Abstract
Ribonucleotide reductases (RNRs) catalyze the conversion of ribonucleotides to deoxyribonucleotides and are essential for de novo DNA synthesis and repair. Streptomyces spp. contain genes coding for two RNRs, either of which is sufficient for vegetative growth. The class Ia RNR is encoded by the nrdAB genes, and the class II RNR is encoded by nrdJ, which is coexpressed with nrdR. We previously showed that the Streptomyces coelicolor nrdR gene encodes a protein, NrdR, which represses transcription of both sets of RNR genes. NrdR is a member of a highly conserved family of proteins that is confined exclusively to prokaryotes. In this report, we describe a physical and biochemical characterization of the S. coelicolor NrdR protein and show that it is a zinc-ATP/dATP-containing protein that binds to the promoter regions of both Streptomyces RNR operons. The NrdR N terminus contains a zinc ribbon motif that is necessary for binding to the upstream regulatory region of both RNR operons. The latter contains two 16-bp direct repeat sequences, termed NrdR boxes, which are located proximal to, or overlap with, the promoter regions. These experiments support the view that NrdR controls the transcription of RNR genes by binding to the NrdR box sequences. We also show that the central NrdR ATP cone domain binds ATP and dATP and that mutations that abolish ATP/dATP binding significantly reduce DNA binding, suggesting that the ATP cone domain may allosterically regulate NrdR binding. We conclude that NrdR is a widely conserved regulator of RNR genes, binding to specific sequence elements in the promoter region and thereby modulating transcription.
Ribonucleotide reductases (RNRs) provide the building blocks for DNA synthesis and repair in all living cells (20). They are essential because they are the only known de novo pathway for the biosynthesis of deoxyribonucleotides, the immediate precursors for DNA synthesis. Three major classes of RNRs are known (8, 15, 19). Class I RNRs are oxygen-dependent enzymes that occur in eukaryotes, bacteria, and some viruses, and class II RNRs are oxygen-independent enzymes confined to bacteria and archaea, while class III RNRs are found predominantly in anaerobic bacteria. Despite significant differences in structure and in cofactor requirements, all three enzymes share similar catalytic mechanisms creating a protein radical that initiates reduction of ribonucleotides, and all allosterically regulate the balanced formation of the four deoxyribonucleotides (15, 19, 24).
Previously we reported that Streptomyces spp., gram-positive aerobic bacteria that produce a remarkable variety of metabolites and possess a complex life cycle (7, 13), contain class Ia and class II RNRs (5). Either type of RNR is sufficient for vegetative growth (4). In Streptomyces coelicolor, the class Ia and class II RNRs are regulated by coenzyme B12 (adenosylcobalamin) in a reciprocal manner. B12 negatively controls transcription of the class Ia RNR nrdABS genes via a riboswitch mechanism and positively controls the activity of the class II RNR via its role as an essential enzyme cofactor (3, 4). We recently showed the existence in S. coelicolor of a second regulatory system that controls transcription of both class Ia nrdABS and class II nrdRJ RNR genes. We observed that deletion of the nrdR gene, which is coexpressed with nrdJ encoding the class II RNR, causes a dramatic increase of transcription of both sets of genes (4). Thus, nrdR encodes a protein that we have termed NrdR that represses transcription of both sets of RNR genes. Bioinformatic analysis was used to predict that NrdR possesses a four-cysteine Zn ribbon-like module and an ATP cone resembling the allosteric effector domain in certain RNRs, suggesting that NrdR functions to regulate transcription by DNA binding (2, 4). A possible binding site for NrdR was indicated by the presence of two imperfect direct repeats positioned immediately upstream of the Streptomyces nrdRJ promoter (4). Subsequent to a comprehensive study by Rodionov and Gelfand (21) that revealed the existence of a highly conserved 16-bp palindromic signal, termed the NrdR box, in the upstream region of many bacterial genes and operons encoding RNRs, we noted that a similar pair of repeats overlapped with the nrdABS promoter in Streptomyces, just upstream of the B12 riboswitch (3). In this report, we describe biochemical and physical characterization of the S. coelicolor NrdR protein and show that it is a zinc-ATP containing protein that binds specifically to the promoter regions of class Ia and class II RNR operons.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Streptomyces coelicolor strain M145 is referred to as the wild type (16). Escherichia coli strain XL1-Blue was used for plasmid constructions, and strain BL21(DE3) was used for protein overexpression.
Culture media and DNA manipulations.
E. coli strains were grown in Luria-Bertani (LB) medium and supplemented with kanamycin or ampicillin (50 μg/ml or 100 μg/ml, respectively) when appropriate. Plasmid DNA was isolated using a High Pure plasmid isolation kit (Roche). DNA restriction digestions and ligations were carried out according to the manufacturer's instructions. DNA linear fragments were isolated from a 0.9% agarose (Sigma) gel using a QIAquick gel extraction kit (QIAGEN). DNA manipulations were as described by Sambrook et al. (22). Electroporation was performed with a Gene Pulsar II apparatus (Bio-Rad Laboratories) according to the manufacturer's instructions.
Cloning of NrdR.
The S. coelicolor nrdR gene was amplified by PCR from M145 genomic DNA as described previously (5) using forward IG-1 (5′-ATATCATATGCACTGCCCCTTCTGC-3′) and reverse IG-2 (5′-TCTCAAGCTTGTCGGCGGCGCCTGCGGG-3′) primers. IG-1 contains an NdeI restriction site, and IG-2 contains a HindIII restriction site (both shown in bold italic). The PCR-amplified fragment was eluted from an agarose gel, ligated to the pGEM-T Easy vector (Promega), and electroporated into E. coli XL1-Blue. Positive transformants were detected by blue-white screening and colony PCR. DNA inserts were sequenced to verify their integrity. The pGEM construct was digested with restriction endonucleases NdeI and HindIII (Fermentas) and electrophoresed, and the small fragment containing the nrdR gene was eluted. The expression vector pET30a(+) (Novagen) was digested with NdeI and HindIII, electrophoresed, and eluted. T4 DNA ligase (Takara) was used to join the two DNA fragments to obtain pET30a(+)::nrdR, which was electroporated into XL1-Blue and transformants selected for kanamycin resistance. The nrdR DNA insert and the adjacent DNA regions were sequenced to verify the correctness of the construct. All pET30a(+)::nrdR constructs described in this work express wild-type and mutant recombinant proteins with six-His tags at their C termini.
