Abstract
Bacterial biofilms at times undergo regulated and coordinated dispersal events where sessile biofilm cells convert to free-swimming, planktonic bacteria. In the opportunistic pathogen Pseudomonas aeruginosa, we previously observed that dispersal occurs concurrently with three interrelated processes within mature biofilms: (i) production of oxidative or nitrosative stress-inducing molecules inside biofilm structures, (ii) bacteriophage induction, and (iii) cell lysis. Here we examine whether specific reactive oxygen or nitrogen intermediates play a role in cell dispersal from P. aeruginosa biofilms. We demonstrate the involvement of anaerobic respiration processes in P. aeruginosa biofilm dispersal and show that nitric oxide (NO), used widely as a signaling molecule in biological systems, causes dispersal of P. aeruginosa biofilm bacteria. Dispersal was induced with low, sublethal concentrations (25 to 500 nM) of the NO donor sodium nitroprusside (SNP). Moreover, a P. aeruginosa mutant lacking the only enzyme capable of generating metabolic NO through anaerobic respiration (nitrite reductase, ΔnirS) did not disperse, whereas a NO reductase mutant (ΔnorCB) exhibited greatly enhanced dispersal. Strategies to induce biofilm dispersal are of interest due to their potential to prevent biofilms and biofilm-related infections. We observed that exposure to SNP (500 nM) greatly enhanced the efficacy of antimicrobial compounds (tobramycin, hydrogen peroxide, and sodium dodecyl sulfate) in the removal of established P. aeruginosa biofilms from a glass surface. Combined exposure to both NO and antimicrobial agents may therefore offer a novel strategy to control preestablished, persistent P. aeruginosa biofilms and biofilm-related infections.
The biofilm mode of growth encompasses surface-associated microbial communities and the extracellular matrix in which they are embedded. Biofilms, which are ubiquitous in aqueous environments, are extremely problematic in industrial settings (9, 46), for example, by acting as reservoirs for pathogens in drinking water systems (47). Biofilms are also associated with many chronic infections in humans. For example, the opportunistic pathogen Pseudomonas aeruginosa causes persistent infections in the lungs of cystic fibrosis patients that are frequently associated with the emergence of antibiotic-resistant subpopulations of bacteria (57).
Using cooperative traits such as cell-cell signaling (quorum sensing), bacteria in biofilms often develop three-dimensional structures known as microcolonies, in which cells become highly differentiated from free-living, planktonic bacteria (65). Microcolonies are generally highly tolerant to standard antimicrobial agents, and previous studies have shown that bacteria embedded within such structures can be 1,000-fold more resistant to antimicrobials than are planktonic cells (7).
Bacteria within biofilms often undergo coordinated dispersal events in which attached biofilm cells convert to free-swimming planktonic bacteria. Dispersal and sloughing events observed during biofilm development are generally thought to benefit bacteria by allowing single organisms to return to the liquid phase and colonize new habitats (54). Strategies to induce biofilm dispersal would have broad applications in industrial, environmental, and medical settings. Several bacterial regulatory systems (e.g., quorum sensing [49]) and active dispersal mechanisms (e.g., expression of matrix-degrading enzymes or surfactants [5, 13]) have been linked to the transition of sessile biofilm organisms to free-swimming bacteria. Changes in nutrient availability have also been linked to biofilm dispersal processes (23, 29, 54). In one commonly observed process of dispersal in P. aeruginosa, organisms evacuate the interior of the microcolonies, leaving behind hollow, shell-like structures (29, 48, 53). The mechanisms underlying these events are poorly understood but have been shown to involve quorum sensing (48) and complex processes of differentiation, including the lysis of a subpopulation of cells within microcolonies (37, 66).
In P. aeruginosa, cell lysis and dispersal were recently linked to both the activation of a prophage and the generation of oxidative or nitrosative stress inside microcolonies (66). Oxidative stress results from either endogenous production of or exogenous exposure to reactive oxygen intermediates (ROI), which include superoxide (O2−), hydrogen peroxide (H2O2), and the extremely reactive hydroxyl radical (HO·). In contrast, another form of stress is termed nitrosative stress. The latter involves production of and ensuing damage from reactive nitrogen intermediates (RNI), which include nitric oxide (NO), peroxynitrite (ONOO−), nitrous acid (HNO2), nitrogen trioxide (N2O3), and others. RNI are small, potentially highly reactive molecules that can be produced continuously in the organisms as by-products of anaerobic respiratory metabolism (24). When the production of ROI and/or RNI overwhelms the capacity of the cell to remove such molecules, damage to DNA, lipids, and proteins may occur. Specifically, Yoon et al. (68) have shown that organisms housed in anaerobic biofilms and lacking the rhl quorum-sensing circuit commit a metabolic suicide via NO intoxication. However, in addition to their damaging properties, ROI and RNI are involved in many signaling and regulatory pathways in both eukaryotic and prokaryotic organisms (43, 62). For example, numerous studies of Escherichia coli have shown that RNI activate global regulatory networks such as the SOS response (36) or the genetic response to oxidative stress controlled by SoxRS and OxyR (16). In particular, NO has been found to have many physiological signaling roles in eukaryotic biology and multicellular organisms, such as within the processes of apoptosis, differentiation, and cell proliferation (40). While the roles of ROI and RNI have been studied extensively in planktonic bacterial physiology in the context of protective mechanisms, there is limited information as to their role in multicellular biofilm development and differentiation processes. It has been suggested that quorum sensing is required for optimal resistance of biofilm bacteria to H2O2 (25) and that H2O2 can trigger mutations of the mucA gene, encoding an anti-sigma factor, leading to mucoid conversion in biofilms (39).
In this study, we examined the role of specific ROI and RNI in triggering differentiation and dispersal in P. aeruginosa biofilms. We showed that anaerobic metabolism can occur inside P. aeruginosa biofilms grown under aerobic conditions and that ONOO− levels are enhanced in mature microcolonies harboring cells that had (i) perished, (ii) differentiated, and/or (iii) dispersed. By exposing P. aeruginosa biofilms to RNI, we also found that NO, the main precursor of ONOO− in vivo (2), is able to induce biofilm dispersal at concentrations that are nontoxic to P. aeruginosa (in the nanomolar range). Furthermore, bacteria that were exposed to low levels of NO were removed from surfaces using combined antimicrobial treatments more effectively than were control biofilms. Our findings suggest a novel application for NO in the control of persistent P. aeruginosa biofilms.
MATERIALS AND METHODS
Bacterial strains and culture media.
P. aeruginosa strains PAO1 and PAO1-GFP, containing a chromosomal mini-Tn7 insertion of the enhanced green fluorescent protein (GFP) gene, were generously provided by Tim Tolker-Nielsen. P. aeruginosa PAO1 transposon insertion mutants ΔnirS and ΔnorCB were obtained from the University of Washington library (strain identification numbers 6761 or 11788 and 4583 or 13703, respectively) (31). We also used other P. aeruginosa strains that had been insertionally inactivated with a gentamicin resistance cassette in the same genes (68). A P. aeruginosa PAO1 nirS::gfp transcriptional reporter strain was constructed by inserting a gfp reporter gene under the regulation of the nirS promoter (details described below). Overnight cultures were grown routinely in Luria-Bertani (LB) medium with shaking at 37°C. Motility assays were performed on LB 0.3% (swimming) and 0.5% (swarming) agar plates as previously described (56). Agar media were supplemented with various concentrations of NO generating and scavenging solutions just prior to pouring the plates on the same day of the experiment. The distance that the cells migrated was determined after 12 to 16 h of incubation at 37°C. Biofilms were grown in modified M9 minimal medium (pH 7.0) (66) containing 48 mM Na2HPO4, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 2 mM MgSO4, 100 μM CaCl2, and glucose at 5 mM for continuous-culture flow cell experiments and at 20 mM for batch microtiter plate experiments. According to the manufacturer's specifications, the medium components used to make M9 medium can contain trace amounts of NO3− as a contaminant. By using Griess reaction nitrite and Nitralizer II kits (World Precision Instruments), we found that M9 medium contains 4.5 ± 1.3 μM NO3−.
