserine, on the other hand, were competitive inhibitors; i.e., they competed with l-ornithine for the same site of the enzyme. The inhibition constants of the latter two compounds were of the same order of magnitude as the apparent Km for l-ornithine, suggesting that these compounds bind relatively well to the enzyme. However, this was not sufficient to stimulate the NADPH oxidase rate in PvdA. l-Serine, which is shorter than l-homoserine by one carbon atom, was not an inhibitor. It appears, therefore, that the presence of a 2-amino group and the length of the carbon chain bearing the terminal amino group are important features that enabled efficient hydride transfer between the two cofactors and subsequent substrate hydroxylation. The results further demonstrate that it is possible to inhibit PvdA's activity with small molecules that compete with l-ornithine for the enzyme's active site.
Complete synthesis of hydroxamate requires further modifications of the hydroxylamine produced by the flavin hydroxylases. For example, PvdF and PvdY are involved in transformation of N5-hydroxyornithine to formyl-hydroxamate or acetyl-hydroxamate in the pyoverdine siderophores of P. aeruginosa (8, 11). The availability of purified PvdA should facilitate the enzymatic synthesis of commercially unavailable N5-hydroxyornithine for use as a substrate to study these downstream enzymes. This will further advance our understanding of these novel siderophore biosynthesis enzymes and may lead to the development of new antibiotics that target the iron acquisition systems in pathogenic microorganisms.
Acknowledgments
We thank G. I. Dmitrienko and T. Viswanatha from the University of Waterloo for helpful discussions.
This research was supported by a grant from the Canadian Institutes of Health Research (MOP-62879) to S. Seah.
REFERENCES
- 1.Ballou, D. P., B. Entsch, and L. J. Cole. 2005. Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases. Biochem. Biophys. Res. Commun. 338:590-598. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Cornish-Bowden, A. 1995. Analysis of enzyme kinetic data. Oxford University Press, New York, N.Y.
- 4.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hissen, A. H., A. N. Wan, M. L. Warwas, L. J. Pinto, and M. M. Moore. 2005. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding l-ornithine N5-oxygenase, is required for virulence. Infect. Immun. 73:5493-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Husain, M., and V. Massey. 1979. Kinetic studies on the reaction of p-hydroxybenzoate hydroxylase. Agreement of steady state and rapid reaction data. J. Biol. Chem. 254:6657-6666. [PubMed] [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.Lamont, I. L., L. W. Martin, T. Sims, A. Scott, and M. Wallace. 2006. Characterization of a gene encoding an acetylase required for pyoverdine synthesis in Pseudomonas aeruginosa. J. Bacteriol. 188:3149-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macheroux, P., H. J. Plattner, A. Romaguera, and H. Diekmann. 1993. FAD and substrate analogs as probes for lysine N6-hydroxylase from Escherichia coli EN 222. Eur. J. Biochem. 213:995-1002. [DOI] [PubMed] [Google Scholar]
- 10.Marrone, L., M. Beecroft, and T. Viswanatha. 1996. Lysine:N-6-hydroxylase: cofactor interactions. Bioorg. Chem. 24:304-317. [Google Scholar]
- 11.McMorran, B. J., H. M. Kumara, K. Sullivan, and I. L. Lamont. 2001. Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa. Microbiology 147:1517-1524. [DOI] [PubMed] [Google Scholar]
- 12.Meyer, J. M., A. Stintzi, D. De Vos, P. Cornelis, R. Tappe, K. Taraz, and H. Budzikiewicz. 1997. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 143(Pt. 1):35-43. [DOI] [PubMed] [Google Scholar]
- 13.Muller, F., and W. J. van Berkel. 1982. A study on p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens. A convenient method of preparation and some properties of the apoenzyme. Eur. J. Biochem. 128:21-27. [PubMed] [Google Scholar]
- 14.Putignani, L., C. Ambrosi, P. Ascenzi, and P. Visca. 2004. Expression of l-ornithine NΔ-oxygenase (PvdA) in fluorescent Pseudomonas species: an immunochemical and in silico study. Biochem. Biophys. Res. Commun. 313:245-257. [DOI] [PubMed] [Google Scholar]
- 15.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Schrettl, M., E. Bignell, C. Kragl, C. Joechl, T. Rogers, H. N. Arst, Jr., K. Haynes, and H. Haas. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokol, P. A., P. Darling, D. E. Woods, E. Mahenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect. Immun. 67:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stehr, M., L. Smau, M. Singh, O. Seth, P. Macheroux, S. Ghisla, and H. Diekmann. 1999. Studies with lysine N6-hydroxylase. Effect of a mutation in the assumed FAD binding site on coenzyme affinities and on lysine hydroxylating activity. Biol. Chem. 380:47-54. [DOI] [PubMed] [Google Scholar]
- 20.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thariath, A., D. Socha, M. A. Valvano, and T. Viswanatha. 1993. Construction and biochemical characterization of recombinant cytoplasmic forms of the IucD protein (lysine:N6-hydroxylase) encoded by the pColV-K30 aerobactin gene cluster. J. Bacteriol. 175:589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thariath, A. M., K. L. Fatum, M. A. Valvano, and T. Viswanatha. 1993. Physico-chemical characterization of a recombinant cytoplasmic form of lysine:N6-hydroxylase. Biochim. Biophys. Acta 1203:27-35. [DOI] [PubMed] [Google Scholar]
- 24.Visca, P., A. Ciervo, and N. Orsi. 1994. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J. Bacteriol. 176:1128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 26.Wille, G., M. Ritter, M. S. Weiss, S. Konig, W. Mantele, and G. Hubner. 2005. The role of Val-265 for flavin adenine dinucleotide (FAD) binding in pyruvate oxidase: FTIR, kinetic, and crystallographic studies on the enzyme variant V265A. Biochemistry 44:5086-5094. [DOI] [PubMed] [Google Scholar]