Construction of mutant NrdR proteins.
Point mutations (Lys50→Asn50, Arg51→Gly51) were created by an overlap PCR procedure adapted from the work of Gov et al. (11). Two PCR fragments were amplified from M145 genomic DNA by use of two nonmutagenic external oligonucleotides, IG-1 and IG-2 (see above), and two complementary mutagenic internal oligonucleotides, IG-3 (5′-GCTCATGGTGGTGAATGGGTCCGGGGTCACCGAAC-3′) and IG-4 (5′-CGGTGACCCCGGACCCATTCACCACCATGAGCGAG-3′). The PCR products of the reaction were purified using a High Pure PCR product purification kit (Roche) and treated with Klenow fragment to remove protruding 3′ A nucleotides, and the DNA fragments containing the mutations were gel purified. The PCR fragments were mixed, denatured, reannealed, and amplified with the two external primers. The mutant DNA fragment was digested with NdeI and HindIII and ligated into the NdeI- and HindIII-digested pET30a(+) vector. The resulting recombinant plasmid was introduced into E. coli BL21(DE3) by electroporation. The DNA insert was sequenced to verify the presence of the expected mutation. The Cys3→Ala3 mutant was obtained by PCR amplification of genomic DNA by use of the forward primer IG-5 (5′-TATACATATGCACGCCCCCTTCTGCAGGCACCCCG-3′; bold italic type indicates a restriction site), which contains the mutation, and the reverse primer IG-2. Truncated NrdR mutants were constructed by PCR amplification of the wild-type nrdR gene with primers flanking the appropriate DNA fragment. An N-terminal deletion (removing residues 1 to 41) was created with the forward primers IG-6 (5′-ATATCATATGGAGACGTGCTCGCTC-3′) and reverse primer IG-2, a C-terminal truncation (removing residues 151 to 182) was created with the forward primer IG-1 and reverse primer IG-7 (5′-TCTCAAGCTTGTGCCCCGTCGTCTCCC-3′), and a truncated mutant lacking both N- and C-terminal domains was created with forward primer IG-6 and reverse primer IG-7.
Protein overexpression.
Overnight cultures of E. coli BL21(DE3)/pET30a(+) bearing wild-type or mutant nrdR genes were diluted to an absorbance at 600 nm of 0.1 in LB containing kanamycin (50 μg/ml) and shaken vigorously at 37°C. At an absorbance at 600 nm of 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) was added to a final concentration of 0.4 mM. The cells were incubated for 3 h at 37°C and harvested by centrifugation at 4,000 × g for 20 min at 4°C. The supernatant was discarded, and the cell pellet was stored at −70°C.
Protein purification.
Frozen cells were thawed and suspended in sonication buffer (50 mM Tris-HCl [pH 8.5], 300 mM NaCl, 5 mM imidazole). Phenylmethylsulfonyl fluoride (PMSF) was added to 1 mM to the cell suspension, and the mixture was sonicated in an ultrasonic processor (Misonics) until clear. The sonicate was centrifuged at 10,000 × g for 45 min at 4°C. The supernatant was loaded on a high-capacity Ni2+-chelate affinity matrix (CAM) resin column (Sigma) equilibrated with the sonication buffer and washed several times with the buffer. Protein was eluted with buffer containing 250 mM imidazole. Protein samples after Ni affinity purification were dialyzed against buffer containing 50 mM Tris-HCl (pH 8.5), 300 mM NaCl, 5 mM dithiothreitol (DTT), 50 μM ZnCl2, 20% glycerol when assayed for DNA binding and against 50 mM Tris-HCl (pH 8.5), 300 mM NaCl, 10% glycerol for spectroscopic analyses and boronate Affi-Gel and Superdex 200 gel filtration chromatography. Anion-exchange chromatography salt was carried with protein samples that had been dialyzed against buffer without salt. Recovery of recombinant protein was monitored by the Bradford assay (6). Purified recombinant proteins were stored at −70°C.
Chromatography.
Gel filtration chromatography was carried out on a Superdex 200HR 10/30 column (Pharmacia) connected to a Pharmacia fast protein liquid chromatography (FPLC) system in 50 mM Tris-HCl (pH 8.5), 300 mM NaCl (unless otherwise stated). MW-GF-200 molecular weight markers (Sigma) were run in parallel. Anion-exchange chromatography was performed on a Mono Q HR 5/5 column (Pharmacia). Protein was loaded on a column equilibrated with buffer consisting of 50 mM Tris-HCl (pH 8) and 10% glycerol. The column was washed with 10 column volumes of equilibration buffer, and the bound protein was eluted with a linear gradient of 50 ml NaCl (0 to 1.0 M NaCl) at a flow rate of 1 ml/min. Fractions (0.5 ml) were collected and analyzed.
Boronate affinity chromatography was performed as described by Shewach (23) with slight changes. Briefly, hydrated Affi-Gel 601 (Bio-Rad) was packed into a 1-ml plastic tuberculin syringe fitted with a frit. The column was equilibrated with 0.15 M ammonium hydrogen carbonate buffer, pH 8.9, containing 15 mM MgCl2 (buffer A). One hundred microliters of the supernatant of the perchloric acid (PCA)-precipitated protein sample was brought to pH 8.5 with KOH, and 2 μl of 1 M MgCl2 was added, followed by the addition of 100 μl of buffer A. The sample was applied to the column, and deoxyribonucleotides were eluted with 2 ml of buffer A and collected in two 1-ml fractions. The column was washed with 2 ml buffer A, and ribonucleotides were eluted with 2 ml of 0.1 M disodium tetraborate buffer, pH 8.9 (buffer B), and then 1-ml fractions were collected. Samples (200 μl) of each fraction were assayed by high-performance liquid chromatography (HPLC) (after acidification to match the pH of the mobile phase) for amounts of nucleotide. ATP and dATP standards were treated in the same way. Boronate chromatography products were quantified using a Waters HPLC system with detection at 260 nm. The 200-μl boronate column eluate was injected onto a Spherisorb S5 SAX column (4.6 × 250 mm; Waters) equilibrated in 0.05 M ammonium dihydrogen phosphate, pH 3. Nucleotides were eluted with a biphasic gradient of ammonium dihydrogen phosphate buffer ranging in concentration from 0. 05 to 1 M (pH 3) at a flow rate of 1 ml/min. Nucleotides were quantified by peak areas compared with standards.