Construction of the nirS reporter strain.
For DNA manipulation, cloning, and reporter strain construction, routine protocols for plasmid and chromosomal DNA purifications were obtained from Sambrook et al. (52). Enzymes were purchased from either Promega (Madison, Wis.) or New England Biolabs (Beverly, Mass.). Oligonucleotides were designed with OLIGO software and purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). To construct the nirS reporter strain, the mini-CTX system (27) was employed to introduce a site-directed, single-copy nirS::gfp transcriptional fusion into the P. aeruginosa PAO1 genome as described previously (18). The nirS promoter was from −226 to +132 with respect to the ATG codon of the nirS gene. This corresponds to bp 580326 to 579968 in the P. aeruginosa genome sequence (61). Briefly, the mini-CTX suicide vector carrying the reporter gene construct integrates into the P. aeruginosa genome at the attB site via an integrase (int)-mediated recombination. Tetracycline-resistant PAO1 colonies were selected, and a second plasmid, pFLP2, was introduced. The pFLP2 construct is a broad-host-range vector that expresses Saccharomyces cerevisiae Flp recombinase leading to excision of DNA sequences included within Flp recombinase target sites. The integrated mini-CTX vector contains two Flp recombinase target sites that flank the Tcr, int, and oriT markers. Therefore, introduction of the pFLP2 construct eliminates these markers from the integrate. After the pFLP2 plasmid is cured from the reporter strain, only the inserted gene fusion, which is also flanked by T4-derived transcriptional terminators, remains. PCR primers were designed to verify integration of the nirS::gfp fusion and excision of the unwanted vector sequences. To verify integrate junctions, PCR products were sequenced with an ABI Prism BigDye kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and an ABI model 310 genetic analyzer (Perkin-Elmer Applied Biosystems).
Chemicals. (i) Detection of ROI and RNI.
We used a series of reactive fluorescent dyes for the detection of specific ROI and RNI that were produced in P. aeruginosa biofilms. The dyes were 4-amino-5-methylamino-2′,7′difluorescein diacetate (DAFFM-DA) (Molecular Probes), 5 mM stock in dimethyl sulfoxide (DMSO), used at 100 μM for the detection of NO (33); dihydrorhodamine 123 (DHR) (Sigma), stock 2.5 mg ml−1 in ethanol, used at 15 μM for the detection of peroxynitrite (ONOO−) (10); 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA) (Molecular Probes), 10 mM stock in DMSO, used at 100 μM for the detection of hydrogen peroxide (H2O2); and hydroethidine (HEt) (Sigma), 1 mg ml−1 stock in 1% DMSO in phosphate-buffered saline (PBS), used at 10 μM for the detection of superoxide radicals (O2−) (4). Stock solutions were kept frozen and covered from light. Final solutions were freshly made in M9 medium before use.
(ii) Nitric oxide generators and scavenger.
Three NO donors were used: sodium nitroprusside (SNP) (Sigma), S-nitroso-l-glutathione (GSNO) (MP Biomedicals), and S-nitroso-N-acetylpenicillamine (SNAP) (Sigma). The NO scavenger 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (PTIO) (Sigma) was also used. Experiments were carried out using freshly made solutions in biofilm medium. Solutions were protected from light and kept no longer than 1 day.
(iii) Antimicrobial compounds.
The aminoglycoside antibiotic tobramycin (Sigma) was used at a final concentration of 100 μM (7), the surfactant sodium dodecyl sulfate (SDS) at 0.05%, and H2O2 at 10 mM. The concentrations of antimicrobials selected for use were those previously established in our laboratory and elsewhere to be effective against planktonic cells but not against P. aeruginosa biofilm cells.
Biofilm experiments. Flow cell (continuous-culture) biofilm experiments.
P. aeruginosa PAO1 was grown in continuous-culture flow cells (channel dimensions, 1 by 4 by 40 mm) at room temperature as previously described (37). Standard silicone tubing was replaced with autoclaved glass tubing (∼10 cm in length, 2.5 mm in diameter) for the collection of flow cell effluent runoff. To investigate cell death during biofilm development, biofilms were stained with a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes). The two stock solutions of the stain (SYTO 9 and propidium iodide) were diluted to 3 μl ml−1 in M9 and injected into the flow channels. Live SYTO 9-stained cells and dead propidium iodide-stained cells were visualized with a confocal laser scanning microscope (Olympus LSM-GB200 confocal BH2) with an argon laser (488 nm) and a helium laser (543 nm), respectively. To detect specific ROI and RNI that are produced in biofilm structures during death and dispersal, the reactive dyes (see “Detection of ROI and RNI” above) were injected into the flow channels and were incubated for 30 min in the dark before confocal laser scanning microscopy was carried out; the argon laser (green fluorescence) was used to visualize DAFFM-stained cells and the helium laser (red fluorescence) to visualize DHR-, H2DCF-, and HEt-stained cells. To assess the metabolic activity of bacteria in mature biofilms, 6-day-old biofilms were stained with 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) at a final concentration of 4 mM (50) and incubated for 30 min in the dark before the cells were visualized using confocal laser scanning microscopy with the helium laser. GFP levels in P. aeruginosa PAO1 nirS::gfp reporter strain biofilms were viewed without further processing by using an epifluorescence microscope (Leica model DMR) and a fluorescein isothiocyanate optical filter. Control experiments were performed by growing P. aeruginosa planktonic cells aerobically in 25 ml of M9 medium. After growth, bacteria were resuspended in 500 μl of M9 and observed using fluorescence microscopy. Nitrite (NO2−) levels in biofilm runoff effluents were detected using Spectroquant nitrite test colorimetric assays (Merck KGaA). Determination of bacteriophage counts in the fluid effluent of the biofilm was performed as previously described (66).
Ninety-six-well plate (batch culture) biofilm experiments.
Aliquots of 100 μl of overnight cultures of PAO1-GFP diluted 1,000 times in M9 were inoculated in 96-well plates (Sarstedt). SNP treatments were added to the wells to final concentrations in the range of 25 nM to 100 mM. Four replicate wells per treatment were used. The plates were incubated for 24 h at 37°C with shaking at 120 rpm. After growth, the planktonic phase was transferred to a new plate for quantifying planktonic growth by fluorescence using a microtiter plate fluorometer (Wallac Victor2) (excitation, 485 nm; emission, 535 nm). For quantification of biofilms in microtiter plates, we used a method similar to that of O'Toole and Kolter (45). Wells containing biofilms were washed twice with PBS and stained for 20 min with 120 μl of crystal violet. The wells were washed again three times with PBS, and the remaining crystal violet was dissolved in 120 μl of absolute ethanol. Biofilm formation was quantified by measurement of the optical density at 540 nm (OD540). The fold increase/decrease in biofilm biomass relative to planktonic biomass was calculated for each concentration of SNP (the ratio of biofilm/planktonic biomass for controls in the absence of SNP was 6.4 × 10−4 [OD540/GFP fluorescence]).
Petri dish (batch culture) biofilm experiments.
We also grew biofilms in petri dishes (90 mm in diameter) containing microscope glass slides (76 by 26 mm). Twenty-five ml of overnight cultures of PAO1-GFP cells diluted 1,000 times in M9 medium was inoculated into the plates and incubated at 37°C with shaking at 50 rpm, allowing biofilm formation on the slides. Biofilms were grown in the presence or absence of the following NO treatments: 500 nM SNP (with or without 1 mM PTIO, a nitric oxide scavenger), 1 μM GSNO, or 1 μM SNAP. After 24 h of growth, the number of planktonic cells was evaluated by measuring fluorescence levels of the planktonic phase, as described above, as well as OD600. Slides were rinsed gently in sterile PBS. The biofilm on the slides was then stained with 250 μl of the LIVE/DEAD BacLight stain for 20 min in a humidified chamber. Seven confocal images per slide were obtained from random locations on the glass surface, and the percentage of the glass surface covered with biofilm was quantified using image analysis (ImageJ software).
For antimicrobial sensitivity assays, all biofilms were first allowed to develop for 24 h in the absence of NO treatment. After 24 h, the planktonic phase was replaced with fresh media with or without 500 nM SNP and the system was then incubated for an additional 24 h. After this time, the slides were rinsed gently in sterile PBS and then incubated with 500 μl of an antimicrobial treatment (H2O2, tobramycin, or SDS) in M9 or with 500 μl of M9 alone as a control for 30 min in a humidified chamber. Slides were then rinsed again with PBS. Biofilm staining, microscopy analysis, and determination of percentage of surface coverage were carried out as described above. Statistical comparison of the percentages of surface covered by biofilms in the different treatments was performed using analysis of variance.
Antimicrobial susceptibility of planktonic cells.
Planktonic cells in the supernatant of the petri dish systems were also tested for antimicrobial sensitivity. After 24 h with or without 500 nM SNP, three aliquots of the petri dish supernatants were diluted 10 times into solutions containing individual antimicrobial compounds and incubated for 2 h at room temperature. CFU were enumerated after plating on LB agar to assess bacterial viability.
RESULTS
Cell death and dispersal in P. aeruginosa biofilms correlate with increased levels of ONOO− in mature microcolonies.
Mature P. aeruginosa biofilms are known to undergo patterns of both cell death and dispersal (Fig. 1A), and these events were previously linked to the accumulation of oxidative or nitrosative stress inside mature microcolonies (66). To investigate the role of specific ROI and RNI in microcolony dispersal, we stained 7-day-old biofilms of P. aeruginosa with fluorescent reporter dyes and examined mature biofilms by using confocal microscopy. As shown in Fig. 1B, we observed fluorescence with two of the dyes assessed: HEt (detects superoxide radicals [O2−]) and, at considerably higher levels of fluorescence, DHR (detects peroxynitrite [ONOO−]) (n = 3). We observed the strongest fluorescence in all mature microcolonies (four to nine per channel) that had undergone death and dispersal events, similar to those shown in Fig. 1A, but not in microcolonies that had not undergone these events. The negative results obtained with H2DCF, used for the detection of hydrogen peroxide (H2O2), are consistent with the previously reported high levels of catalase A (KatA), a scavenger of H2O2, in P. aeruginosa biofilms (18, 60). Because ONOO− is produced mainly from the direct reaction of O2− with NO in vivo (2), it was surprising not to detect NO with DAFFM (data not shown). However, the addition of a NO donor (SNP) into the biofilm medium also did not generate detectable fluorescence with DAFFM (data not shown), suggesting that the dye was unable to detect NO in P. aeruginosa biofilms. Overall, our observations suggest that, in P. aeruginosa biofilms, mature microcolonies undergo nitrosative stress and that RNI, to a greater extent than ROI, play a role in cell death and dispersal processes. Therefore, we investigated in more detail the role of RNI in P. aeruginosa biofilm dispersal.
FIG. 1.
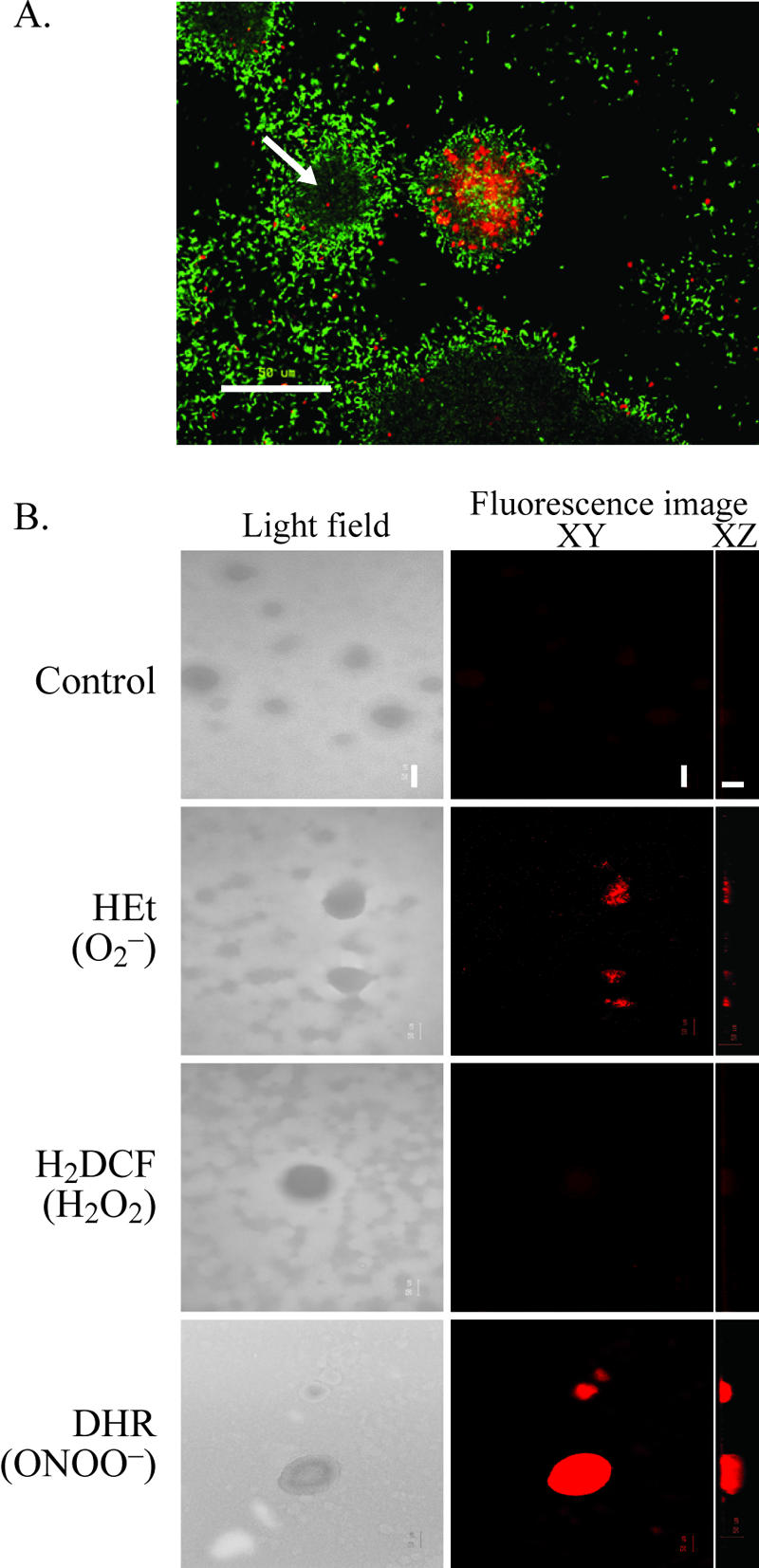
Cell death and dispersal events in P. aeruginosa biofilms correlate with enhanced levels of ONOO− inside microcolony structures. (A) Cell death inside biofilm structures occurs simultaneously with biofilm dispersal, as indicated by the formation of hollow biofilm structures (arrow). The image shows a 7-day-old biofilm stained with LIVE/DEAD BacLight stain. Live cells are green, and dead cells are red. (B) Analysis of ROI and RNI in 7-day-old biofilms by use of reactive fluorescent dyes. Images were taken using a confocal microscope; the left panel shows light-field images, and the right panel shows laser scanning fluorescence images showing XY (top) and XZ (side) views. Bacteria in the biofilm showed a low level of autofluorescence, as revealed by the control treatments. Bar, 50 μm.
Anaerobic respiratory metabolism in aerobic P. aeruginosa biofilms.