Spectroscopic methods.
Inductively coupled plasma atomic emission spectroscopy was performed at the Analytical Laboratory of Hebrew University. Batches (200 to 500 mg) of samples and 1-ml volumes of 65% nitric acid were transferred into 50-ml polypropylene tubes with scandium added as internal standard. The tubes were heated in a water bath for 5 hours at 90°C, and the volume was made up to 10 ml with water. A blank with just nitric acid was processed in parallel. Amounts of zinc bound after deducting the value of the blank value from that of the protein sample are reported below (see “NrdR is a zinc binding protein”). Blank values were 2 to 4% of that present in wild-type NrdR protein samples. Analyses were conducted on portions of the solutions versus standards from Merck. Elements were determined by inductively coupled plasma atomic emission spectrometry with an inductively coupled plasma atomic emission spectrometer (Spectroflame Modula E; Spectro GMBH, Kleve, Germany). UV absorption spectra were recorded with an Ultrospec 2100 pro UV/visible spectrophotometer (Amersham Biosciences) by use of 1-cm quartz cuvettes. Circular dichroism spectroscopy was carried out with a model 202 spectrometer (Aviv Instruments, Inc.).
Electrophoretic gel mobility shift assay.
S. coelicolor DNA fragments of 84 bp spanning the nrdABS and nrdRJ NrdR box sequences were generated by PCR and purified by elution from an agarose gel. Purified fragments were labeled at the 3′ end with digoxigenin (DIG)-ddUTP by use of a terminal transferase kit (Roche, Mannheim, Germany). Binding assays were carried out in a final volume of 20 μl containing labeled DNA (6 fmoles), binding buffer (20 mM Tris-HCl, pH 8.5, 5% [vol/vol] glycerol, 1 mM MgCl2, 40 mM KCl, 1 mM DTT), purified wild-type or mutant recombinant NrdR (1 to 10 μg protein), 1 μg salmon sperm DNA, and 0.1 μg bovine serum albumin. An S. coelicolor 84-bp cydA promoter PCR fragment served as a negative control. Following a 30-min incubation at 30°C, the binding reaction products were separated on a native 6% polyacrylamide gel in 0.5× TB (Tris-borate buffer, pH 8.5). The gel was contact blotted onto a Hybond-N+ membrane (Amersham Biosciences). The chemiluminescent detection was performed following the manufacturer's instructions (Roche, Mannheim, Germany). The membrane was exposed to X-ray film (FUJI) for 15 to 40 min at 37°C.
Bioinformatics and DNA and protein sequence analysis.
Primary sequences of NrdR-like proteins were identified in unannotated genomic databases via the NCBI site (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) by use of the tBLASTn algorithm (1). More-detailed BLAST searches of Streptomyces coelicolor A3(2), Streptomyces avermitilis MA-4580, and Streptomyces scabies 87.22 genomes were prepared by use of the websites http://www.sanger.ac.uk/Projects/S_coelicolor/, http://avermitilis.ls.kitasato-u.ac.jp/, and http://www.sanger.ac.uk/Projects/S_scabies, respectively. Pairwise and multiple sequence alignments were made with the ClustalW program, version 1.84 (12), by use of the EMBL ClustalW server (http://www.ebi.ac.uk/clustalw/) and its mirror (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html). Sequence entry, primary analysis, and open reading frame (ORF) searches were performed using the NCBI server ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and the Clone Manager 7 program (Scientific & Educational Software, Durham, NC).
Other methods.
Protein concentration was determined by the method of Bradford (6) with bovine serum albumin as the standard. Sodium-dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (17). Nucleotide sequencing was determined using an AB1 Prism 3100 genetic analyzer (Applied Biosystems) and a BigDye Terminator cycle sequencing kit (Applied Biosystems) as recommended by the manufacturer, except that 5% dimethyl sulfoxide was added to each reaction mixture. PCA precipitation of NrdR was performed by adding perchloric acid to the protein solution to a final concentration of 1 M, the protein was then left on ice for 15 min, and the solution was centrifuged for 30 min at 4°C at 14,000 rpm. The supernatant was adjusted to neutral pH and used for spectroscopic analysis and boronate affinity chromatography.
RESULTS
NrdR is an oligomeric protein.
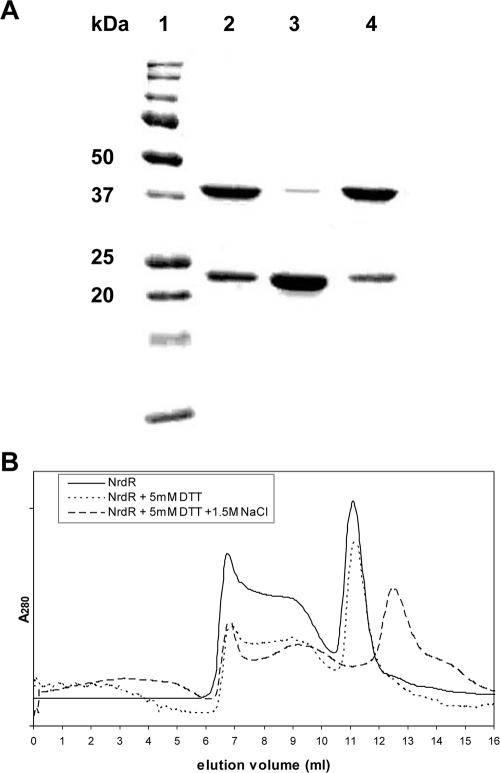
A T7 promoter vector was used to express soluble recombinant wild-type and mutant S. coelicolor NrdR six-His-tagged proteins. Figure 1A shows SDS-PAGE profiles of purified recombinant NrdR obtained after IPTG treatment of cultures of E. coli BL21(DE3) containing plasmid pET30a::nrdR and after Ni-CAM acid affinity chromatography. To assess size and purity, NrdR was run on denaturing gels. Two main bands were observed with sizes of approximately 23 kDa and 44 kDa, corresponding to the monomer (deduced molecular mass of the His-tagged NrdR is 22.1 kDa) and dimer species, respectively. To assess the stability of the dimer species to heat, the protein sample was heated to 100°C for 10 min prior to loading; this resulted in the almost complete disappearance of the 44-kDa species. Addition of up to 50 mM DTT, a powerful disulfide reducing agent, did not decrease the amount of the 44-kDa species, indicating that the dimer species was not due to disulfide bond formation.