RNI production in P. aeruginosa biofilms implies anaerobic respiratory metabolism. This process uses nitrate (NO3−, i.e., denitrification), nitrite (NO2−), or nitrous oxide (N2O) as the terminal electron acceptor (24). In our experiments, biofilms were grown under aerobic conditions. However, steep gradients in oxygen availability can occur within aerobically grown biofilms (15), and biofilms often exhibit gene expression profiles consistent with an anaerobic mode of growth (26, 54, 68). To investigate whether anaerobic denitrification may occur in our biofilm system, we examined the effluent runoff of biofilms and detected NO2− levels of 0.4 ± 0.1 μM in 6-day-old P. aeruginosa biofilm effluent but not in the effluent of young, 2-day-old biofilms or in the control medium (below the detection limit). We further constructed a P. aeruginosa reporter strain, which expressed GFP when the anaerobic gene nirS (encoding a NO2− reductase) is expressed. We observed bright GFP fluorescence in microcolonies of mature 5-day-old biofilms but not in 1-day-old biofilms or aerobically grown planktonic P. aeruginosa (Fig. 2). These data, therefore, provide evidence for denitrification in our aerobically grown P. aeruginosa biofilms and suggest that NO2− and NO are produced within microcolonies from the NO3− available in the M9 biofilm medium.
FIG. 2.
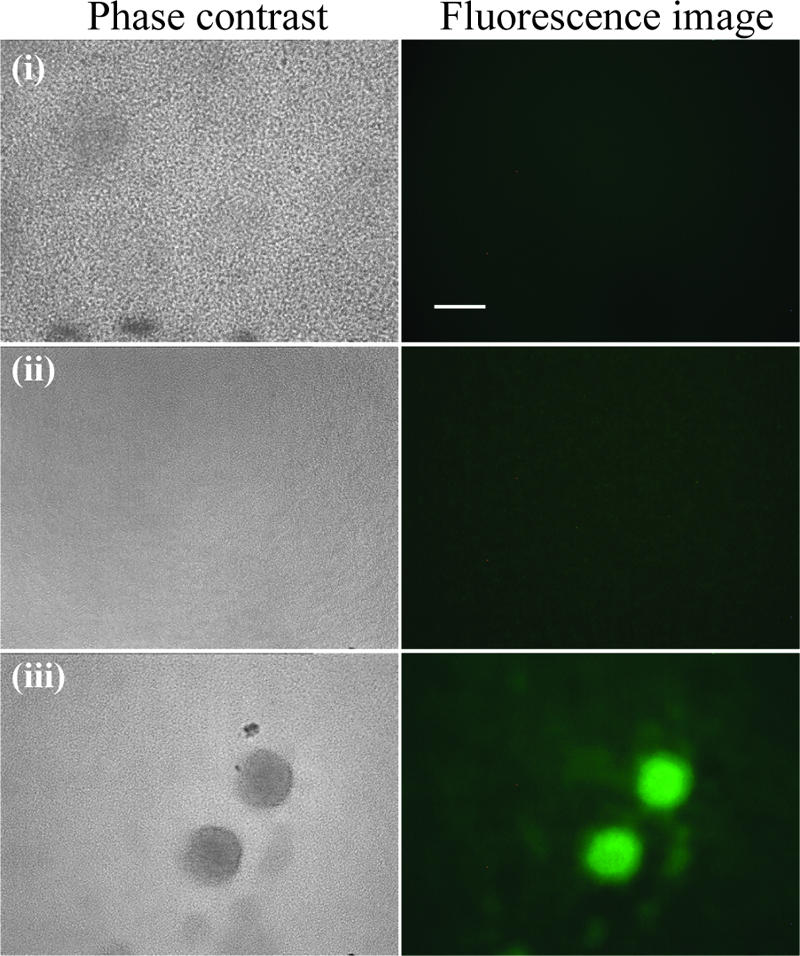
Expression of nirS in P. aeruginosa biofilms. Expression of GFP under the control of the nirS (NO2− reductase) promoter was viewed using epifluorescence microscopy; the left panel shows phase-contrast images, and the right panel shows green fluorescence (GFP) images. (i) Aerobically grown planktonic cells (control), (ii) 1-day-old biofilm showing confluent layer of cells on the glass substratum, and (iii) mature biofilms harboring microcolonies are shown. Bar, 50 μm.
Endogenously produced NO or downstream reactive species mediate cell death and dispersal events in P. aeruginosa biofilms.
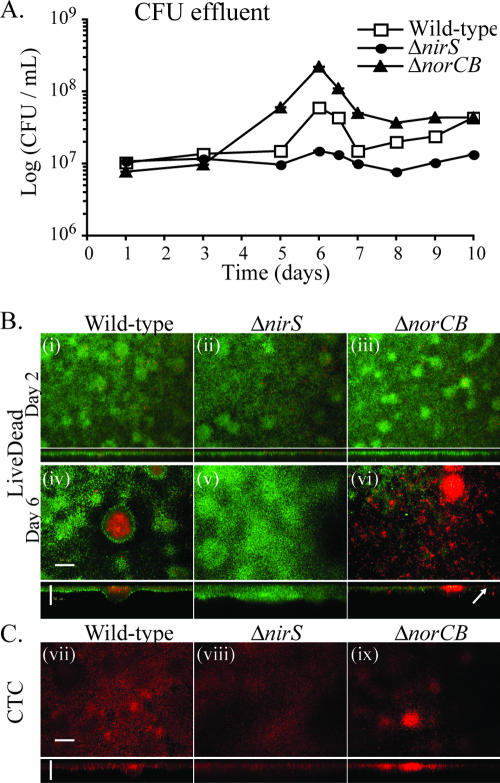
Because NO is the main precursor of ONOO− and is highly diffusible and broadly used as a signal molecule in biological systems (3), we examined whether this molecule may play a role in P. aeruginosa biofilm dispersal. We compared the levels of biofilm development of P. aeruginosa wild-type, NO2− reductase-deficient (ΔnirS, unable to produce metabolic NO), and NO reductase-deficient (ΔnorCB) mutant strains in glass flow cells (Fig. 3) (n = 3). After 2 days of growth, all three strains exhibited normal biofilm development with no major dispersal events. However, as the biofilms matured, significant differences in dispersal behavior were observed for the ΔnirS and ΔnorCB mutants compared to the wild-type strain. The wild-type biofilms displayed increased numbers of viable cells in their effluent runoff at the same time as the onset of cell death in biofilm microcolonies (Fig. 3A and B). The ΔnirS mutant biofilms did not show enhanced dispersal after 6 days (Fig. 3A) and were thicker and confluent over the entire surface, and we did not observe patterns of cell death or lysis in this strain (Fig. 3B). In contrast, the ΔnorCB mutant biofilms contained higher numbers of viable dispersed cells in their effluent (Fig. 3A), displayed numerous hollow voids within biofilm structures, and exhibited enhanced patterns of cell death (Fig. 3B). Cell death and lysis in mature P. aeruginosa biofilms were previously linked to the appearance of infectious bacteriophage in the biofilm runoff (66). In this study, we observed more bacteriophage (typically 1 log higher, 108 PFU/ml) in the spent medium effluent from mature 6-day-old biofilms of the P. aeruginosa ΔnorCB strain than in that from the wild type (107 PFU/ml) and fewer in that from the ΔnirS strain (typically 1 log lower, 106 PFU/ml). Possibly, cells that survive bacteriophage lysis benefit from the nutrients released from their dead siblings during dispersal from the biofilm matrix (38).
FIG. 3.
Biofilm development and dispersal of P. aeruginosa wild-type, nitrite reductase-deficient mutant (ΔnirS, unable to produce NO), and NO reductase-deficient mutant (ΔnorCB) strains. (A) Viable cells in the effluent runoff of the wild-type, ΔnirS mutant, and ΔnorCB mutant strain biofilms were enumerated by performing CFU counts. (B) Biofilms were grown in flow cells and stained with LIVE/DEAD staining. Live cells are green, and dead cells are red. Images were collected using a confocal microscope; the upper panel depicts XY (top) views, and the lower panel reveals the XZ (side) axis. After 2 days, all strains show normal biofilm development (i, ii, and iii). After 6 days, the ΔnirS mutant (v) shows a thick biofilm without major dispersal events or cell death, and the ΔnorCB mutant (vi) exhibits extensive detachment and dispersal from the glass surface and numerous hollow voids within the biofilm structure (arrow). Cell death was also greatly enhanced in the ΔnorCB mutant (vi) compared to the wild type (iv). (C) Six-day-old biofilms were stained with the metabolic dye CTC and observed using confocal microscopy. The wild-type strain (vii) and, to a greater extent, the ΔnorCB mutant strain (ix) exhibit high levels of fluorescence in mature microcolonies, whereas ΔnirS mutant biofilm cells (viii) show uniform fluorescence. These images suggest that a subpopulation of cells continue to be active after cell lysis in the microcolonies. Bar, 50 μm.
To further investigate the role of NO in differentiation and dispersal events in P. aeruginosa biofilms, wild-type and ΔnirS and ΔnorCB mutant strain biofilms were stained with the metabolic dye CTC in flow cell experiments. We observed increased fluorescence in localized regions inside mature microcolonies in the wild-type strain and, to a greater extent, in the ΔnorCB mutant strain at the same time and location as the appearance of cell death (Fig. 3C). This observation was not the result of nitrosylation of CTC by NO, as the NO donor SNP failed to increase fluorescence levels of CTC in solution (data not shown). In contrast, the ΔnirS mutant biofilm exhibited uniform fluorescence levels (Fig. 3C). These results suggest that a subpopulation of bacteria continue to be active after others lyse in microcolonies within P. aeruginosa biofilms.
NO modulates the ratio of biofilm to planktonic biomass.
We tested a range of concentrations of the NO donor SNP for effects on both planktonic and biofilm biomass in P. aeruginosa biofilm cultures. The use of such a compound has the advantage of established steady-state levels of NO, which mimics endogenous NO production during bacterial denitrification (34). At low concentrations, in the micromolar and nanomolar ranges, we observed a decrease in biofilm biomass and an increase in planktonic biomass. The greatest effect was observed repeatedly with 500 nM SNP, with a 10-fold decrease in the ratio of biofilm to planktonic cells (Fig. 4A). Therefore, we used this concentration for further experiments. In contrast, at high (millimolar) concentrations, SNP caused an increase in biofilm biomass and a decrease in planktonic biomass (Fig. 4A). For example, the biofilm biomass in the presence of 75 mM SNP was fourfold higher than that of control biofilms, resulting in a sixfold-higher ratio of biofilm to planktonic cells. Possible explanations for this increase in biofilm biomass may include a rapid conversion of NO to NO2− and NO3− at a slightly acidic pH (67), which could lead to enhanced anaerobic metabolism and biomass production within the biofilm. The observed increase in biofilm biomass may also result from an adaptive response of P. aeruginosa cells to the much higher and possibly toxic levels of NO or its downstream products. It has been shown previously that the presence of antibiotic compounds can enhance biomass production in biofilms (28).
FIG. 4.
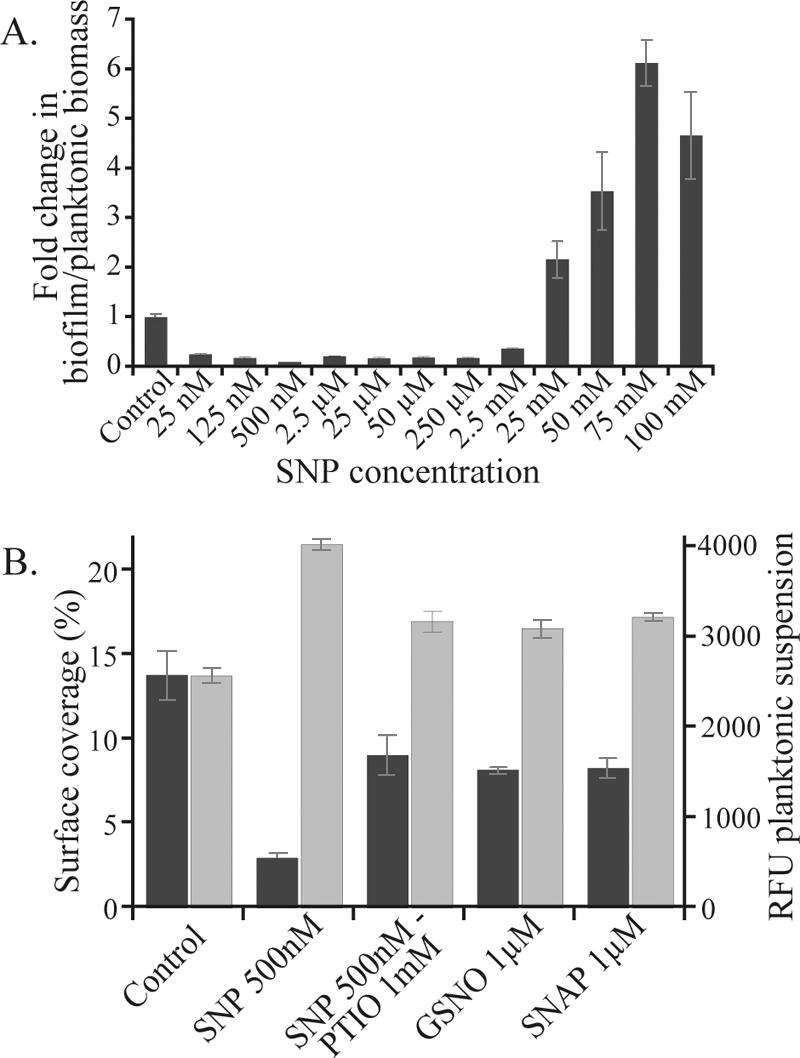
NO mediates a transition from biofilm to planktonic mode of growth in P. aeruginosa. (A) PAO1-GFP grown for 24 h in 96-well plates in the presence of SNP. The number of planktonic cells is quantified by fluorescence measurement and biofilm biomass by crystal violet staining. (B) PAO1-GFP grown for 24 h in petri dishes containing microscope slides in the presence of different NO donors (SNP, GSNO, and SNAP) and the scavenger PTIO. Planktonic growth (light-gray bars) was assessed by measuring RFU of the supernatant, and biofilm growth (dark-gray bars) was assessed by measuring the percentage of surface coverage. Error bars indicate standard deviations.
Other NO donors also prevent initial biofilm formation.
We investigated the effect of SNP and other NO donors in a second laboratory model biofilm system that allowed for microscopic observations of the biofilms. Biofilms were allowed to form on glass slides in petri dishes for 24 h in the presence or absence of low concentrations of NO donors. SNP treatment (500 nM) resulted in a fivefold decrease in the percentage of surface coverage of the glass surface with the biofilm and a significant increase in the number of planktonic cells (relative fluorescence units [RFU] = 4,030 ± 58, OD600 = 0.491 ± 0.011) compared to results for untreated controls (RFU = 2,581 ± 82, OD600 = 0.383 ± 0.010, P ≤ 0.05) (Fig. 4B). We also tested the NO donors GSNO and SNAP, as well as the NO scavenger PTIO, in the same experimental system. With added PTIO, the SNP effect was reduced by 40% or more in both planktonic and biofilm phenotypes. Both additional NO donors, although less effective than SNP, gave rise to a reduction in biofilm formation by 40% in the surface coverage, and SNAP also significantly increased the number of planktonic cells (RFU = 3,230 ± 43, OD600 = 0.422 ± 0.004, P ≤ 0.05) compared to the control (Fig. 4B).
Low concentrations of NO induce biofilm dispersal.