FIG. 1.
Molecular size and oligomeric properties of Ni-CAM affinity-purified recombinant NrdR. (A) SDS-PAGE results. Lanes: 1, protein molecular mass standards; 2, NrdR without any treatment; 3, NrdR heated to 100°C for 10 min; 4, NrdR with 50 mM DTT. (B) FPLC Superdex 200 gel filtration chromatography. Solid line, NrdR containing 0.3 M NaCl; dotted line, NrdR containing 5 mM DTT and 0.3 M NaCl; dashed line, NrdR containing 5 mM DTT and 1.5 M NaCl. Elution profiles monitored by the absorbance at 280 nm.
FPLC gel filtration was used to assess the oligomeric state of NrdR under nondenaturing conditions. Column fractions were monitored for protein by SDS-PAGE. When NrdR was chromatographed in 1.5 M NaCl and 5 mM DTT on a Superdex 200 column, the main fraction corresponded to a molecular mass of about 85 kDa, suggesting a tetrameric form, although a significant amount of protein eluted in fractions corresponding to higher-molecular-weight species (Fig. 1B). In 0.3 M NaCl and 5 mM DTT, the predominant species had a molecular mass of about 180 kDa. Similarly, an N-terminally truncated protein in which the first 41 residues were deleted also formed multimers. Nondenaturing gel electrophoresis confirmed that NrdR exists in a multimeric state (data not shown).
NrdR is a zinc binding protein.
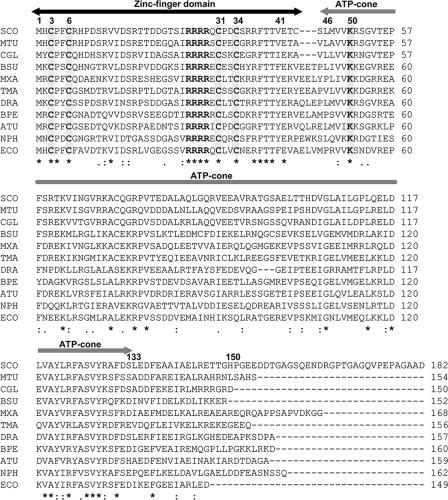
Streptomyces NrdR proteins are hydrophilic proteins comprising 167 to 187 amino acids. Systematic analysis of genomic databases revealed that NrdR orthologs exist in most eubacteria but not in certain species of ɛ-Proteobacteria and Mycoplasma. Fig. 2 shows the amino acid sequence of S. coelicolor NrdR and those of representative members from diverse eubacteria. Inspection of more than 400 eubacterial genomes revealed that all contained a single copy of an nrdR-like gene. Interestingly, just 1 of 32 archaeal genomes that were available for analysis at the NCBI website, that of halobacterium Natronomonas pharaonis DSM 2160, contains an nrdR-like gene, which happens to be linked to the putative nrdBA RNR genes (10). Computational analysis of NrdR proteins reveals a highly conserved 42-amino-acid N-terminal domain containing two pairs of vicinal cysteines separated by 24 (or rarely 22 or 23) residues and a sequence of four consecutive arginines, a central ATP cone domain that is present in the allosteric effector site of class Ia and class III RNRs (2), and a C-terminal domain of variable length and charge. The arrangement of the cysteines in the N-terminal domain suggests that it may form a Zn ribbon with the zinc atom coordinated to each of four cysteines. To determine whether NrdR binds zinc, recombinant six-His-tagged NrdR was purified by Ni affinity chromatography and analyzed by atomic absorption spectroscopy. Our results show that wild-type NrdR contains 0.7 to 0.8 mol zinc per mole NrdR, while amounts of zinc bound by NrdRΔN and NrdR Cys3→Ala3 were 0.01 and 0.1 mol zinc atoms per mol protein, respectively, after deducting the value of the blank value from that of the protein sample. The value for the wild-type NrdR is the average of three measurements made with independent protein preparations having values of 1.1, 0.5, and 0.7 mol zinc per mol protein. We suppose that the value of less than 1 (based on the average of three independent determinations) may be due to insufficient zinc in the culture broth (no zinc salts were included in the broth) or possibly due to a loss of zinc during purification. No other metals were present in any significant amount.
FIG. 2.
Multiple sequence alignment and domain organization of deduced NrdR proteins from representative members of major groups of bacteria. The proposed positions of the N-terminal zinc finger (residues 1 to 44) and the central ATP cone (residues 46 to 133) domains are indicated by overhead arrows. Conserved cysteine residues that define the zinc finger motif are highlighted (residues 3, 6, 31, and 34 by S. coelicolor numbering), as is a sequence of four consecutive arginine residues. Residues in the ATP cone domain presumed to be responsible for nucleotide binding are shown in Fig. 3. Symbols: asterisks, full conservation; colons, strongly similar; periods, weakly similar. Abbreviations: SCO, Streptomyces coelicolor (Actinobacteria); MTU, Mycobacterium tuberculosis (Actinobacteria); CGL, Corynebacterium glutamicum (Actinobacteria); BSU, Bacillus subtilis (Firmicutes, low GC content); MXA, Myxococcus xanthus (δ-Proteobacteria); TMA, Thermotoga maritima (Thermotogales); DRA, Deinococcus radiodurans; BPE, Bordetella pertussis (β-Proteobacteria); ATU, Agrobacterium tumefaciens (α-Proteobacteria); NPH, Natronomonas pharaonis (Euryarchaeota); ECO, Escherichia coli (γ-Proteobacteria).
To show that the N-terminal portion of NrdR is responsible for zinc binding, we constructed a mutant in which residues 1 to 41 were deleted. Our results show that the truncated protein, denoted NrdRΔN, is unable to bind zinc (see “NrdR is a zinc binding protein” above). We next constructed a mutant in which one of the four conserved cysteines, Cys3, which is presumed to coordinate the zinc atom, was replaced with alanine. The Cys3Ala mutant showed a reduction of about sevenfold in the amount of zinc bound. These results establish that the N-terminal domain is responsible for zinc binding.
NrdR is a nucleotide binding protein.