We also investigated the effect of exposure to low doses of NO on preestablished biofilms in petri dish cultures. While untreated biofilms exhibited normal development and microcolony formation in this system, exposure of 1-day-old biofilms to 500 nM SNP for 24 h reversed biofilm formation and caused detachment and dispersal of cells from the surface. Only a few cells remained attached to the slide, resulting in an 80% reduction in biofilm surface coverage (see Fig. 6). We again observed an increase in the number of planktonic cells after addition of SNP, strongly suggesting that decreases in biofilm biomass were caused by dispersal events. Moreover, in flow cell experiments, in which the volume of the flow chamber and the flow rate are known, calculations show that the increases in cells in the effluent after addition of SNP (data not shown) can derive only from a dispersal event rather than from growth of planktonic cells. These effects were observed using sublethal concentrations of NO. Although we have not yet elucidated the exact amount and location of NO liberated in vivo within biofilms from SNP, by using a NO analyzer (Apollo 4000 with ISO-NOP electrode; World Precision Instruments) and a solution of 1 mM SNP in M9 medium, we measured NO concentrations in the range of 100 nM to 1 μM in vitro (data not shown). This suggests that the effective concentration of NO delivered to the cells may be over 1,000 times lower than the concentration of SNP used herein. At 500 nM SNP, treatment did not cause any reduction in CFU in P. aeruginosa planktonic cultures compared to CFU in untreated cells (see Fig. 7). We also observed that biofilm bacteria remaining on the surface after SNP treatment were 100% viable when examined by using the LIVE/DEAD staining kit (data not shown).
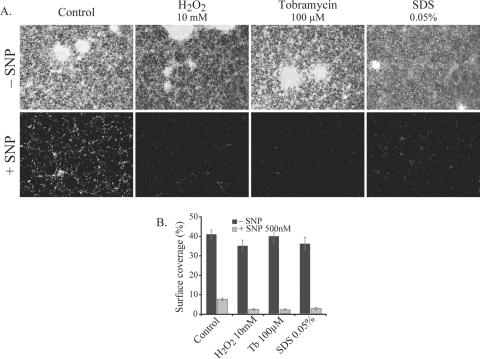
FIG. 6.
NO treatment reverses biofilm formation in P. aeruginosa. Cells remaining on the surface are easily removed by various antimicrobials (tobramycin [Tb], H2O2, and SDS). P. aeruginosa PAO1 was grown in petri dishes containing glass microscope slides. Preestablished biofilms that were grown for 24 h without SNP were allowed to grow for an additional 24 h with (+) or without (−) 500 nM SNP; then, the biofilms on the slides were treated for 30 min with the antimicrobial agents, stained with LIVE/DEAD staining to allow analysis using fluorescence microscopy, and quantified (percent surface coverage) using digital image analysis. (A) The images show microscopic pictures of the biofilms on the glass slides after the combinatorial treatments. (B) The bars show the levels of biofilm biomass after antimicrobial treatment when grown without or with 500 nM SNP, and error bars indicate standard errors.
FIG. 7.
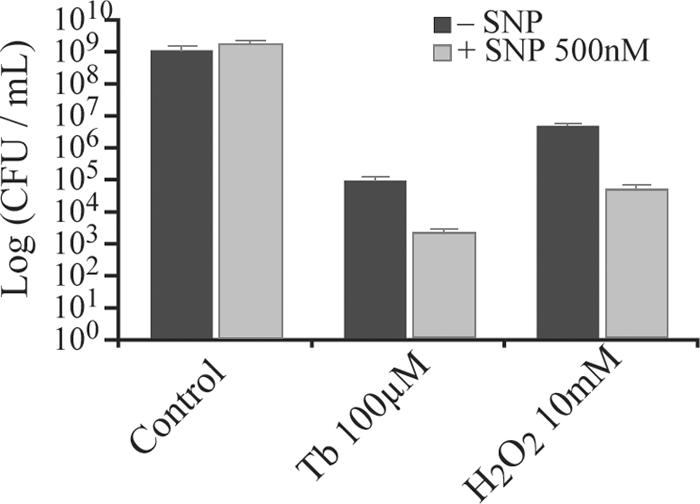
NO treatment increases the sensitivity of dispersed P. aeruginosa planktonic cells to antimicrobials (tobramycin [Tb] or H2O2). After 24 h of growth in petri dishes without (−) or with (+) 500 nM SNP, planktonic cells in the supernatant were treated for 2 h with the antimicrobial solutions; then, CFU plate counts were performed to assess the viability of the bacteria. Error bars indicate standard errors.
Low concentrations of NO enhance swimming and swarming motilities in P. aeruginosa.
Previous studies have suggested that dispersal of single organisms from biofilms is linked to the motility of a subpopulation of cells (32, 48, 53). Because low doses of NO seem to promote dispersal of sessile cells, we investigated the effect of NO donors on the motility behavior of P. aeruginosa cells. The addition of 500 nM SNP in the agar resulted in a 25% increase in swimming motility and a 77% increase in swarming motility compared to results for the controls (P ≤ 0.005), and in the presence of 1 μM GSNO, swimming was enhanced by 39% and swarming by 116% compared to results for the controls (P ≤ 0.005) (Fig. 5). These effects were abolished at higher concentrations of SNP (12.5 μM and above) or GSNO (20 μM and above) or in the presence of the NO scavenger PTIO.
FIG. 5.
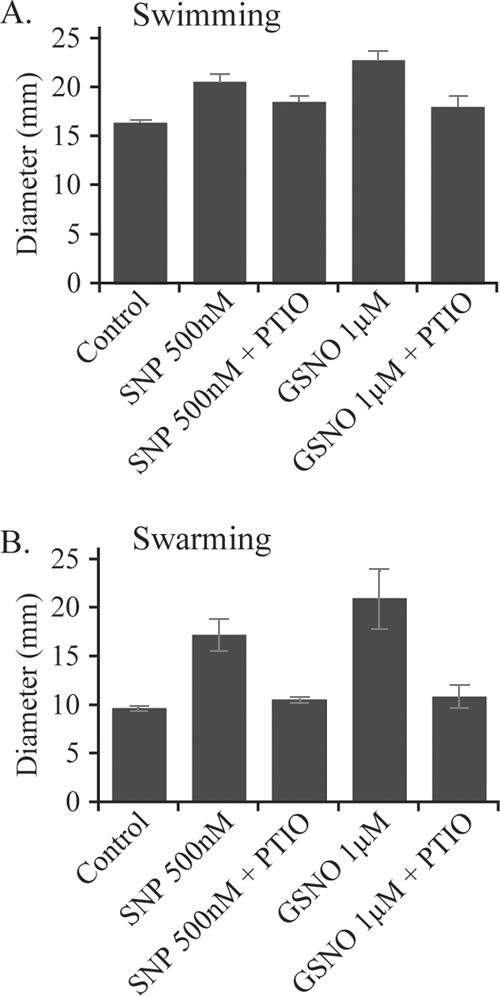
NO effects on motility behavior in P. aeruginosa. Low concentrations of NO donors (500 nM SNP and 1 μM GSNO) and scavengers (1 mM PTIO) were diluted in motility assay agar plates in triplicate. Migration pattern diameters were measured after 12 to 16 h of swimming (A) and swarming (B). Error bars indicate standard errors.
In summary, the findings reported here suggest that NO or a downstream reactive intermediate(s) is able to modulate the ratio of biofilm biomass to planktonic biomass in P. aeruginosa biofilms. Moreover, low concentrations of NO prevented initial biofilm formation and induced dispersal in existing biofilms. This suggests that, at low concentrations, NO induces the transition of sessile biofilm cells to free-swimming planktonic cells.
Combined NO and antimicrobial treatment of P. aeruginosa biofilms.
We tested various antimicrobial agents against P. aeruginosa biofilms and planktonic cells in the presence or absence of the NO donor SNP. When 48-h biofilms were treated in the absence of SNP, none of the antimicrobial compounds exerted a significant effect on the percentage of surface area covered by the biofilm (P ≥ 0.4) (Fig. 6). In contrast, after SNP treatment, the biofilm cells remaining on the surface were easily removed and killed by the addition of the antimicrobial compounds. For example, after NO treatment the addition of tobramycin in fresh medium caused the removal of 80% of the cells remaining on the surface. Combined treatments with both SNP and the antimicrobials caused almost-complete removal of the biofilm in all cases (Fig. 6).
To further investigate the effect of SNP on the sensitivity of P. aeruginosa cells towards antimicrobial agents, we also tested the effect of SNP treatment on planktonic cells. We found that treatment of planktonic cells with 500 nM SNP, which alone did not affect viable CFU counts in P. aeruginosa, dramatically increased the sensitivity of these cells to other antimicrobials. Combined exposure of planktonic cells to both NO and each of the antimicrobials H2O2 and tobramycin caused an additional 2-log decrease in CFU counts compared to exposure to the antimicrobial treatment alone (Fig. 7). Thus, combined treatments using low concentrations of NO together with antimicrobials were highly effective in removing P. aeruginosa biofilms, and our data suggest that NO may increase sensitivity of P. aeruginosa cells to these compounds.
DISCUSSION
NO and P. aeruginosa biofilm development.
In the present study, we observed that NO and/or its downstream derivatives play a role in P. aeruginosa biofilm development and dispersal. Our data suggest that anaerobic respiratory metabolism occurred inside microcolonies within biofilms grown under aerobic conditions. We also demonstrated that endogenous RNI production was intimately correlated with differentiation and dispersal events in situ in P. aeruginosa biofilms. Moreover, we showed that exogenous NO, at sublethal concentrations, was able to induce a transition from biofilm to planktonic phenotype and increase the antimicrobial efficacy against P. aeruginosa.
Because of the impact of biofilms in ubiquitous environments, interest in understanding the physiological changes that occur in biofilm cells during biofilm formation has increased in recent years. P. aeruginosa biofilms predominantly exhibit gene expression profiles that most closely resemble those of bacteria that are in stationary-phase (nutrient-limited), anaerobic, and iron-limited modes of growth (24, 26). Considerable overlap also occurs between genes expressed in the biofilm mode of growth and genes controlled by quorum sensing (26, 55, 63). Interestingly, NO is intimately associated with each of these biofilm-relevant processes. NO is produced as a by-product of the anaerobic reduction of nitrite (NO2−) and is involved in the regulation of anaerobic processes. NO exposure was found to activate genes for anaerobic metabolism in P. aeruginosa, a process which involves the anaerobic regulator ANR (21). P. aeruginosa was reported previously to form more-robust biofilms under anaerobic conditions than under aerobic conditions. In such anaerobic biofilms, if NO is not reduced by NO reductase to N2O, it may accumulate and its toxicity could compromise the viability of the biofilm (68). NO is also closely linked with iron acquisition and metabolism. Microarray studies have revealed a large overlap in genes upregulated upon exposure to NO and those expressed under iron-limiting conditions (21, 44). Furthermore, in diverse bacterial species, NO is known to inhibit DNA binding by the ferric uptake regulator (Fur), leading to upregulation of genes required for iron acquisition (12, 41). This is likely due to the inherent iron-stripping properties of NO, which include its reactivity with virtually all iron- and heme-containing proteins (11). NO levels in P. aeruginosa biofilms are also regulated by quorum sensing. Microarray studies have revealed that both nirS (nitrite reductase) and norCB (NO reductase) are highly expressed in biofilms compared to planktonic cells and that this overexpression is quorum sensing dependent (26, 54, 63). Thus, NO is involved in many processes known to be important during biofilm formation.
Several lines of evidence within the literature support our data that low levels of NO induce a transition from the biofilm mode of growth to the planktonic, free-living form in P. aeruginosa. First, microarray studies have revealed that genes involved in adherence are downregulated in P. aeruginosa upon exposure to NO (21). This suggests that a mechanism exists by which bacteria detach from the biofilm, leading to a reduced biofilm biomass and an increased number of planktonic organisms, as observed in this study. Second, the transition from sessile to motile P. aeruginosa is known to be regulated by GGDEF and EAL protein domains that are involved in the turnover of cyclic di-GMP (c-di-GMP) (56). Several signal transduction pathways are known to regulate the activities of these GGDEF and EAL domains, including sensing of oxygen, pH, temperature, and other environmental stimuli (22, 51, 56). Intriguingly, Iyer and colleagues (30) have also found that NO-sensing proteins, called heme nitric oxide binding (HNOB) domains, are frequently associated with GGDEF and EAL domains in diverse bacteria, suggesting a link between NO sensing and c-di-GMP turnover. Further studies in our laboratory will establish whether NO-mediated biofilm dispersal in P. aeruginosa involves the GGDEF and EAL domains and variation in c-di-GMP levels, and candidate gene products have already been identified for prioritization.
In this study, we observed that exposure to sublethal concentrations of NO can reduce bacterial attachment, cause biofilm dispersal, and enhance motility behavior in P. aeruginosa. We also observed that exposure to low levels of NO significantly increased the efficacy of a range of antimicrobial compounds to both biofilm and planktonic cells. Taken together, these results suggest a more general effect of NO on P. aeruginosa physiology. Sessile biofilm cells display a decreased metabolic activity, and their physiology resembles that of stationary-phase cells (26, 59). Many antimicrobial treatments are known to be more efficient against metabolically active cells; for example, the aminoglycoside tobramycin and β-lactam antibiotics are effective only at killing actively dividing cells (20). Thus, one possibility is that low concentrations of NO induce a transition to a planktonic physiology that is more characteristic of actively growing cells in P. aeruginosa.
We detected ONOO− inside microcolonies in P. aeruginosa biofilms, and ONOO− is formed from NO oxidation only in the presence of reactive oxygen intermediates (e.g., O2−). Therefore, it is not clear how ONOO− is produced inside microcolonies, which are thought to be largely anaerobic (15). However, it is also clear that O2 gradients can occur inside microcolonies, and simultaneous O2 and NO3− respiration has recently been demonstrated for P. aeruginosa populations (8). Thus, it is feasible that ONOO− could be produced inside microcolonies. ONOO− is a potent antimicrobial agent that can induce DNA damage and increased mutagenesis (19). Enhanced levels of this molecule in mature microcolonies, together with an increased sensitivity of cells exposed to endogenously produced NO, may eventually lead to the induction of lytic bacteriophage and cell lysis as previously reported for P. aeruginosa biofilms (66).
Biofilm control.
One practical application of understanding biofilm dispersal processes is in the removal of detrimental biofilms. In this study we used low, nontoxic doses of NO to induce dispersal of biofilm cells, and by further treatment with antimicrobial agents, we demonstrated almost-complete removal of P. aeruginosa biofilms. Although previous studies have demonstrated that NO can reduce initial bacterial attachment to surfaces, the mechanism for reduced attachment was assumed to be that of NO toxicity and of nitrosative stress (21, 42). The present investigation is the first to demonstrate a role for NO in dispersal events from mature biofilms and the first to suggest that NO may be involved in regulated processes of differentiation within multicellular biofilms.
The mechanism by which exposure to NO influences the effect of antimicrobials against biofilms is not yet fully understood. However, these effects were observed using several different classes of antimicrobial compounds, suggesting that a generalized mechanism of tolerance of antimicrobial stress is affected. Several factors have been suggested to contribute to increased resistance of biofilm cells, including reduced metabolic activity and growth rates of the sessile cells (58), limited antibiotic penetration due to the protective structure of the biofilm (9, 14), and phenotypic diversification of biofilm cells (17, 35). Because NO treatment alone (in the absence of antibacterial compounds) induced dispersal and caused significant reduction in biofilm biomass, the ability of antimicrobials to penetrate the remaining biofilm is likely to be increased. However, for some antibiotics (including tobramycin), limited penetration of the biofilm does not appear to be the cause of biofilm tolerance (64). As discussed earlier, at low concentrations, NO may induce a transition to a physiology characteristic of growing cells. Thus, upon NO exposure, both cells remaining attached to the surface and planktonic cells are more sensitive to antimicrobials. Further research in our laboratory will be directed towards a more detailed understanding of this process and towards delivery of relevant concentrations of NO to bacterial biofilms in environmental and medical settings.
Evolutionary perspectives.
Recent analyses of microbial genomes have suggested that homologous NO-sensing receptor domains are common to both prokaryotic and eukaryotic regulatory proteins (1, 30). In eukaryotes, NO signaling is known to play an important role in cyclic GMP turnover and the regulation of diverse processes, including apoptosis, cell proliferation, and differentiation. Intriguing similarities exist between the signaling role of NO in eukaryotes and its control of biofilm cell differentiation, death, and dispersal, as demonstrated in this study. Biofilms are thought to exhibit developmental analogies with multicellular eukaryotes (6, 65), and therefore it may be relevant to examine these bacterial biofilm populations for primitive forms of key regulatory processes observed to occur in more-complex organisms. Our data on NO-mediated control of biofilm development in P. aeruginosa may point to a conserved role for NO signaling in the regulation of differentiation and developmental events across prokaryotic and eukaryotic physiology.