The Streptomyces NrdR central domain comprises approximately 100 amino acid residues that share significant sequence conservation with the N-terminal allosteric activity domain of the NrdA subunit of class Ia RNRs (Fig. 3). The crystal structure of the E. coli NrdA protein, containing a bound ATP analog, shows that the activity site lies in the N-terminal portion of the molecule, which forms a cleft with a four-helix bundle covered by a three-stranded mixed β-sheet (9, 25). The ATP analog is bound at the top of this structure below the β-sheet. Following Aravind et al. (2) we refer to this structure as the ATP cone domain. Comparison of the E. coli NrdA and NrdD N-terminal sequences and the S. coelicolor NrdR ATP cone domain shows that many of the residues that bind the analog are conserved in NrdR. Thus, the adenine base of the analog is hydrogen bonded to the main chain and is surrounded by the hydrophobic residues Val7 and Ile22, corresponding in S. coelicolor NrdR to Val48 and Val63; three of four positively charged residues which surround the negatively charged phosphates at the entrance to the cleft, Lys9, Arg10, and Lys21, correspond in S. coelicolor NrdR to Lys50, Arg51, and Lys62; and Thr55, which binds to the 5′ oxygen, most likely corresponds in S. coelicolor NrdR to Thr95 (Fig. 3). These residues are mostly conserved in the E. coli NrdR (Fig. 3).
FIG. 3.
Comparison of the amino acid sequence of the E. coli NrdA allosteric activity domain with that of the S. coelicolor and E. coli NrdR ATP cone domains to show conservation of ATP binding amino acid residues. NrdA residues reported to interact with an ATP analog (see text) are numbered; NrdA residues reported to interact with an ATP analog and which are conserved in S. coelicolor and E. coli NrdR are shown shaded. Two S. coelicolor NrdR residues that were mutated, Lys50 and Arg51, are shown boxed. The ATP cone domain consensus based on the deduced sequences of 250 NrdR proteins is shown at the bottom. Fully conserved NrdR residues are shown in bold and underlined. Symbols and abbreviations are as in the legend to Fig. 2. The function of the conserved sequence in the C-terminal region of the NrdR ATP cone domain, DEVAYVRFASVYRSF, is unknown.
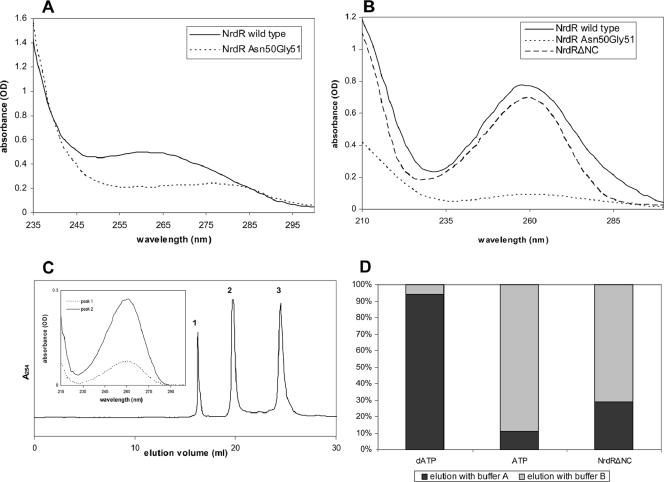
We examined the UV absorption spectra of the S. coelicolor NrdR to determine whether it contains bound nucleotide (Fig. 4A). The absorption profile shows a broad absorption band with a maximum around 260 nm. We supposed that the unusual profile resulted from NrdR containing a bound nucleotide, probably ATP or dATP. We calculated the approximate molar extinction coefficients of NrdR on the basis of it containing two tyrosines and six phenylalanines as a λ275 of 2.8 × 103 and a λ260 of 1.8 × 103, which would yield a spectra with a shallow maximum at 270 to 275 nm. Using these numbers and the known molar extinction coefficient of ATP, a λ259 of 15.4 × 103, we estimate, from the experimental absorption data presented in Fig. 4A, that NrdR contains approximately 0.7 mol ATP or (dATP) per mole NrdR (molecular mass, 21.2 kDa). We conclude that NrdR likely contains a single nucleotide binding site and that the departure of the calculated value of less than one nucleotide per molecule may reflect a partial loss of the nucleotide during protein purification.
FIG. 4.
UV absorption spectra of wild-type and mutant NrdR proteins. (A) Spectra of 0.04 mM solutions of the wild type and the Lys50Asn Arg51Gly double mutant. (B) Spectra of the supernatant obtained after perchloric acid treatment of 0.08 mM solutions of wild-type NrdR, NrdRΔNC, and NrdR Lys50Asn Arg51Gly double mutant. (C) Mono Q chromatography of NrdRΔNC. Peak fractions 1 and 2 did not contain protein and exhibited spectra matching that of adenine nucleotides (insert; see text for further details). (D) Affi-Gel boronate chromatography of the supernatant obtained after perchloric acid treatment of the NrdRΔNC mutant containing only the ATP cone domain. Standard solutions of dATP (left column), ATP (middle column), and the perchloric acid supernatant of NrdRΔNC (right column) were loaded onto an Affi-Gel column, and nucleotides were eluted, in succession, with buffer A to elute deoxyribonucleotides and with buffer B to release bound ribonucleotides. Amounts of released nucleotide were determined by HPLC and absorbance at 260 nm. OD, optical density.
Free nucleotide was detected after treatment of wild-type NrdR with PCA, which precipitated the protein and released material which gave a well-defined absorption band with a maximum at 258 nm characteristic of ATP and dATP in acid solution (Fig. 4B). We estimated that most of the presumably bound ATP was released by this treatment. To demonstrate that the ATP cone domain is responsible for nucleotide binding, a deletion mutant, NrdRΔNC, which lacks both the N-terminal (residues 1 to 41) and C-terminal (residues 151 to 182) domains, was constructed and then treated with PCA. The amount and the absorption profile of the released nucleotide material were essentially the same as those of the wild-type NrdR (Fig. 4B). Confirmation that the ATP cone domain is responsible for nucleotide binding and identification of specific nucleotide binding residues were obtained by constructing a mutant in which residues Lys50 and Arg51, corresponding to Lys9 and Arg10 in E. coli NrdA, which participate in binding ATP/dATP in the allosteric activity site, were replaced with asparagine and glycine, respectively. Figure 4A shows that the absorption profile of the Lys50→Asn50 Arg51→Gly51 mutant differs markedly from that of wild-type NrdR; its absorption is much reduced in the range of 255 to 275 nm, and it lacks a maximum at 260 nm. The broadness of the profile is due to six phenylalanines, which have a maximum absorption at 257 nm, and two tyrosines, which have a maximum absorption at 275 nm. Moreover, in contrast to wild-type NrdR, no significant 260-nm-absorbing material was released following treatment of the Lys50→Asn50 Arg51→Gly51 mutant with PCA (Fig. 4B).
We were also able to detect free nucleotide in preparations of NrdR and the NrdRΔNC mutant that had been stored at −70°C by Mono Q anion-exchange chromatography. Figure 4C shows results for the NrdRΔNC mutant protein, and similar results were obtained with wild-type NrdR (not shown). Three fractions that absorbed at 254 nm were obtained; one contained protein as judged by Bradford assay, and the two other fractions lacked protein and had a spectrum (Fig. 4C, insert) that corresponded to adenine nucleotides. The peak fractions were identified as the monophosphate (peak 1) and diphosphate (peak 2) species by comparison with known standards and presumably are the result of hydrolysis of the triphosphate, which could be detected in other experiments by HPLC (data not shown). These results further support the view that NrdR copurifies with a nucleotide and that the NrdR central ATP cone domain is necessary for nucleotide binding.
Finally, evidence that NrdR contains both ATP and dATP was obtained from boronate Affi-Gel chromatography of the material released after PCA treatment of the NrdRΔNC mutant. The Affi-Gel column is unable to bind deoxyribonucleotides which are first eluted with buffer A, and ribonucleotides are subsequently eluted with buffer B (Fig. 4D) (23). The amounts of nucleotides, assumed to be ATP/dATP, that eluted with each buffer were determined by HPLC and absorption spectroscopy. The results show that approximately 75% of the released nucleotides were ATP and 25% were dATP.
NrdR binds to the promoter regions of nrdABS and nrdRJ operons.
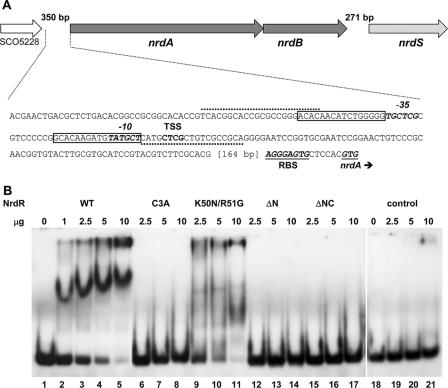
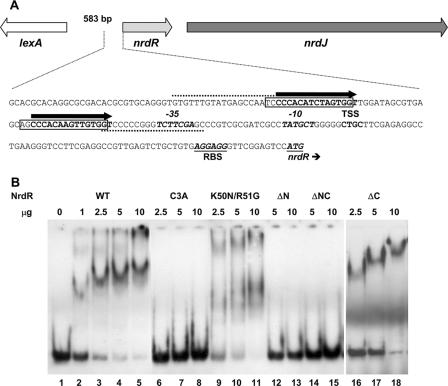
The 5′ upstream regions of the S. coelicolor nrdABS and nrdRJ operons contain two 16-bp motifs separated by 15 bp, which partially overlap with or are positioned just upstream of the −10 and −35 promoter recognition elements. We supposed that the repeats were possible binding sites for NrdR based on the observations that similar motifs, termed NrdR boxes, are present in front of other bacterial ribonucleotide reductase genes (21). Also, we showed that deletion of the S. coelicolor NrdR caused upregulation of transcription of both RNR operons (4). To assess whether NrdR binds to the repeat sequences, we initially performed gel electrophoretic mobility shift assays with purified NrdR and PCR-amplified DIG-labeled ∼200-bp DNA probes that span the two NrdR boxes. These experiments showed that NrdR binds to both nrdABS and nrdRJ probes (data not shown). Subsequently, we assessed the ability of NrdR to bind to shorter probes of 84 bp which consist of the two 16-bp NrdR boxes, the intervening 15-bp sequence, and the flanking 18-bp sequences (Fig. 5A and 6A). Wild-type NrdR binds to both DNA probes (Fig. 5B and 6B). A single protein-DNA complex was formed over a 10-fold range in NrdR concentration. The relatively high amounts of protein used in the gel shift experiments may reflect the tendency of NrdR to aggregate. NrdR did not bind to a control cydA DNA probe at any of the NrdR concentrations used (Fig. 5B).
FIG. 5.
Binding of NrdR to the DNA region upstream of the S. coelicolor class Ia nrdABS RNR operon. (A) Nucleotide sequence of the DNA region containing the 84-bp probe used in binding assays. Primers used for preparing the PCR probe are indicated by dotted lines. The two NrdR boxes, shown by open rectangular frames, are located near to the transcription start site (TSS) and partially overlap with the promoter recognition element shown in bold italic. The putative ribosome binding site (RBS) and translation initiation codon GTG are shown in bold italic and underlined. (B) Binding of wild-type and mutant proteins to the nrdABS probe. DIG labeling of DNA probes and electrophoretic mobility shift assays were performed as described in Materials and Methods. Lanes: 1 to 5, NrdR wild type (WT) (0, 1, 2.5, 5, and 10 μg protein); 6 to 8, NrdR Cys3Ala (2.5, 5, and 10 μg protein); 9 to 11, NrdR Lys50Asn Arg51Gly (2.5, 5, and 10 μg protein); 12 to 14, NrdRΔN (2.5, 5, and 10 μg protein); 15 to 17, NrdRΔNC (2.5, 5, and 10 μg protein); 18 to 21, NrdR wild type (0, 2.5, 5, and 10 μg protein with the negative control cydA probe [84 bp]).
FIG. 6.
Binding of NrdR to the DNA region upstream of the S. coelicolor class II nrdRJ RNR operon. (A) Nucleotide sequence of the DNA region containing the 84-bp probe used in binding assays. Primers used for preparing the PCR probe are indicated by dotted lines. The two NrdR boxes are shown by open rectangular frames according to the work of Rodionov and Gelfand (21), and our prediction of NrdR binding sites is indicated with solid arrows above the nucleotides shown in bold. The putative ribosome binding site (RBS) and translation initiation codon ATG are shown in bold italic and underlined. (B) Binding of wild-type and mutant proteins to the nrdRJ probe. DIG labeling of DNA probes and electrophoretic mobility shift assays were performed as described in Materials and Methods. Lanes: 1 to 5, NrdR wild type (WT) (0, 1, 2.5, 5, and 10 μg protein); 6 to 8, NrdR Cys3Ala (2.5, 5, and 10 μg protein); 9 to 11, NrdR Lys50Asn Arg51Gly (2.5, 5, and 10 μg protein); 12 and 13, NrdRΔN (5 and 10 μg protein); 14 and 15, NrdRΔNC (5 and 10 μg protein); 16 to 18, NrdRΔC (2.5, 5, and 10 μg protein).