Acknowledgments
We thank our colleagues at the University of New South Wales and the Environmental Biotechnology CRC for their support.
This work was partially supported by grants from the Australian Research Council, the Centre for Marine Biofouling and Bio-innovation, and the Environmental Biotechnology CRC.
REFERENCES
- 1.Aravind, L., V. Anantharaman, and L. M. Iyer. 2003. Evolutionary connections between bacterial and eukaryotic signaling systems: a genomic perspective. Curr. Opin. Microbiol. 6:490-497. [DOI] [PubMed] [Google Scholar]
- 2.Beckman, J. S., T. W. Beckman, J. Chen, P. A. Marshall, and B. A. Freeman. 1990. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. USA 87:1620-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckman, J. S., and W. H. Koppenol. 1996. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am. J. Physiol. 271:C1424-C1437. [DOI] [PubMed] [Google Scholar]
- 4.Bindokas, V. P., J. Jordan, C. C. Lee, and R. J. Miller. 1996. Superoxide production in rat hippocampal neurons: selective imaging with hydroethidine. J. Neurosci. 16:1324-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda, S. S., and R. Kolter. 2004. Multicellularity and biofilms, p. 20-29. In M. Ghannoum and G. A. O'Toole (ed.), Microbial biofilms. ASM Press, Washington, D.C.
- 7.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, F., Q. Xia, and L. K. Ju. 2006. Competition between oxygen and nitrate respirations in continuous culture of Pseudomonas aeruginosa performing aerobic denitrification. Biotechnol. Bioeng. 93:1069-1078. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 10.Crow, J. P. 1997. Dichlorodihydrofluorescein and dihydrorhodamine 123 are sensitive indicators of peroxynitrite in vitro: implications for intracellular measurement of reactive nitrogen and oxygen species. Nitric Oxide 1:145-157. [DOI] [PubMed] [Google Scholar]
- 11.Cruz-Ramos, H., J. Crack, G. Wu, M. N. Hughes, C. Scott, A. J. Thomson, J. Green, and R. K. Poole. 2002. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Autreaux, B., D. Touati, B. Bersch, J. M. Latour, and I. Michaud-Soret. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. USA 99:16619-16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114-122. [DOI] [PubMed] [Google Scholar]
- 15.De Beer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 16.Demple, B. 1991. Regulation of bacterial oxidative stress genes. Annu. Rev. Genet. 25:315-337. [DOI] [PubMed] [Google Scholar]
- 17.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 18.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epe, B., D. Ballmaier, I. Roussyn, K. Briviba, and H. Sies. 1996. DNA damage by peroxynitrite characterized with DNA repair enzymes. Nucleic Acids Res. 24:4105-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, D. J., M. R. Brown, D. G. Allison, and P. Gilbert. 1990. Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J. Antimicrob. Chemother. 25:585-591. [DOI] [PubMed] [Google Scholar]
- 21.Firoved, A. M., S. R. Wood, W. Ornatowski, V. Deretic, and G. S. Timmins. 2004. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa. J. Bacteriol. 186:4046-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 23.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7:894-906. [DOI] [PubMed] [Google Scholar]
- 24.Hassett, D. J., J. Cuppoletti, B. Trapnell, S. V. Lymar, J. J. Rowe, S. S. Yoon, G. M. Hilliard, K. Parvatiyar, M. C. Kamani, D. J. Wozniak, S. H. Hwang, T. R. McDermott, and U. A. Ochsner. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Delivery Rev. 54:1425-1443. [DOI] [PubMed] [Google Scholar]
- 25.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 26.Hentzer, M., L. Eberl, and M. Givskov. 2005. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms 2:37-61. [Google Scholar]
- 27.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171-1175. [DOI] [PubMed] [Google Scholar]
- 29.Hunt, S. M., E. M. Werner, B. Huang, M. A. Hamilton, and P. S. Stewart. 2004. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 70:7418-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer, L. M., V. Anantharaman, and L. Aravind. 2003. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics 4:5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Justice, S. S., C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer, and S. J. Hultgren. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 101:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kojima, H., Y. Urano, K. Kikuchi, T. Higuchi, Y. Hirata, and T. Nagano. 1999. Fluorescent indicators for imaging nitric oxide production. Angew. Chem. Int. Ed. Engl. 38:3209-3212. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski, A. V., and J. P. Shapleigh. 1996. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Biol. Chem. 271:24382-24388. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, K. 2005. Persister cells and the riddle of biofilm survival. Biochemistry (Moscow) 70:267-274. [DOI] [PubMed] [Google Scholar]
- 36.Lobysheva, I. I., M. V. Stupakova, V. D. Mikoyan, S. V. Vasilieva, and A. F. Vanin. 1999. Induction of the SOS DNA repair response in Escherichia coli by nitric oxide donating agents: dinitrosyl iron complexes with thiol-containing ligands and S-nitrosothiols. FEBS Lett. 454:177-180. [DOI] [PubMed] [Google Scholar]
- 37.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mai-Prochnow, A., J. S. Webb, B. C. Ferrari, and S. Kjelleberg. 2006. Ecological advantages of autolysis during the development and dispersal of Pseudoalteromonas tunicata biofilms. Appl. Environ. Microbiol. 72:5414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Hoiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 40.Moncada, S., E. Higgs, and G. Bagetta. 1998. Nitric oxide and the cell: proliferation, differentiation and death. Portland Press, London, United Kingdom.
- 41.Moore, C. M., M. M. Nakano, T. Wang, R. W. Ye, and J. D. Helmann. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 186:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nablo, B. J., and M. H. Schoenfisch. 2003. Antibacterial properties of nitric oxide-releasing sol-gels. J. Biomed. Mater. Res. 67:1276-1283. [DOI] [PubMed] [Google Scholar]
- 43.Nathan, C. 2003. Specificity of a third kind: reactive oxygen and nitrogen intermediates in cell signaling. J. Clin. Investig. 111:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 45.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 46.Parsek, M. R., and C. Fuqua. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piriou, P., S. Dukan, Y. Levi, and P. A. Jarrige. 1997. Prevention of bacterial growth in drinking water distribution systems. Water Sci. Technol. 35:283-287. [Google Scholar]
- 48.Purevdorj-Gage, B., W. J. Costerton, and P. Stoodley. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151:1569-1576. [DOI] [PubMed] [Google Scholar]
- 49.Rice, S. A., K. S. Koh, S. Y. Queck, M. Labbate, K. W. Lam, and S. Kjelleberg. 2005. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 187:3477-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez, G. G., D. Phipps, K. Ishiguro, and H. F. Ridgway. 1992. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl. Environ. Microbiol. 58:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauer, K., M. C. Cullen, A. H. Rickard, L. A. Zeef, D. G. Davies, and P. Gilbert. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 57.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 58.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart, P. S., F. Roe, J. Rayner, J. G. Elkins, Z. Lewandowski, U. A. Ochsner, and D. J. Hassett. 2000. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 66:836-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 62.Thannickal, V. J. 2003. The paradox of reactive oxygen species: injury, signaling, or both? Am. J. Physiol. Lung Cell. Mol. Physiol. 284:L24-L25. [DOI] [PubMed] [Google Scholar]
- 63.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webb, J. S., M. Givskov, and S. Kjelleberg. 2003. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 6:578-585. [DOI] [PubMed] [Google Scholar]
- 66.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoon, S. S., R. Coakley, G. W. Lau, S. V. Lymar, B. Gaston, A. C. Karabulut, R. F. Hennigan, S.-H. Hwang, G. Buettner, M. J. Schurr, J. E. Mortensen, J. L. Burns, D. Speert, R. C. Boucher, and D. J. Hassett. 2006. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J. Clin. Investig. 116:436-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]