To identify the protein domain responsible for binding, experiments were performed with mutant proteins. NrdRΔN lacks the putative zinc finger domain, NrdRΔC lacks the C-terminal domain, and NrdRΔNC lacks N- and C-terminal domains. Both N-terminally truncated proteins failed to bind the nrdABS and nrdRJ DNA probes (Fig. 5B and 6B), whereas the C-terminally truncated protein was able to bind both probes to the same extent as wild-type NrdR (Fig. 6B shows binding to the nrdRJ probe). These results establish that the N-terminal domain is necessary for binding to each probe. To show that the binding properties of the N terminus are due to it containing a zinc finger motif, we tested the ability of the Cys3→Ala3 mutant, which lacks one of four cysteines necessary for forming a zinc finger, to bind DNA. Figures 5B and 6B show that Cys3Ala mutant protein is unable to bind to either the nrdABS or the nrdRJ probe. To assess whether the ATP cone domain is required for binding, we tested the binding of the Lys50→Asn50 Arg51→Gly51 double mutant. The mutant protein exhibited weak, ill-defined binding to both probes. This result suggests that the ATP cone domain may function to recruit ATP/dATP to elicit a conformational change in NrdR that is necessary for binding. Comparison of the far UV circular dichroism spectra of the wild type and of the Lys50→Asn50 Arg51→Gly51 mutant, however, failed to show any significant difference in conformation (data not shown).
DISCUSSION
Previously we reported that in S. coelicolor, NrdR represses transcription of class Ia and class II RNR genes (4). Prokaryotic transcription regulatory proteins possess distinct binding domains, such as the helix-turn-helix, winged helix, and, less commonly, zinc finger. The S. coelicolor NrdR contains three domains (Fig. 2 and 3), an N-terminal domain containing two pairs of vicinal cysteines that might form a zinc ribbon module (or alternatively function as redox sensors), a central domain predicted to contain an ATP cone motif, and a C-terminal acidic domain (2, 4). In this paper, we show that the S. coelicolor NrdR (i) contains an N-terminal zinc finger domain that permits binding of one zinc atom per molecule, (ii) possesses a central ATP cone domain that confers binding of ATP/dATP, and (iii) requires functional zinc finger and ATP cone domains for binding to the promoter regions of both sets of RNR genes. In the nrdABS operon, the putative NrdR binding sites (NrdR boxes) overlap with the promoter, whereas in the nrdRJ operon the NrdR boxes lie just upstream of the promoter. The fact that an intact ATP cone is necessary for DNA binding suggests that binding of ATP/dATP to NrdR triggers a conformational change in the zinc finger domain, thereby modulating its interaction with its cognate NrdR box. We note that NrdR box motifs have been identified in most bacterial operons encoding all three classes of RNRs and that they therefore appear to provide a common mechanism for the regulation of RNR genes (21).
NrdR is the first example of a protein that contains both a C4-type zinc finger and an ATP cone domain. ORFs encoding an ATP cone (Pfam03477) have been identified in the genomes of most eubacteria and in those of a few archaea. In eubacteria, they occur in NrdR, in the N-terminal regions of class Ia and class III RNRs (in some cases in more than one copy), and in the N-terminal region of the Deinococcus radiodurans 2-phosphoglycerate kinase; in archaea, they are found in certain class II and class III RNRs (19) and in 2-phosphoglycerate kinase (2). An extensive search of the genome databases detected related ORFs that consist only of the ATP cone motif. Thus, streptococcal species contain ORFs encoding ATP cone proteins with sizes of 112 to 115 amino acids, which are most similar to the ATP cone present in class III RNR NrdD subunits. Similarly sized ATP cone ORFs were also identified in a few archaea. Inspection of the sequences of some 250 NrdR homologs showed that the NrdR N terminus is highly conserved and that the long central domains are less so, while the C-terminal domain is considerably more variable both in length and sequence. We noted in the 250-NrdR consensus sequence a conserved motif in the C-terminal portion of the ATP cone, an octapeptide, YV/I/LRFASVY, containing the only two tyrosines in the molecule (Fig. 3). The tyrosines may serve as a sensor to external signals.
Several lines of evidence indicate that NrdR is a regulator of all three classes of RNR genes. The gene encoding NrdR was first identified for Streptomyces spp. as part of an operon containing nrdJ, which encodes the class II RNR; subsequently, it was shown that NrdR represses transcription of class Ia and class II RNR operons (4). Homologous nrdR genes are widespread in eubacteria, where they are found linked to a variety of different genetic loci (4, 21). In some cases nrdR is linked to RNR genes. For example, nrdR is frequently linked to nrdJ (encoding the class II RNR) in G+C-rich Actinobacteria; it is linked to nrdDG (encoding the class III RNR) in certain extreme thermophiles, such as Dictyoglomus thermophilum; and in Natronomonas pharaonis (the only archaeal species out of 32 examined to have an nrdR gene), it is plasmid borne and linked to nrdB (encoding one subunit of the putative class Ia RNR). In many Firmicutes/Bacilli species, nrdR is linked to the DNA replication gene cluster (dnaB-dnaI), while in certain members of Proteobacteria it is denoted ybaD or ribX, which forms part of the riboflavin biosynthetic gene cluster.
We recently proposed a model for the regulation of Streptomyces RNR gene expression in which binding of NrdR to tandem repeat sequences in the nrdRJ promoter region controls expression of the class II RNR operon (4). At that time, we failed to notice the presence of a similar repeat in front of the nrdABS promoter and assumed that NrdR activated a hypothetical gene whose product was able to bind and regulate nrdABS expression. In addition to being reported for Streptomyces, conserved sequences in the upstream regions of nrd operons were earlier reported for E. coli and Salmonella enterica serovar Typhimurium (14) and for Staphylococcus aureus (18), but their significance then was unknown. More recently, Rodionov and Gelfand (21) performed a systematic search of bacterial genomes and extracted a highly conserved palindromic 16-bp sequence present in the upstream regions of most RNR genes and operons, which they termed an NrdR box. The experiments reported here strongly suggest that the Streptomyces NrdR binds to such NrdR boxes that overlap with or are proximal to the promoter regions of class Ia and class II RNR operons, respectively (Fig. 5A and 6A). In the nrdRJ operon, the two 16-bp repeats lie just upstream of the promoter −35 recognition element and are separated by 15 bp. We have adjusted the positions of the two repeats by two bases, as proposed by Rodionov and Gelfand (21), for a better match between pairs of repeats (Fig. 6A). In the nrdABS operon, the two repeats overlap with the promoter −35 and −10 sequences. We suppose that the very low level of transcription of class Ia RNR genes in vegetative growth is a consequence of two effects, i.e., NrdR binding to NrdR boxes that overlap with the promoter elements and B12 riboswitch repression (3, 5); in contrast, class II RNR genes are well expressed in vegetative growth and are likely to be less tightly controlled by NrdR, since the NrdR boxes are located a short distance upstream of the promoter elements (Fig. 5A and 6A). In other studies to be reported elsewhere, we have shown that the S. coelicolor NrdR is able to bind to probes containing NrdR boxes present in the promoter regions of each of the E. coli class Ia nrdAB, class Ib nrdIEF, and class III nrdDG RNR operons.
Our working model for NrdR regulation of RNR genes is that it functions to regulate transcription by binding to NrdR box sequences in or near to the promoter, thereby interfering with recruitment or activation of RNA polymerase. The experiments reported here show that the N-terminal zinc finger domain is responsible for DNA binding. What then is the role of the ATP cone domain? The NrdR ATP cone domain resembles the E. coli NrdA (R1) allosteric effector activity site (9, 25). The latter is located in the N-terminal region of the subunit and binds ATP or dATP to regulate overall NrdAB RNR activity. When ATP is bound, enzyme activity is stimulated, and when dATP is bound, enzyme activity is inhibited, presumably due to conformational changes in the NrdA subunit (15, 19). ATP and dATP may function in an analogous way to control the transcription of Streptomyces RNR genes by causing a conformational change in NrdR. We suppose that binding of ATP/dATP to NrdR causes it to undergo a conformational change that modulates its ability to bind to its cognate NrdR box and thus to inhibit transcription of downstream genes. This view is supported by the properties of an NrdR mutant that is deficient in ATP/dATP and is impaired in DNA binding.
Acknowledgments
We thank Mordechay Schonfeld for assistance with atomic absorption spectroscopy measurements.
We thank the Israel Science Foundation for financial support (1189/04). I.G. is supported by a fellowship from the NoE EuroPathoGenomics (EPG) program.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., Y. I. Wolf, and E. V. Koonin. 2000. The ATP-cone: an evolutionarily mobile, ATP-binding regulatory domain. J. Mol. Microbiol. Biotechnol. 2:191-194. [PubMed] [Google Scholar]
- 3.Borovok, I., B. Gorovitz, R. Schreiber, Y. Aharonowitz, and G. Cohen. 2006. Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J. Bacteriol. 188:2512-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borovok, I., B. Gorovitz, M. Yanku, R. Schreiber, B. Gust, K. Chater, Y. Aharonowitz, and G. Cohen. 2004. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 54:1022-1035. [DOI] [PubMed] [Google Scholar]
- 5.Borovok, I., R. Kreisberg-Zakarin, M. Yanko, R. Schreiber, M. Myslovati, F. Aslund, A. Holmgren, G. Cohen, and Y. Aharonowitz. 2002. Streptomyces spp. contain class Ia and class II ribonucleotide reductases: expression analysis of the genes in vegetative growth. Microbiology 148:391-404. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 8.Eklund, H., U. Uhlin, M. Farnegardh, D. T. Logan, and P. Nordlund. 2001. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77:177-268. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson, M., U. Uhlin, S. Ramaswamy, M. Ekberg, K. Regnstrom, B. M. Sjoberg, and H. Eklund. 1997. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure 5:1077-1092. [DOI] [PubMed] [Google Scholar]
- 10.Falb, M., F. Pfeiffer, P. Palm, K. Rodewald, V. Hickmann, J. Tittor, and D. Oesterhelt. 2005. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 15:1336-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gov, Y., I. Borovok, M. Korem, V. K. Singh, R. K. Jayaswal, B. J. Wilkinson, S. M. Rich, and N. Balaban. 2004. Quorum sensing in Staphylococci is regulated via phosphorylation of three conserved histidine residues. J. Biol. Chem. 279:14665-14672. [DOI] [PubMed] [Google Scholar]
- 12.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 13.Hopwood, D. A. 1988. The Leeuwenhoek lecture, 1987. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc. R. Soc. Lond. B 235:121-138. [DOI] [PubMed] [Google Scholar]
- 14.Jordan, A., I. Gibert, and J. Barbe. 1995. Two different operons for the same function: comparison of the Salmonella typhimurium nrdAB and nrdEF genes. Gene 167:75-79. [DOI] [PubMed] [Google Scholar]
- 15.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Masalha, M., I. Borovok, R. Schreiber, Y. Aharonowitz, and G. Cohen. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J. Bacteriol. 183:7260-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681-706. [DOI] [PubMed] [Google Scholar]
- 20.Reichard, P. 1993. From RNA to DNA, why so many ribonucleotide reductases? Science 260:1773-1777. [DOI] [PubMed] [Google Scholar]
- 21.Rodionov, D. A., and M. S. Gelfand. 2005. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Shewach, D. S. 1992. Quantitation of deoxyribonucleoside 5′-triphosphates by a sequential boronate and anion-exchange high-pressure liquid chromatographic procedure. Anal. Biochem. 206:178-182. [DOI] [PubMed] [Google Scholar]
- 24.Torrents, E., P. Aloy, I. Gibert, and F. Rodriguez-Trelles. 2002. Ribonucleotide reductases: divergent evolution of an ancient enzyme. J. Mol. Evol. 55:138-152. [DOI] [PubMed] [Google Scholar]
- 25.Uhlin, U., and H. Eklund. 1994. Structure of ribonucleotide reductase protein R1. Nature 370:533-539. [DOI] [PubMed] [Google Scholar]